Introduction
3β, 16β, 17α-trihydroxycholest-5-en-22-one
16-O-(2-O-4-methoxybenzoyl-β-D-xylopyranosyl)-(1→3)-(2-O-acetyl-α-L-arabinopyranoside)
(OSW-1) is located in the bulbs of Ornithogalum
saudersiae(1) and its
anticancer effect is 10–100 times greater than that of doxorubicin,
camptothecin or paclitaxel (2).
Non-malignant cells are significantly less sensitive to OSW-1 than
cancer cell lines, with concentrations that cause a 50% loss of
cell viability of 40–150-fold greater than that observed in
malignant cells. More significantly, OSW-1 can lead to a loss of
mitochondrial transmembrane potential, an increase in cytosolic
calcium and the activation of calcium-dependent apoptosis in human
leukemia and pancreatic cancer cells (3). However, the method by which OSW-1
exerts its anticancer activity is complex and the exact mechanisms
responsible for such selectivity remain unclear. To investigate the
mechanism of this unnatural anticancer activity, gene expression
analysis was used to examine the potential changes in the gene
expression of a hepatocellular carcinoma (HCC) cell line (Hep3B)
that had been incubated with OSW-1 in vitro. A network
extension of the signaling pathways that participate in apoptosis
and necroptosis mediated by OSW-1 was then created.
Materials and methods
Cell cultures
Hep3B was obtained from the Chinese Academy of
Sciences Cell Bank and the cell line was maintained in Dulbecco’s
modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA)
supplemented with 10% fetal bovine serum (Gibco Life Technologies,
Carlsbad, CA, USA). A monoclonal cell line was acquired using a
limiting dilution assay (LDA) and maintained in DMEM with 20% fetal
bovine serum. A humidified incubator was set at 37°C with 5%
CO2. The Hep3B monoclonal cell line was treated with 200
ng/ml OSW-1 for 24 h.
RNA isolation
A total of 7×106 monoclonal cells were
used for total RNA isolation. The total RNA was isolated using a
TRIzol reagent according to the manufacturer’s instructions
(Invitrogen, Hong Kong, China). The RNA concentration was
determined by measuring the absorbance at 260 nm using the NanoDrop
ND1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and
the purity of the RNA was estimated using the OD260/280 ratio. The
RNA integrity was assessed by standard denaturing agarose gel
electrophoresis, then the RNAs were used for labeling and array
hybridization.
cDNA synthesis and labeling
In total, 10 μg of the RNA was processed and labeled
according to the standard NimbleGen instructions. Briefly, the RNA
was converted into cDNA using a Superscript Double-Stranded cDNA
Synthesis kit (Invitrogen). The double-stranded cDNA was
random-prime labeled with Cy3 converted via an oligo-dT using a
NimbleGen One-Color DNA Labeling kit subsequent to RNase cleanup
and cDNA precipitation.
Expression profiling using
microarrays
The labeled cRNAs were hybridized to the NimbleGen
Human Gene Expression 12×135K microarray using the following steps:
i) Reverse transcription with the Invitrogen Superscript ds-cDNA
Synthesis kit; ii) ds-cDNA labeling with the NimbleGen One-Color
DNA Labeling kit; iii) array hybridization using the NimbleGen
Hybridization System followed by washing with the NimbleGen Wash
Buffer kit; and iv) array scanning using the Axon GenePix 4000B
microarray scanner (Molecular Devices Co., Sunnyvale, CA, USA).
Results
Data analysis
After washing, the slides were scanned with an Axon
GenePix 4000B scanner. The data were extracted and normalized using
NimbleScan v2.5 Software. The raw signal intensities were
normalized using the RMA method with NimbleScan v2.5 and the low
intensity genes were filtered (genes that had at least 2 out of 2
samples with values ≥ the lower cut-off; 50.0 were chosen for
further analysis). The quality of the gene data was assessed using
box and scatter plots subsequent to filtering. The differentially
expressed genes that passed fold change filtering (fold change,
≥2.0) and the final data were used to create a heat map, then the
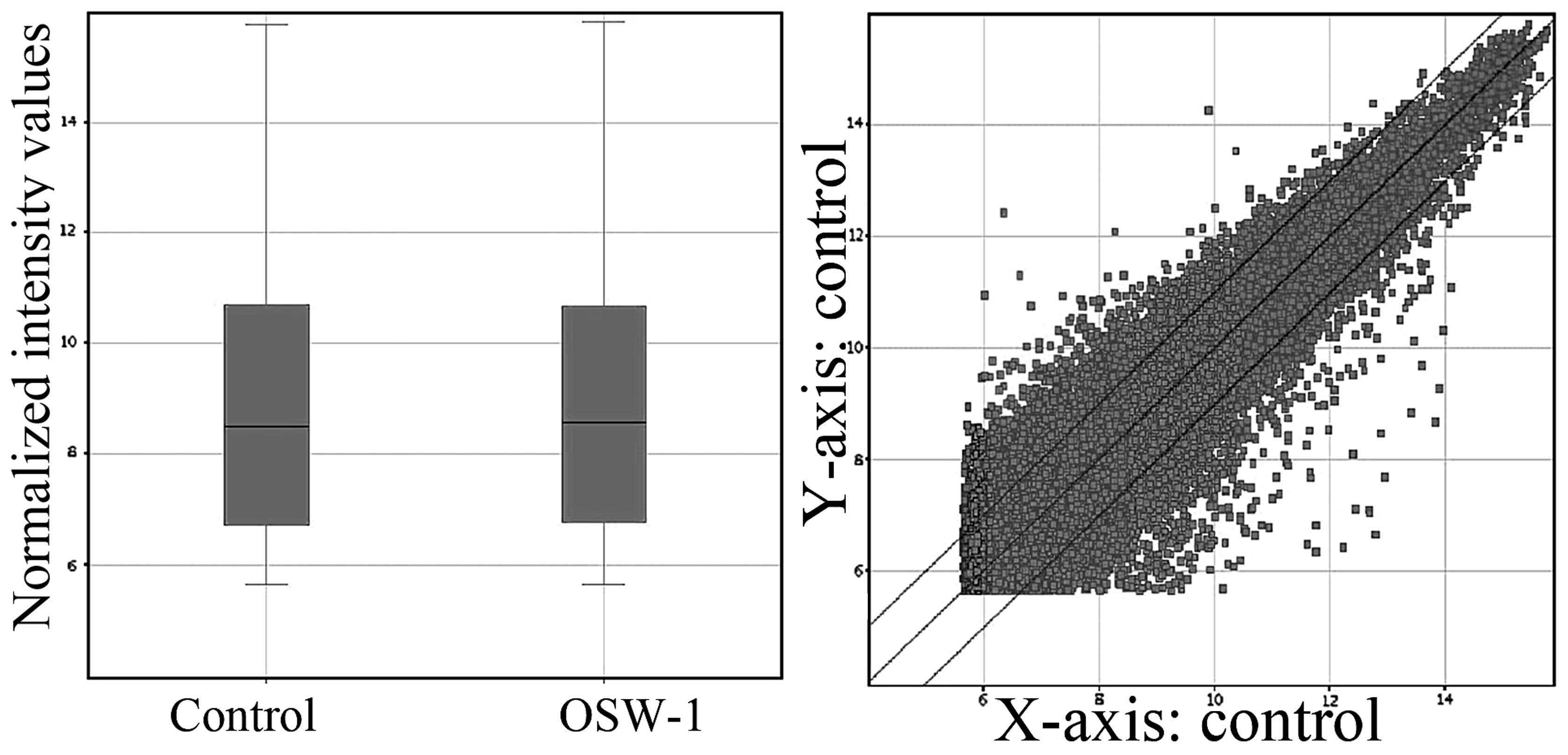
hierarchical clustering pathway analysis was completed (Fig. 1). The box plot shown in Fig. 1 is a convenient way to quickly
visualize the distributions of a dataset. It is commonly used for
comparing the distributions of the intensities from all samples.
After normalization, the distributions of log2-ratios among all
tested samples are nearly the same. The scatterplot shown in
Fig. 1 is a visualization method
used for assessing the expression variation (or reproducibility)
between two groups. The values of X and Y axes in the scatterplot
are the normalized signal values of each sample (log2 scaled) [or
averaged normalized signal values of each group (log2 scaled)]. The
three lines are fold change lines (the fold change value given is
2.0). Genes above the top line and below the bottom line indicated
more than 2.0-fold change of genes between two groups.
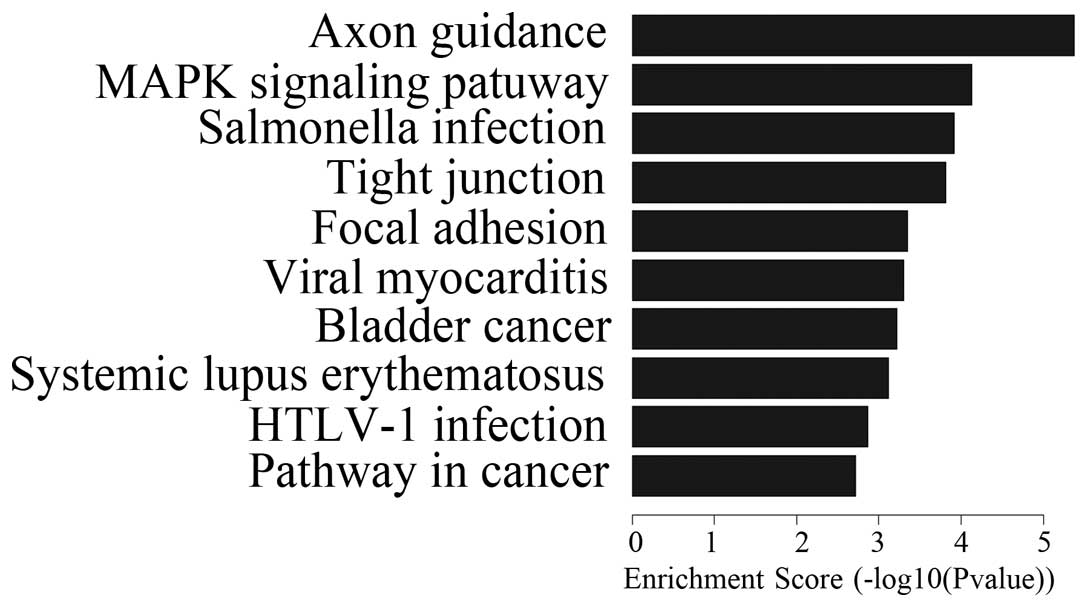
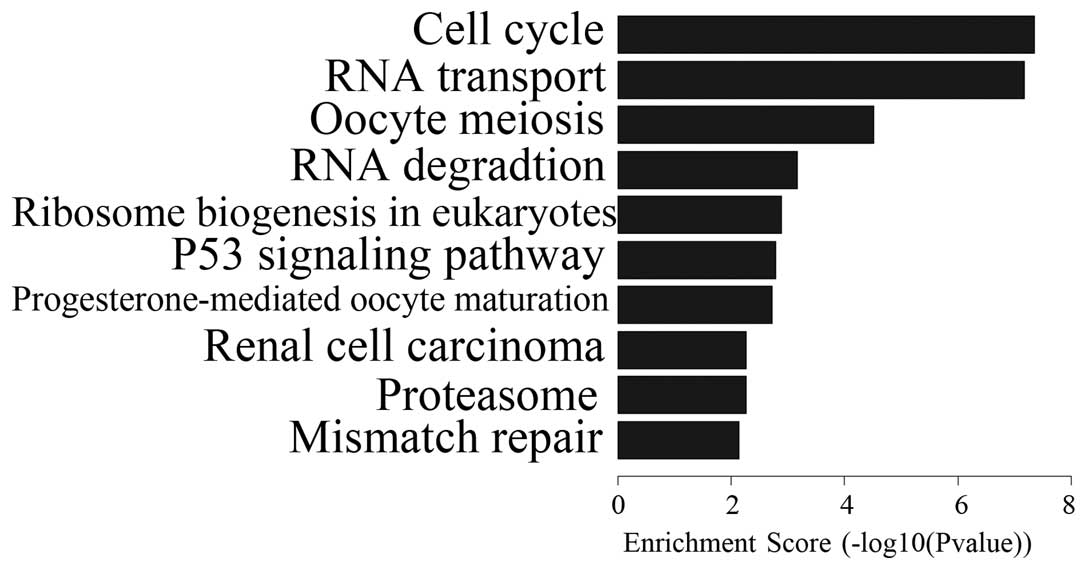
Pathway analysis
OSW-1 affected the expression of numerous genes
in vitro. Despite the fact that OSW-1 was at a nanomolar
concentration, 570 genes were downregulated and 341 genes were
upregulated. Differential expression analysis of the genes was
performed in the Hep3B cells that had been treated with OSW-1 and
included the analysis of PDGF, Ras, c-Myc, MEK1, WNT, TCF/LEF,
PI3K, caspase-8,9, FADD, CytC and Chk1/2. These genes are involved
in certain pathways, including those for axon guidance, MAPK, tight
junctions, WNT, the cell cycle and P53 (Figs. 2 and 3; Tables
I and II). These results
indicated that OSW-1 OSW-1 inhibits cell invasion, angiogenesis,
cell adhesion and destruction of cell polarity.
 | Table IGenes from the various signaling
pathways that were downregulated by OSW-1. |
Table I
Genes from the various signaling
pathways that were downregulated by OSW-1.
| Signaling
pathway | Downregulated
genes |
|---|
| Wnt |
CCND1/CCND2/CSNK1E/FRAT2/FZD2/FZD9/JUN/LRP5/MYC/NFATC1/NKD2/PPP2R5B/RAC3/TCF7L1/WNT10A/WNT10B/WNT5B/WNT7B/WNT9A |
| MAPK |
ARRB1/BDNF/CD14/DUSP1/DUSP2/DUSP4/DUSP5/DUSP9/FGF11/FGF21/FGF8/FGFR1/FGFR4/FLNB/FLNC/HSPA1A/HSPA6/IL1B/JUN/MAP2K1/MAP2K3/MAP2K6/MAP3K12/MAP3K6/MAPK13/MAPKAPK2/MKNK2/MRAS/MYC/PAK1/PDGFB/PLA2G12A/PPP5C/RAC3/RPS6KA1/RRAS/SRF |
| VEGF |
CASP9/MAP2K1/MAPK13/MAPKAPK2/NFATC1/NOS3/PIK3CD/PLA2G12A/PLCG2/RAC3 |
 | Table IIGenes from the various signaling
pathways that were upregulated by OSW-1. |
Table II
Genes from the various signaling
pathways that were upregulated by OSW-1.
| Signaling
pathway | Upregulated
genes |
|---|
| P53 |
CASP8/CCNB1/CCNB2/CHEK1/CYCS/GTSE1/PERP/PMAIP1/PPM1D/RRM2/SERPINB5/SIAH1/THBS1 |
| Cell cycle |
ANAPC1/ANAPC7/BUB1/BUB3/CCNA2/CCNB1/CCNB2/CDC16/CDC23/CDC25B/CDC25C/CDC25C/CDC6/CHEK1/CUL1/E2F2/E2F3/EP300/HDAC2/PCNA/PRKDC/PTTG1/RAD21/SKP2/SMAD4/TGFB2/WEE1/YWHAB/YWHAQ/YWHAZ |
Discussion
In the present study, the NimbleGen expression
values correlated strongly with the TaqMan expression values, the
current ‘gold standard’ for gene expression quantitation,
demonstrating the accuracy of the data obtained through using this
technology. In addition to highly reproducible and accurate
expression values, the unique combination of NimbleGen high-density
arrays, long oligos and flexible design abilities provides superior
results. The raw signal intensities were normalized using the RMA
method with NimbleScan v2.5 and the low intensity genes were
filtered, followed by the differentially expressed genes that
passed fold change filtering.
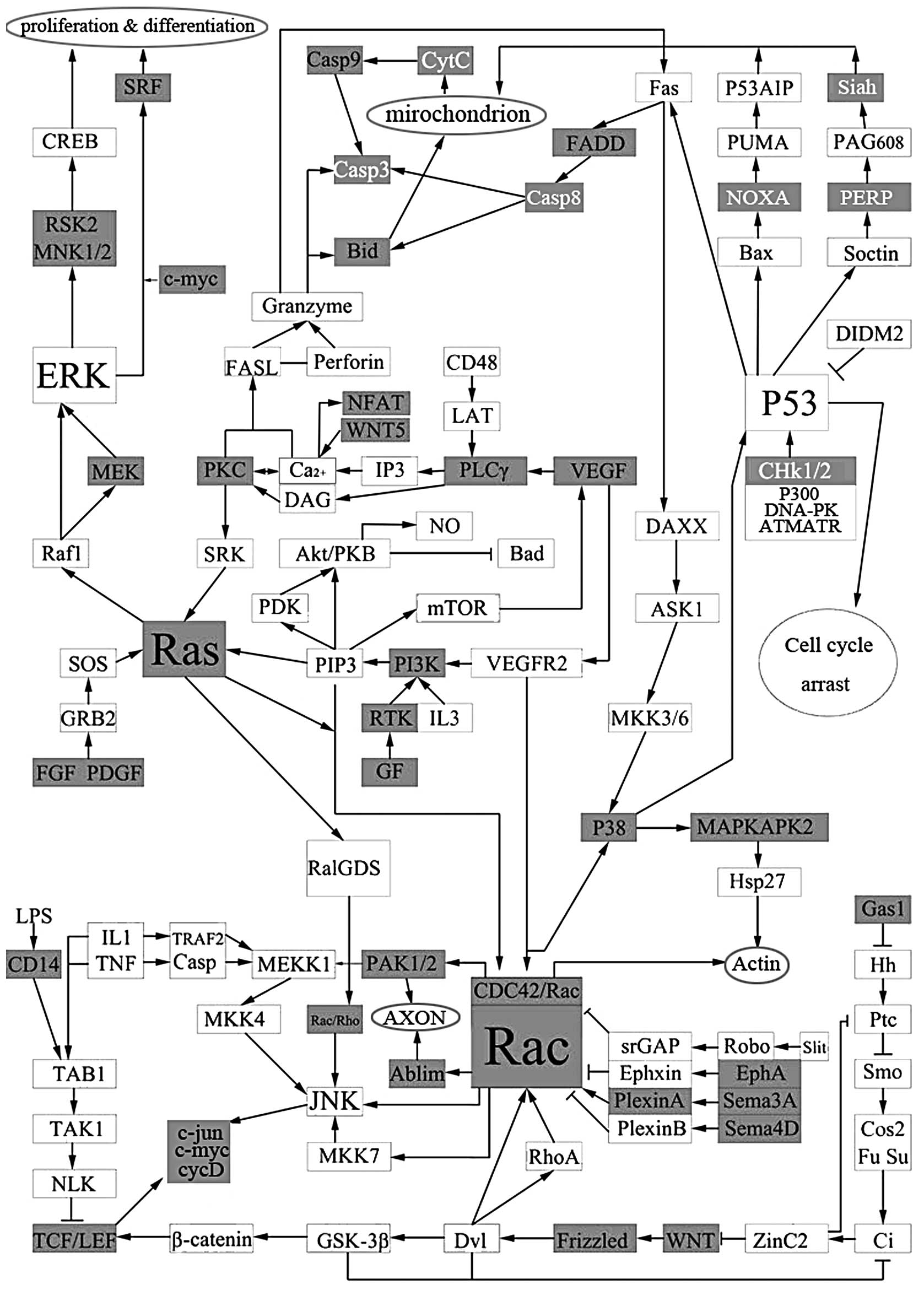
To investigate the role of OSW-1 on Hep3B, the
differentially expressed genes that were associated with OSW-1 were
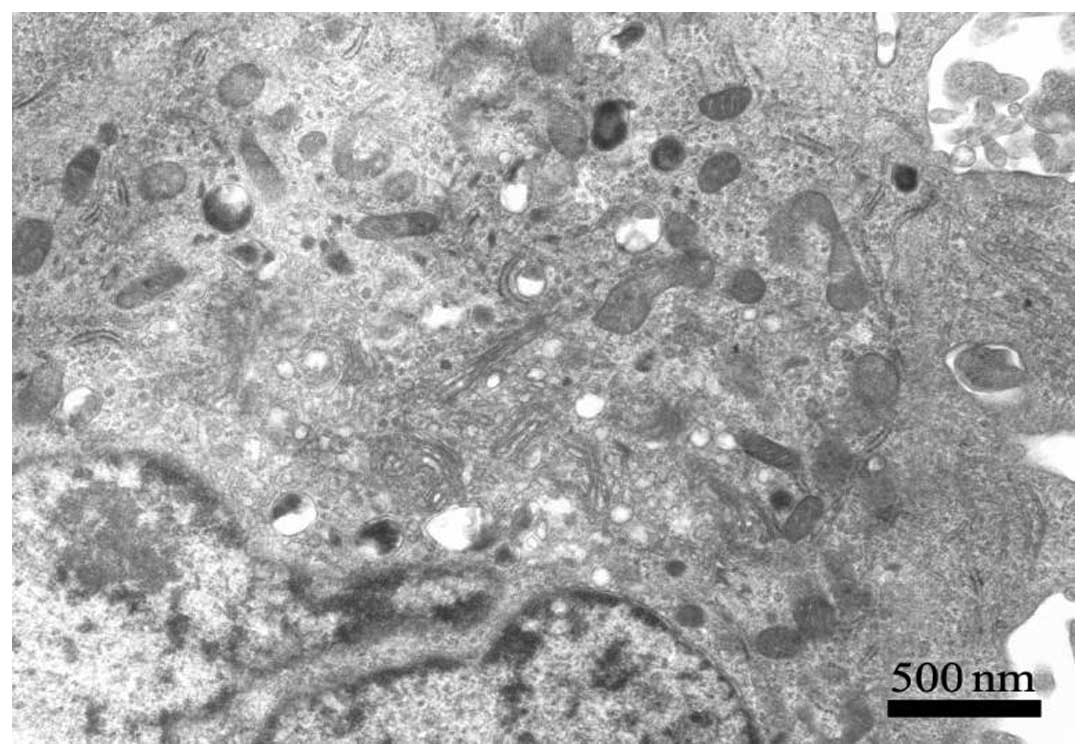
collated into a comprehensive pathway (Fig. 4). Mitochondria play a critical role
in mediating the apoptotic signal transduction pathway which
includes mitochondrial swelling and disruption of the mitochondrial
outer membrane and cristae when treated by OSW-1 (Fig. 5).
Once the classical WNT signaling pathway is
activated, it may cause target gene overexpression. c-myc, cyclinD1
and c-jun have been determined as a few of the proto-oncogenes that
are the target genes affecting cell fate, migration and polarity
(4). These genes are all WNT
family glycoproteins with conserved 22–24 Cys residues that are
downregulated by OSW-1 in the WNT signaling pathway. More
prominently, the Frizzled and TCF/LEF genes demonstrate the same
characteristics. This suggests that OSW-1 may act on WNT proteins
directly, with its effects visible in the cytoplasm and cell
nucleus.
The ERK signaling pathway plays a key role in
several steps of tumorigenesis (5). Ras activates the ERK pathway through
receptor-mediated activation of the small G-protein (6). Ras is activated through the exchange
of bound GDP into GTP as a membrane-bound protein. OSW-1 is able to
prevent this progress specifically. It has been demonstrated that
Ras/Raf-independent activation of MEK1/2 is inhibited downstream.
ERK1/2-mediated signaling plays a significant role in mediating
cell survival. The activated alleles of MEK1/2 promote cell
survival independently of other survival factors (7) and OSW-1 downregulates MEK1
expression. RSK, which is also downregulated by OSW-1, activates
the transcription factor CREB, which promotes cell survival through
transcriptional upregulation of the antiapoptotic Bcl family of
proteins (8,9). Taken together, these OSW-1-based
findings and those obtained from expression profiling in the
present study, may support the results of a previous study
(10). The induction of apoptosis
in several cell lines requires the activation of the p38 MAPK
pathway (11). In the present
study, OSW-1 acted on Hep3B, MKK3 MKK6 and P38, whose transcription
levels were lower than the control groups without the activation of
an upstream gene in the control groups. From observing the
enrichment score of the MAPK signaling pathway (-Log10P-value) this
pathway may be considered as one of the most significant
mechanisms. In KRAS mutants and RAS/RAF wild-type tumors, RAF
inhibitors activate the RAF-MEK-ERK pathway in a RAS-dependent
manner, thus enhancing tumor growth in certain engraft models
(m14). Non-upstream gene activation mediated by OSW-1 indicates
that it may direct the conformational effects of the inhibitors on
the RAF kinase domain as the RAS and mitochondrial apoptosis
effects mediate RAS-GTP signaling.
Previous studies have demonstrated that the
VEGF/RTK/PI3K/AKT pathway plays a key role in regulating migration,
angiogenesis, proliferation and survival (12,13).
Downregulation of eNOS increases the sensitivity of Ca2+
for apoptosis induced by the mitochondria. This observation may
demonstrate, through a complex signaling pathway, the fact that the
VEGF signaling pathway has a key role in regulating tumor
angiogenesis and an auxiliary role in apoptosis of the mitochondria
which is induced by OSW-1 in HCC. However, VEGF may promote tumor
growth and be inhibited by other reagents (14). This may be evidence of the
particular anticancer mechanisms that increase the cytosolic
calcium in the mitochondria and downregulate other Ca2+
apoptosis-related genes to increase the sensitivity of
Ca2+ and antitumoral activities. OSW-1 inhibits the
VEGF/RTK/PI3K/AKT pathway to reduce angiogenesis and tumor growth
in HCCs.
The p53 gene, as a tumor suppressor that is best
characterized as a transcription factor which binds to specific DNA
sequences, has a major role in the cell response to DNA damage. The
BH3-only gene, Noxa, is a direct transcriptional target of p53 and
has a role in DNA damage-induced p53-mediated apoptosis (15). OSW-1 transactivates and upregulates
BH3, which is a member of the Bcl-2 family. Noxa has a significant
affinity for p53 (16) and may be
essential to apoptosis during cellular stress (17). P53 induces apoptosis by the
transcriptional induction of Puma and Noxa, which encode the
proapoptotic BH3-only Bcl-2 family of proteins. However, at the
molecular level, the mechanisms of action of the Puma and Noxa
proteins remain poorly defined. Although certain studies have
previously hypothesized that p53 induces apoptosis largely via Puma
(18), the present study observed
that OSW-1 upregulated Noxa only and not Puma when inducing
apoptosis in the hepatoma cells. Additionally, the
apoptosis-associated targets of p53, PERP and Siah, were
upregulated by OSW-1.
The Bcl-2 family members and the mitochondria are
significant targets of p53 (19).
Activated extensively in the p53 signaling pathway, p53 also alters
the function of the mitochondria and subsequently mediates the
release of cytochrome-c (20).
Perforation of the mitochondrial membrane results in the release of
several death-promoting factors, which ultimately neither caspase
dependently or independently execute cell death (21). Even more noteworthy was the fact
that caspase9 was downregulated in OSW-1 induced apoptosis in the
present study. Also, FADD and Bid were expressed in the opposite
manner to the classical apoptosis theory. Although
mitochondria-regulated cell death program was activated, apoptosis
induced by caspase was inhibited. In the extrinsic pathways, the
cell-surface receptor Fas promotes cell death through caspase-8
(22). However, Fas appears to be
dispensable in p53-dependent apoptosis (23). Therefore, OSW-1 had the ability to
activate the extrinsic and intrinsic apoptotic pathways. At the
same time, in the present study, this special type of mitochondrial
apoptosis, which featured caspase inhibition induced by OSW-1,
suggested that necroptosis was involved.
Necroptosis is a novel form of caspase-independent
cell death. The initiation of programmed necrosis, necroptosis, by
death receptors requires the kinase activity of
receptor-interacting proteins and its execution involves the active
disintegration of mitochondrial, lysosomal and plasma membranes
(24). In a study by Han et al
(25), necroptosis did not involve
caspase, Bcl-2 or the release of cytochrome-c from the
mitochondria. Other previous studies (9,26)
are in agreement with the present study results which showed the
release of cytochrome-c and the cleavage of Bcl-2 dependents by
caspase8, OSW-1 target oxysterol-binding protein (OSBP) and its
closest paralog, OSBP-related protein 4L (ORP4L). Additionally,
FADD and caspase 9 were downregulated and the expression of RIPK2
was upregulated in the Hep3B OSW-1-treated cells. Of note was the
fact that BH3 was required for death receptor-induced necroptosis
(27) and that the characteristics
of the upregulation of Noxa were expounded in the p53 pathway in
the present study.
Acknowledgements
This study was financially supported by the National
Natural Science Foundation of China (Grant No. 81160529) and the
Jilin Province Science and Technology Development Project (grant
no. 200905207)
References
|
1
|
Kubo S, Mimaki Y, Terao M, Sashida Y,
Nikaido T and Ohmoto T: Acylated cholestane glycosides from the
bulbs of Ornithogalum saudersiae. Phytochemistry.
31:3969–3973. 1992. View Article : Google Scholar
|
|
2
|
Mimaki Y, Kuroda M, Kameyama A, Sashida Y,
Hirano T and Oka K: Cholestane glycosides with potent cytostatic
activities on various tumor cells from Ornithogalum
saundersiae bulls. Bioorg Med Chem Lett. 7:633–636. 1997.
View Article : Google Scholar
|
|
3
|
Zhou Y, Garcia-Prieto C, Carney DA, et al:
OSW-1: a natural compound with potent anticancer activity and a
novel mechanism of action. J Natl Cancer Inst. 97:1781–1785. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cai FG, Xiao JS and Ye QF: Effects of
ischemic preconditioning on cyclinD1 expression during early
ischemic reperfusion in rats. World J Gastroenterol. 12:2936–2940.
2006.PubMed/NCBI
|
|
5
|
Kim EK and Choi EJ: Pathological roles of
MAPK signaling pathways in human diseases. Biochim Biophys Acta.
1802:396–405. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McKay MM and Morrison DK: Integrating
signals from RTKs to ERK/MAPK. Oncogene. 26:3113–3121. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ballif BA and Blenis J: Molecular
mechanisms mediating mammalian mitogen-activated protein kinase
(MAPK) kinase (MEK)-MAPK cell survival signals. Cell Growth Differ.
12:397–408. 2001.PubMed/NCBI
|
|
8
|
Bonni A, Brunet A, West AE, Datta SR,
Takasu MA and Greenberg ME: Cell survival promoted by the Ras-MAPK
signalling pathway by transcription-dependent and -independent
mechanisms. Science. 286:1358–1362. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu J, Xiong L, Yu B and Wu J: Apoptosis
induced by a new member of saponin family is mediated through
caspase-8-dependent cleavage of Bcl-2. Mol Pharmacol. 68:1831–1838.
2005.PubMed/NCBI
|
|
10
|
Porras A, Zuluaga S, Black E, et al: P38
alpha mitogen-activated protein kinase sensitizes cells to
apoptosis induced by different stimuli. Mol Biol Cell. 15:922–933.
2004. View Article : Google Scholar
|
|
11
|
Grethe S and Pörn-Ares MI: p38 MAPK
regulates phosphorylation of Bad via PP2A-dependent suppression of
the MEK1/2-ERK1/2 survival pathway in TNF-alpha induced endothelial
apoptosis. Cell Signal. 18:531–540. 2006. View Article : Google Scholar
|
|
12
|
Zhong Q, Zhou Y, Ye W, Cai T, Zhang X and
Deng DY: Hypoxia-inducible factor 1-α-AA-modified bone marrow stem
cells protect PC12 cells from hypoxia-induced apoptosis, partially
through VEGF/PI3K/Akt/FoxO1 pathway. Stem Cells Dev. 21:2703–2717.
2012.
|
|
13
|
Adya R, Tan BK, Punn A, Chen J and Randeva
HS: Visfatin induces human endothelial VEGF and MMP-2/9 production
via MAPK and PI3K/Akt signalling pathways: novel insights into
visfatin-induced angiogenesis. Cardiovasc Res. 78:356–365. 2008.
View Article : Google Scholar
|
|
14
|
Chiang IT, Liu YC, Wang WH, et al:
Sorafenib inhibits TPA-induced MMP-9 and VEGF expression via
suppression of ERK/NF-κB pathway in hepatocellular carcinoma cells.
In Vivo. 26:671–681. 2012.PubMed/NCBI
|
|
15
|
Villunger A, Michalak EM, Coultas L, et
al: p53- and drug-induced apoptotic responses mediated by BH3-only
proteins puma and noxa. Science. 302:1036–1038. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Park SY, Jeong MS and Jang SB: In vitro
binding properties of tumor suppressor p53 with PUMA and NOXA.
Biochem Biophys Res Commun. 420:350–356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang DC and Strasser A: BH3-only proteins
- essential initiators of apoptotic cell death. Cell. 103:839–842.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Michalak EM, Villunger A, Adams JM and
Strasser A: In several cell types tumour suppressor p53 induces
apoptosis largely via Puma but Noxa can contribute. Cell Death
Differ. 15:1019–1029. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vaseva AV and Moll UM: The mitochondrial
p53 pathway. Biochim Biophys Acta. 1787:414–420. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leu JI, Dumont P, Hafey M, Murphy ME and
George DL: Mitochondrial p53 activates Bak and causes disruption of
a Bak-Mcl1 complex. Nat Cell Biol. 6:443–450. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gogvadze V, Orrenius S and Zhivotovsky B:
Mitochondria as targets for cancer chemotherapy. Semin Cancer Biol.
19:57–66. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nagata S and Golstein P: The Fas death
factor. Science. 267:1449–1456. 1995. View Article : Google Scholar
|
|
23
|
O’Connor L, Harris AW and Strasser A: CD95
(Fas/APO-1) and p53 signal apoptosis independently in diverse cell
types. Cancer Res. 60:1217–1220. 2000.PubMed/NCBI
|
|
24
|
Vandenabeele P, Galluzzi L, Vanden Berghe
T and Kroemer G: Molecular mechanisms of necroptosis: an ordered
cellular explosion. Nat Rev Mol Cell Biol. 11:700–714. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han W, Li L, Qiu S, et al: Shikonin
circumvents cancer drug resistance by induction of a necroptotic
death. Mol Cancer Ther. 6:1641–1649. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Burgett AW, Poulsen TB, Wangkanont K, et
al: Natural products reveal cancer cell dependence on
oxysterol-binding proteins. Nat Chem Biol. 7:639–647. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hitomi J, Christofferson DE, Ng A, Yao J,
Degterev A, Xavier RJ and Yuan J: Identification of a molecular
signalling network that regulates a cellular necrotic cell death
pathway. Cell. 135:1311–1323. 2008. View Article : Google Scholar : PubMed/NCBI
|