Introduction
Cholangiocarcinoma (CCA) is a lethal malignancy
derived from the epithelial cells (i.e. cholangiocytes) of the bile
duct. CCA exhibits a considerable variety of symptoms commonly at
the later stages of disease and therefore treatment for CCA is
extremely difficult. CCA is grossly divided into mass forming (MF),
periductal infiltrating and intraductal papillary subtypes
(1). Gross pathological
classifications of CCA are important in clinical practice and
further translational investigations due to the distinct
characteristics and outcomes following hepatectomy (2). The incidence of CCA exhibits
considerable geographical variation but generally accounts for
5–30% of primary liver cancer (3).
Previous studies have reported that the incidence and mortality
rates of CCA have been increasing worldwide, particularly
intrahepatic CCA (4–6). CCA is caused by a number of risk
factors, including parasitic infections, primary sclerosing
cholangitis, choledochal cysts, hepatolithiasis and carcinogen
exposure, which leads to the significant variance in incidence
rates of CCA worldwide (7–9).
Clinically, CCA remains extremely challenging as
patients do not typically exhibit clear symptoms until the disease
is quite advanced and therefore it is difficult to diagnose in its
early stages. In addition to surgical treatments (2,10–14),
radiation therapy and current chemotherapeutic protocols have not
been found to significantly improve the long-term survival rates of
CCA patients (8,15). In our previous study, a
thioacetamide (TAA)-induced CCA rat model was established to
analyze the molecular and morphological behavior of CCA, aiming to
generate a powerful preclinical platform to provide insights into
therapeutic and chemopreventative strategies for human CCA
(16). Since the model
recapitulates the dysplasia-carcinoma sequence of human CCA, it is
likely to be crucial for the identification of the genetic basis of
cholangiocellular neoplasia.
A number of previous studies have aimed to determine
the molecular alterations involved in cholangiocarcinogenesis;
however, these processes remain largely unknown (17–19).
At present, gene expression profiling by DNA microarray represents
a promising technique for understanding the molecular abnormalities
involved in cancer development. In our previous study, MUC4
overexpression was identified in rat CCA (carcinogenesis caused by
TAA) compared with non-tumor liver tissue (20). In the present study, a whole genome
rat cDNA microarray was used to determine whether the gene
expression profile for CCA reflects a specific etiological agent,
with the aim to improve the understanding of the molecular events
associated with CCA. In addition, this study compared the molecular
profiles in non-cancerous liver to TAA-induced CCA to gain insight
into changes in gene expression associated with cholangiocellular
carcinogenesis and to identify potential diagnostic biomarkers. The
investigation of the molecular pathophysiology associated with CCA
is becoming increasingly important and necessary.
Materials and methods
Animals, treatment and CCA samples
The experimental animal ethics committee of Chang
Gung Memorial Hospital (Linkou, Taiwan, R.O.C.) approved all animal
protocols in this study. This study conformed to the US National
Institute of Health guidelines for the care and use of laboratory
animals (21). Seven adult male
Sprague-Dawley (SD) rats (330–370 g) were used in these
experiments. Rats were housed in an animal room under a 12:12-hour
light-dark cycle (light between 08:00 a.m. and 08:00 p.m.) at an
ambient temperature of 22±1°C, with food and water available ad
libitum. Seven experimental rats were administered 300 mg/l TAA
in their drinking water daily until week 24. CCA was collected over
the 24-week TAA treatment. Only CCA was used for array analysis to
avoid variations in expression arising from histologically
different tumor progression. Each carcinoma used in this study was
obtained from a separate rat.
RNA isolation
Total RNA was isolated using TRIzol (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions. The integrity of RNA was checked using an agarose
gel.
Expression array
The Whole Rat Genome oligo-microarray (P/N G4131A;
Agilent Technologies, Santa Clara, CA, USA) was used for microarray
experiments. RNA sample preparation for microarray analysis was
performed according to the manufacturer’s instructions. In brief,
20 μg total RNA was used for cyanine 3-dUTP (Cy3; test) and
Cy5-dUTP (reference) labeling. Labeling was performed by
oligo(dT)-primed polymerization using SuperScript II reverse
transcriptase (Life Technologies, Grand Island, NY, USA) and the
labeled Cy3 and Cy5 cDNA probes were purified using a Qiagen PCR
QIAquick PCR Purification kit (#28104; Qiagen, Hilden, Germany).
Array hybridization was performed at 60°C for 14–16 h. Following
hybridization, the array was washed and dried using the Agilent
washing kit. The array image was captured using the Axon GenePix
4000 laser scanner and probe intensity was calculated with GenePix
Pro 6.0 software (Molecular Devices, Sunnyvale, CA, USA). The raw
data was further examined using Nexus Expression Software
(BioDiscovery, Hawthorne, CA, USA).
Data processing and analysis
Microarray data analysis was performed as described
previously with specific modifications (22). Image analysis was performed with
GenePix Pro software. Automatic and manual flagging were used to
localise absent or extremely weak spots (<2-fold higher than
background), which were excluded from the analysis. The signal from
each spot was calculated as the average intensity minus the average
local background. Expression ratios of Cy5/Cy3 (or Cy3/Cy5 in case
of dye-swap) were normalized using a method that accounts and
corrects for intensity-dependent artefacts in the measurements; the
LOWESS method in the SMA package. SMA is an add-on library written
in the public domain statistical language, R. Three independent
microarray experiments were performed. Following data
normalization, genes with a 2-fold change in expression compared
with the control sample were considered as differentially expressed
genes between samples. All genes with a log2 ratio ≥1 or
≤-1 were considered to be statistically significant. Specific
differentially expressed genes were grouped based on information
from the KEGG database (23,24),
NCBI, Gene Ontology and DAVID (25,26)
(Tables I and II). Specific genes were annotated for
several functions; however, genes were assigned to one group only
(Tables I and II).
 | Table ITop 50 significantly upregulated
genes with biological process ontologies. |
Table I
Top 50 significantly upregulated
genes with biological process ontologies.
| Ontology | Gene ID | Gene symbol | Gene
description | Fold change |
|---|
| Cell adhesion
molecules | NM_031521 | Ncam1 | Neural cell
adhesion molecule 1 |
3.92235410303046 |
| NM_012705 | Cd4 | CD4 antigen |
3.85832760624149 |
| NM_172067 | Spon1 | Spondin 1 |
3.83860585169924 |
| NM_053909 | Nfasc | Neurofascin |
3.44243163237158 |
| Cell death | NM_001007735 | Sertad1 | Serta
domain-containing 1 |
4.75153117961431 |
| NM_171988 | Bcl2l11 | Bcl2-like 11
(apoptosis facilitator; Bcl2l11), transcript variant 3 |
4.34809778573906 |
| Cell growth | AF454371 | Ahnak | Ahnak-related
protein |
4.14343358370448 |
| NM_057211 | Bteb1 | Basic transcription
element binding protein 1 |
3.63347765601812 |
| NM_199267 | v-rel | V-rel
reticuloendotheliosis viral oncogene homolog A (avian; Rela) |
3.53615002143989 |
| NM_012817 | Igfbp5 | Insulin-like growth
factor binding protein 5 |
3.38491249464085 |
| Deoxyribonuclease I
activity | NM_013097 | Dnase1 | Deoxyribonuclease
I |
3.8275058577354 |
| Fibrosis | XM_342827 | GliPR2 | Similar to
chromosome 9 open reading frame 19; 17 kD fetal brain protein
(LOC362509) |
4.42322128503159 |
| NM_031050 | Lum | Lumican |
3.45085666984717 |
| Hematopoietic cell
lineage | NM_001008884 | RT1-Db1 | RT1 class II, locus
Db1 |
3.36846228092561 |
| Metabolic
pathways | NM_022525 | Gpx3 | Glutathione
peroxidase 3 |
3.93130723332512 |
| NM_012879 | Slc2a2 | Solute carrier
family 2 (facilitated glucose transporter), member 2 |
3.8236468641884 |
| NM_153300 | Aldh1a3 | Aldehyde
dehydrogenase family 1, subfamily |
3.74838767192997 |
| NM_175869 | Plod2 | Procollagen lysine,
2-oxoglutarate 5-dioxygenase 2 |
3.42998378780718 |
| Neuroendocrine
secretory pathway | NM_019279 | Pcsk1n | Proprotein
convertase subtilisin/kexin type 1 inhibitor |
3.43934389253231 |
| Neuronal
differentiation | NM_053369 | Tcf4 | Transcription
factor 4 |
3.35972972064775 |
| Osteoporosis | XM_213440 | COLIA1 | Similar to collagen
α1 (LOC287636) | 3.580131772724 |
| Protein digestion
and absorption | XM_216399 | LOC298069 | Collagen, type XV,
α1 (Col15a1) |
3.33229268844969 |
| NM_031341 | Slc7a7 | Solute carrier
family 7 (cationic amino acid transporter, y+ system), member
7 |
3.33134108780008 |
| RNA transport | NM_017063 | Kpnb1 | Karyopherin
(importin) β1 |
3.51437343518773 |
| Signal transduction
pathways | NM_001007005 | Arhgdia | Rho gdp
dissociation inhibitor (GDI) α |
3.9113860940851 |
| NM_013127 | Cd38 | Cd38 antigen |
3.91115383379829 |
| NM_057116 | Ppp2r2c | Protein phosphatase
2 (formerly 2A), regulatory subunit B (PR 52), γ isoform |
3.40886940369163 |
| NM_024129 | Dcn | Decorin |
3.36137935471163 |
| Structural
proteins | NM_181089 | Mast1 | Microtubule
associated serine/threonine kinase 1 |
4.11144062655087 |
| Others | U06751 | pSMC | Fisher 344
pre-sialomucin complex |
6.57759198187846 |
| XM_345756 | LOC366769 | Similar to Ig heavy
chain precursor V region (IdB5.7) |
4.80829731905218 |
| XM_341923 | LOC361644 | Similar to pyruvate
kinase, M1 isozyme (pyruvate kinase muscle isozyme) |
4.24360287494609 |
| XM_225043 | LOC306628 | Similar to collagen
α 2(IV) chain precursor |
4.20372195431914 |
| XM_223569 | LOC305482 | Similar to
myotubularin-related protein 3 |
4.15416630589892 |
| XM_233686 | LOC313722 | Similar to SPRY
domain-containing SOCS box protein SSB-1 |
4.08502203156001 |
| XM_223781 | LOC305679 | Similar to vinculin
(metavinculin) |
3.90695618191964 |
| NM_139041 | MUC-13 | Putative cell
surface antigen (LOC207126) |
3.89086963314564 |
| XM_236535 | LOC300920 | Similar to
claudin-2 |
3.84946284637836 |
| XM_214861 | LOC292699 | Similar to casitas
B-lineage lymphoma c |
3.77729841356227 |
| XM_223944 | LOC305824 | Similar to α
enolase (2-phospho-D-glycerate hydro-lyase) (Non-neural enolase;
NNE; Enolase 1) |
3.69235152157205 |
| XM_242992 | LOC313536 | Similar to
β-1,4-galactosyltransferase II |
3.67034914805361 |
| XM_343901 | LOC363605 | Similar to RIKEN
cDNA 2210407C18 |
3.66158192578579 |
| XM_342245 | LOC361945 | Similar to
osteoblast specific factor 2 precursor |
3.66054009489806 |
| XM_237497 | LOC316717 | Similar to protein
phosphatase 1 |
3.60236548308193 |
| XM_344268 | LOC364208 | Similar to
DKFZP566K1924 protein |
3.51188899584009 |
| XM_214386 | LOC290856 | Similar to defensin
5 precursor (RD-5; Enteric defensin) |
3.45619469144432 |
| XM_227388 | LOC310614 | Similar to
transcription repressor p66 |
3.42529996437158 |
| XM_346200 | LOC367530 | Similar to RIKEN
cDNA 4933431D05 |
3.41118393261051 |
| XM_243652 | Plxnb2 | Similar to KIAA0315
(LOC315217) |
3.40547098028761 |
| XM_233386 | LOC313499 | Similar to
hypothetical protein DKFZp566D1346 |
3.40381726129247 |
 | Table IITop 50 significantly downregulated
genes with biological process ontologies. |
Table II
Top 50 significantly downregulated
genes with biological process ontologies.
| Ontology | Gene ID | Gene symbol | Gene
description | Fold change |
|---|
| Bile secretion | NM_133616 | Sult2a1 | Hydroxysteroid
preferring 2 (Sth2) |
−4.9054121581746 |
| NM_017047 | Slc10a1 | Solute carrier
family 10 (sodium/bile acid cotransporter family), member 1 |
−3.14433787788225 |
| Cell
differentiation | XM_223053 | Usher syndrome
2A | Similar to usherin
(LOC289369) |
−6.40569654980506 |
| Complement and
coagulation cascades | NM_022257 | Masp1 | Mannose-binding
protein associated serine protease-1 |
−2.85305822741612 |
| Lipid metabolic
process | NM_139192 | Scd1 | Stearoyl-coenzyme A
desaturase 1 |
−3.31761447196781 |
| XM_341791 | Sult2a2 | Similar to alcohol
sulfotransferase (hydroxysteroid sulfotransferase; ST; ST-60;
LOC361510) |
−3.18536619753157 |
| NM_053923 | Pik3c2g |
Phosphatidylinositol 3-kinase, C2 domain
containing, γ polypeptide (Pik3c2g) | −
3.01034237267081 |
| NM_144750 | LOC246266 |
Lysophospholipase |
−2.92586164948934 |
| NM_012737 | Apoa4 | Apolipoprotein
A-IV |
−2.84424543169138 |
| Metabolic
pathways | NM_017158 | Cyp2c7 | Cytochrome P450,
family 2, subfamily c, polypeptide 7 |
−3.66306395998859 |
| NM_138904 | Gls2 | Glutaminase 2
(liver, mitochondrial) |
−3.34037650319876 |
| NM_012540 | Cyp1a1 | Cytochrome P450,
family 1, subfamily a, polypeptide 1 |
−3.32079040147379 |
| NM_017193 | Aadat | Aminoadipate
aminotransferase |
−3.01966588124673 |
| NM_175760 | Cyp4a3 | Cytochrome P450,
family 4, subfamily a, polypeptide 3 |
−2.87595624954108 |
| NM_017159 | Hal | Histidine ammonia
lyase |
−2.84715718276888 |
| NM_053902 | Kynu | kynureninase
(L-kynurenine hydrolase) |
−2.71604891669278 |
| NM_030850 | Bhmt |
Betaine-homocysteine
methyltransferase |
−2.68923042042384 |
| NM_031835 | Agxt2 | Alanine-glyoxylate
aminotransferase 2 (Agxt2) |
−2.64435761342884 |
| NM_012541 | Cyp1a2 | Cytochrome P450,
family 1, subfamily a, polypeptide 2 (Cyp1a2) |
−2.64005746673363 |
| NM_001013057 | LOC291283 | Aldo-keto reductase
family 1, member C2 (Akr1c2) |
−2.62748122574613 |
| NM_198784 | Mup4 | Major urinary
protein 4 (Mup4) |
−2.57882737786856 |
| Olfactory
transduction | NM_001000888 | Olr1692 | Olfactory receptor
gene |
−4.83343644893506 |
| NM_001000696 | Olr1845 | Olfactory receptor
gene |
−3.1610886895668 |
| NM_001000386 | Olr415 | Olfactory receptor
gene |
−2.75631096513904 |
| Peroxisome
biogenesis | NM_031587 | Pxmp2 | Peroxisomal
membrane protein 2 (Pxmp2) |
−2.79776944314083 |
| Signaling
pathways | NM_024352 | Mst1 | Macrophage
stimulating 1 (hepatocyte growth factor-like) |
−3.01686336303489 |
| NM_012630 | Prlr | Prolactin receptor
(Prlr) |
−2.78270670754098 |
| NM_012799 | Nmbr | Neuromedin B
receptor (Nmbr) |
−2.72210897797388 |
| Trypsin
inhibitor | NM_152936 | LOC266602 | Serine peptidase
inhibitor, Kazal type 1 (Spink1) |
−2.72988934134003 |
| Others | TC500715 | TC500715 | Unknown |
−4.99924740959998 |
| XM_226197 | LOC291810 | Similar to cDNA
sequence BC033409 |
−4.85708838735576 |
| XM_224106 | LOC290148 | Similar to T-cell
receptor α chain precursor V and C regions (TRA29)-rat
(fragment) |
−4.47997612759399 |
| NM_001001799 | RSEP4 | Spinal cord
expression protein 4 |
−3.92583197177519 |
| TC462695 | TC462695 | I52849 alcohol
sulfotransferase |
−3.75483258125134 |
| U33847 | Gucy2g | ksGC mRNA, complete
cds |
−3.68824766043768 |
| XM_228610 | LOC302446 | Similar to
expressed sequence AW011752 |
−3.59095310312283 |
| AY383691 | AY383691 | LRRGT00036 mRNA,
complete cds |
−3.20650059581476 |
| AF010442 | AF010442 | MARRLC7A mRNA |
−3.17415725515125 |
| TC490222 | TC490222 | AB027125 aldo-keto
reductase AKR1C13 |
−3.12591316822196 |
| XM_224468 | LOC290458 | Similar to
tripartite motif-containing 52 |
−3.12295823834706 |
| XM_222983 | LOC289295 | Similar to putative
pheromone receptor (Go-VN2) |
−3.0085660426128 |
| XM_230584 | LOC311387 | Similar to
CG1090-PB (Drosophila melanogaster) |
−2.85086212260344 |
| XM_341007 | LOC360734 | Similar to dnaJ
(Hsp40) homolog, subfamily B, member 11 |
−2.82980905422533 |
| XM_233818 | LOC313840 | Similar to
hypothetical protein |
−2.79863476103598 |
| XM_344625 | LOC364771 | Similar to
aldo-keto reductase family 1 member C3
(Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase) (Chlordecone
reductase homolog HAKRb) (HA1753) (Dihydrodiol dehydrogenase, type
I) (Dihydrodiol dehydrogenase 3; DD3) |
−2.795863390899 |
| XM_342422 | LOC362120 | Similar to
complement C5 precursor |
−2.70059226884497 |
| NM_012674 | Spink3 | Serine peptidase
inhibitor, Kazal type 3 (Spink3) |
−2.67592384788853 |
| AY387049 | AY387049 | LRRGT00063 |
−2.65932082072998 |
| NM_147215 | Obp3 | α-2u globulin PGCL4
(Obp3) |
−2.65115608927097 |
| XM_235065 | LOC299735 | Similar to
hypothetical protein MGC35366 (LOC299735) |
−2.64919760822929 |
Quantitative real-time PCR (qPCR)
qPCR was performed using SYBR Green Super mix
(Bio-Rad, Hercules, CA, USA) in a 20 μl total volume and a Bio-Rad
iCycler iQ Real-Time Detection System according to the
manufacturer’s instructions. Primers were designed using Beacon
Designer software (Premier Biosoft International, Palo Alto, CA,
USA) and are presented in Table
III. PCR was performed in triplicate and relative gene
expression levels in normal and tumor tissue were calculated by
normalizing against β-actin expression levels using the comparative
CT method. CT represents the cycle numbers at
which the amplification reaches a threshold level selected in the
exponential phase of all PCR. Data were analyzed using the iCycle
iQ system software. Significance of expression difference was
identified by the t-test calculator in Graph pad software (GraphPad
Software, Inc., La Jolla, CA, USA).
 | Table IIIPrimer sequences used for qPCR
validation. |
Table III
Primer sequences used for qPCR
validation.
| Accession no. | Gene | Gene name | Primers
(forward/reverse) | Annealing
temperature (°C) |
|---|
| XM_217689 | Clca3 | Chloride channel
calcium activated 3 | 5′-AAG GTG GCC TAC
CTC CAA GT-3′
5′-GAG AAT AGG CGA GGC TCC TT-3′ | 58 |
| NM_053356 | Col1a2 | Procollagen, type
I, α2 | 5′-TTG ACC CTA ACC
AAG GAT GC-3′
5′-CAC CCC TTC TGC GTT GTA TT-3′ | 60 |
| NM_024129 | Dcn | Decorin | 5′-CAA TAG CAT CAC
CGT TGT GG-3′
5′-CCG GAC AGG GTT GCT ATA AA-3′ | 60 |
| XM_342827 | Glipr2 | GLI
pathogenesis-related 2 | 5′-GAA TGT CCC ACC
TCC AAA GA-3′
5′-TCA CAG GAG ATG CTC ACA GG-3′ | 60 |
| XM_213954 | Nid 1 | Nidogen 1 | 5′-CCA CCC ACA TAA
GCA TAC CC-3′
5′-ACT CCC AAG GTG TTG TCA GG-3′ | 60 |
| NM_017158 | Cyp2c7 | Cytochrome P450,
family 2, subfamily c, polypeptide 7 | 5′-ACG GGG AGA AGT
TTT CTG GT-3′
5′-TGT GCT TCC TCT TGA ACA CG-3′ | 60 |
| NM_017047 | Slc10a1 | Solute carrier
family (sodium/bile acid cotransporter family), member 1 | 10 5′-GGT GCC CTA
CAA AGG CAT TA-3′
5′-TGA TGA CAG AGA GGG CTG TG-3′ | 60 |
| Reference | | | 60 |
| β-actin | | 5′-GAC AGG ATG CAG
AAG GAG AT-3′
5′-CTG CTT GCT GAT CCA CAT CT-3′ | |
Immunohistochemical analysis
Rat CCA tissues embedded in paraffin were cut into
5-mm sections. The sections were dewaxed in Bioclear (Bio-Optica,
Milan, Italy) and rehydrated in decreasing concentrations of
ethanol. Paraffin sections were pre-treated in 0.01 M citrate
buffer in a microwave oven. Normal horse serum was used as a
blocking agent. The sections were then incubated with antibodies
against GLIpr2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, US)
and SLC10A1 (Abnova, Walnut, CA, USA). Following washing in TBS
containing 0.1% Tween-20, the sections were exposed to a secondary
antibody. Next, the slides were incubated with horseradish
peroxidase-avidin-biotin complex (Vectastain ABC Elite; Vector
Laboratories, Burlingame, CA, USA). The complex-binding site was
visualized by 3,3′-diaminobenzidine (Vector Laboratories). Sections
were counterstained with hematoxylin and dehydrated prior to
mounting with Pertex (Histolab Products AB, Gothenburg, Sweden) and
observed under a microscrope (Olympus, Yuan Li Instrument, Taipei,
Taiwan).
Results
Systemic effects of TAA administration
and tumor detection rate
No instances of TAA-induced mortality were observed
during the 20-week study period. TAA-fed rats were observed to
exhibit significantly lower levels of body weight gain compared
with the control rats beginning at 8 weeks post-treatment. Our
previous biochemical analysis revealed that levels of total
protein, albumin, aspartate aminotransferase, alkaline phosphatase
(ALK), bilirubin and prothrombin time (PT) were similar in both
groups. According to necropsy and histological results, the
incidence of TAA-induced CCA was 100% (16).
Comparative expression profiling of
TAA-induced CCA and non-cancerous liver tissue
Microarray gene expression profiling identified
10,427 differentially expressed genes (8,318 for ≥2-fold
upregulation, 3,489 for ≤0.5-fold downregulation) in CCA compared
with the non-cancerous liver tissue. Fisher 344 pre-sialomucin
complex, LOC366769 (similar to Ig heavy chain precursor V region),
Serta domain-containing 1, LOC362509 (GliPR 2), Bcl2-like 11
(apoptosis facilitator), pyruvate kinase muscle isozyme (similar to
pyruvate kinase, M1 isozyme) and LOC306628 were predominantly
overexpressed at high levels in CCA tissues; however, usher
syndrome 2A [similar to usherin (LOC289369)], TC500715,
hydroxysteroid preferring 2 (sult2a1), LOC291810, olfactory
receptor gene (Olr1692), LOC290148 (similar to T-cell receptor α
chain precursor V and C regions (TRA29)-rat (fragment) and spinal
cord expression protein 4 (RSEP4) were markedly downregulated
(Tables I and II). The top 50 upregulated and
downregulated genes were selected and classified based on their
functional involvement as demonstrated in Tables I and II.
Association of differentially expressed
genes with significant molecular processes
The top 50 genes were selected to determine their
functional involvement. Molecular databases, including KEGG and
NCBI, were used to identify the role of each gene with different
pathways. The top most differentially expressed genes in CCA were
found to play a significant role in controlling cellular metabolism
(Tables I and II). Upregulated genes were largely
classified in groups associated with cellular metabolism,
extracellular regions and ECM organization/biosynthesis,
tumorigenic cascades and other important pathways associated with
liver disorders, including fibrosis. Similarly, pathway analysis
was performed for downregulated genes. The majority of the
downregulated genes were grouped under different pathways of
various processes involved in metabolism. Specifically, Sult2a1 and
Slc10a1 were classified under roles in bile secretion.
Gene expression validation by qPCR
A number of genes, including Clca3, Col1a2, Dcn,
Glipr2 and Nid1 were selected from the microarray expression
profile based on roles associated with liver disorders and the
observed increased expression was validated. In addition, Cyp2c7
and Slc10a1 were selected to confirm significant alteration of the
expression of these genes in the tumor when compared with the
non-tumor liver samples. qPCR was performed using total RNA
extracted from CCA tissues and normal tissue samples. β-actin was
used as an internal control.
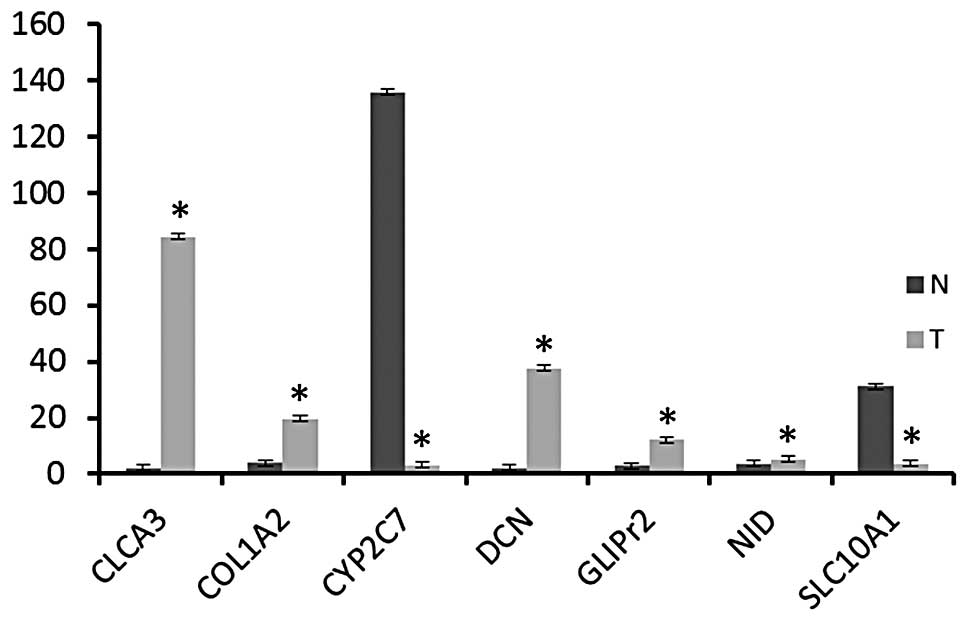
Consistent with microarray expression profiling
data, Clca3, Col1a2, Dcn, Glipr2 and Nid1 were found to be
upregulated in all rat tumor tissues compared with normal rat
tissues (Fig. 1). However,
expression of Slc10a1 and Cyp2c7 was lower in rat CCA tissues
compared with normal rat tissues (Fig.
1). These expression patterns were found to be statistically
significant (P<0.05).
Validation of GLIpr2 and SLC10A1
expression by immunohistochemical analysis
The mRNA expression levels of GLIpr2 and SLC10A1
were identified by microarray and qPCR analysis. To determine their
protein expression in CCA tissues, immunohistochemical analysis was
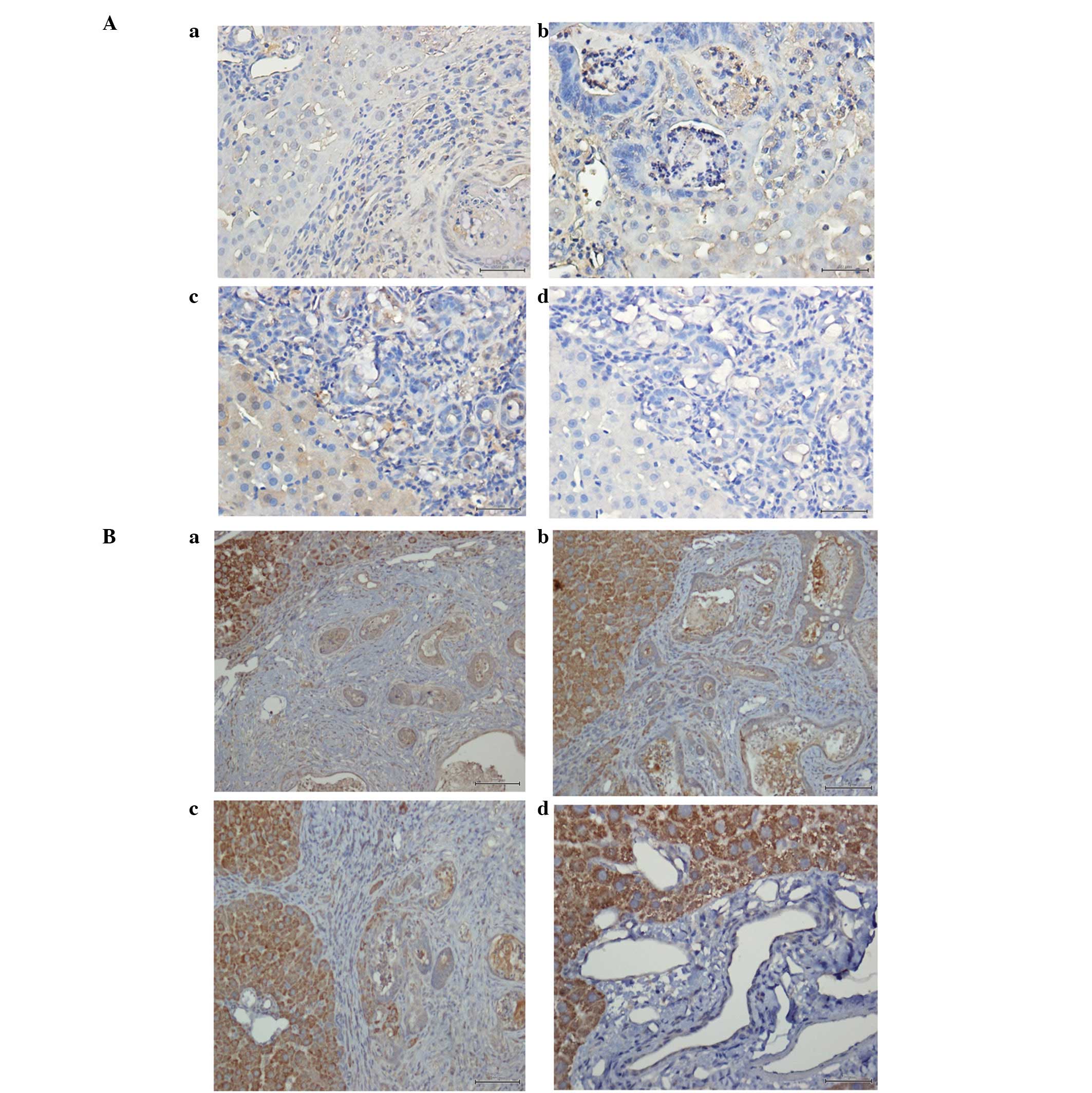
performed. GLIpr2 was observed as diffusely expressed in the
cytoplasm and at the membrane in rat CCA samples; however,
expression was absent in normal liver tissue (Fig. 2A). This observation was consistent
with mRNA expression levels obtained by microarray where GLIpr2
expression was upregulated in rat CCA compared with normal liver
tissue. Immunohistochemical validation was also performed for
SLC10A1. However, protein expression levels were observed to be
inconsistent with results obtained in the microarray;
immunohistochemical analysis revealed upregulation of SLC10A1
protein levels in rat CCA compared with normal liver tissue
(Fig. 2B), whereas, SLC10A1 mRNA
levels were identified to be downregulated.
Discussion
CCA is a malignant neoplasm which develops through a
multistep process, affecting thousands of individuals worldwide.
TAA is used as a preservative for oranges; however, it is also
considered to be a hepatotoxin and carcinogen, and requires
metabolic activation by mixed-function oxidases (27–30).
Cytochrome (CY) P450 2B, 2E1 and flavin monooxygenase metabolize
TAA into its toxic metabolites (30). Previous studies have identified a
number of TAA-induced liver diseases, including hyperplastic liver
nodules, liver cell adenomas, hepatocarcinomas, liver cirrhosis and
tumors (31–35). In our previous study, male SD rats
were administered with 300 mg/l TAA in drinking water to construct
an easy and reproducible animal model recapitulating the
multi-stage progression of human CCA. The TAA rat model may serve
as an important preclinical platform for the development of
therapeutic strategies in invasive CCA and the evaluation of
rational chemoprevention strategies in the dysplastic biliary
epithelium. Yield of invasive CCA in the model rats was 100% at
week 22 and at week 25, the yield of CCA and cirrhosis was 100%
(16).
Although TAA-induced hepatic pathology is well
characterized, a limited number of studies have analyzed the
molecular alterations in the development of CCA. For example,
alterations in the kinases, c-erb-B2 and c-met, together with
possible aberrant autocrine expression of hepatocyte growth
factor/scatter factor (HGF/SF), may play a significant role in the
development and/or progression of human CCA (17,19,36).
In addition, in our previous study the role of MUC4 as a marker of
poor prognosis in mass-forming cholangiocarcinoma (MF-CCA) patients
undergoing hepatectomy was investigated (20). The aim of the present study was to
characterize the molecular alterations associated with TAA-induced
rat CCA through cDNA microarray analysis and to identify
significantly expressed genes as distinct diagnostic biomarkers for
CCA. cDNA microarray analysis was used to identify the most common
upregulated and downregulated genes of TAA-induced CCA. The
majority of the genes were identified to play important roles in
the control of various metabolic pathways.
The liver is the major drug metabolizing organ where
several drug-metabolizing enzymes are present, including CYP450.
CYP450 is a multi-gene family of important drug-metabolizing
enzyme-encoding genes. P450 plays a key role in the metabolism of
drugs, steroids, fatty acids and environmental pollutants (37). In the present microarray analysis,
altered expression of members of the CYP450 family, including
CYP2C7, CYP1A1, CYP4A3 and CYP1A2 (Tables I and II) was identified, consistent with the
hypothesis that CYP450 family members are important for the
metabolism of carcinogens. Similar to other hepatotoxins (e.g.,
diethylnitrosamine and carbon tetrachloride), TAA resulted in a
significant reduction in the expression of CYP2C7. In agreement
with previous studies (38,39),
downregulation of CYP2C7 was found in male rats in the current
analysis. In addition, increased expression of a number of other
genes was identified, including glutathione peroxidase 3, solute
carrier family 2, aldehyde dehydrogenase family 1, procollagen
lysine and 2-oxoglutarate 5-dioxygenase 2, which are associated
with various metabolic processes. These observations indicated
that, to support the active function of cells in the CCA
environment, genes involved in the metabolism of cells must be
upregulated.
In addition, decreased expression of the
Na+-dependent taurocholate co-transporting protein
(SLC10A1; Fig. 1) was observed, a
protein responsible for the majority of hepatocellular uptake of
bile salt-coupled chemotherapeutics (40). Previously, downregulation of Ntcp1
(Slc10a1) protein levels has been implicated in cholestasis
(41). Reduced expression of
Sult2a1 and Slc10a1, genes important for bile secretion (Table III), may play an important role
in CCA aetiopathogenesis and those specific proteins may represent
future biomarkers.
Increased expression of CLCA3, COL1A2, DCN, GLIpr2
and NID1 was further validated by qPCR (Fig. 1). DCN is a member of the small
leucine-rich repeat proteoglycan family and is a major component of
the extracellular matrix (42).
DCN has been reported to mediate a number of functions, including
proliferation, migration and differentiation of human keratinocytes
by interacting with the epidermal growth factor receptor, ErbB2
(43), TGFβ (44) and cytokines. In addition to its
well-known role in extracellular matrix organization, previous
studies have also reported abnormal expression in a number of types
of cancer, including oral cancer (45). In the present study, DCN was found
to be differentially expressed in CCA, indicating its appearance
and overexpression as a possible biological marker of CCA
progression.
Nid is an important constituent of basement
membranes, which forms a defined supramolecular complex between the
extracellular matrix molecules, laminin-1 and type IV collagen
(46). Previously, Nid and
specific laminin chains were revealed to play a crucial role in
determining the outcome of hepatic injury, in a study involving
partial hepatectomy. Increased expression of Nid1 may be involved
in the concomitant correlation between TAA-induced rat CCA and
liver cirrhosis.
In a previous study, increased GLIpr-2 expression in
the kidney was hypothesized to contribute to the development of
fibrosis by increasing the pool of activated fibroblasts, possibly
through the induction of epithelial-mesenchymal transition
(47). The biological function of
GLIpr-2 remains poorly understood. The enhanced expression of
GLIpr-2 in TAA-induced CCA may play a pivotal role in liver
fibrosis and represent an additional molecular target which must be
analyzed further.
In conclusion, the extensive information gained from
the gene expression profiling of TAA-induced CCA performed in the
present study is likely to provide important insights into the
genes involved in the development of CCA. Further studies must be
performed to develop a further understanding of the cellular
activities of differentially expressed genes during CCA
progression.
Acknowledgements
The present study was supported by grants from the
Chang Gung Medical Research Program (no. CMRPG3B0531, CMRPG3B0532,
CMRPG3B0361 and CMRPG3B0362).
References
|
1
|
Liver Cancer Study Group of Japan.
Classification of Primary Liver Cancer. 1st English edition.
Kanehara Press Co; Tokyo: 1997
|
|
2
|
Yeh CN, Yeh TS, Chen TC, Jan YY and Chen
MF: Gross pathological classification of peripheral
cholangiocarcinoma determines the efficacy of hepatectomy. J
Gastroenterol. Sep 25–2012.(Epub ahead of print).
|
|
3
|
Chen MF: Peripheral cholangiocarcinoma
(cholangiocellular carcinoma): clinical features, diagnosis and
treatment. J Gastroenterol Hepatol. 14:1144–1149. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shaib YH, Davila JA, McGlynn K and
El-Serag HB: Rising incidence of intrahepatic cholangiocarcinoma in
the United States: a true increase? J Hepatol. 40:472–477. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Patel T: Worldwide trends in mortality
from biliary tract malignancies. BMC Cancer. 2:102002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chang KY, Chang JY and Yen Y: Increasing
incidence of intrahepatic cholangiocarcinoma and its relationship
to chronic viral hepatitis. J Natl Compr Canc Netw. 7:423–427.
2009.PubMed/NCBI
|
|
7
|
Shaib Y and El-Serag HB: The epidemiology
of cholangiocarcinoma. Semin Liver Dis. 24:115–125. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
DeGroen PC, Gores GJ, LaRusso NF,
Gunderson LL and Nagorney DM: Biliary tract cancers. N Engl J Med.
341:1368–1378. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Albores-Saavedra J, Henson DE and Sobin
LH: The WHO histological classification of tumors of the
gallbladder and extrahepatic bile ducts. A commentary on the second
edition. Cancer. 70:410–414. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weber SM, Jarnagin WR, Klimstra D,
DeMatteo RP, Fong Y and Blumgart LH: Intrahepatic
cholangiocarcinoma: resectability, recurrence pattern and outcomes.
J Am Coll Surg. 193:384–391. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Uenishi T, Hirohashi K, Kubo S, et al:
Histologic factors affecting prognosis following hepatectomy for
intrahepatic cholangiocarcinoma. World J Surg. 25:865–869. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jarnagin WR, Fong Y and DeMatteo RP:
Staging, resectability and outcome in 225 patients with hilar
cholangiocarcinoma. Ann Surg. 234:507–519. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Patel T: Increasing incidence and
mortality of primary intrahepatic cholangiocarcinoma in the United
States. Hepatology. 33:1353–1357. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shirabe K, Shimada M and Harimoto N:
Intrahepatic cholangiocarcinoma: Its mode of spreading and
therapeutic modalities. Surgery. 131:S159–S164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bathe OF, Pacheco JT and Ossi PB:
Management of hilar bile duct carcinoma. Hepatogastroenterology.
48:1289–1294. 2001.PubMed/NCBI
|
|
16
|
Yeh CN, Maitra A, Lee KF, Jan YY and Chen
MF: Thioacetamide-induced intestinal-type cholangiocarcinoma in
rat: an animal model recapitulating the multi-stage progression of
human cholangiocarcinoma. Carcinogenesis. 25:631–636.
2004.PubMed/NCBI
|
|
17
|
Terada T, Nakanuma Y and Sirica AE:
Immunohistochemical demostration of MET overexpression in human
intrahepatic cholangiocarcinoma and in hepatolithiasis. Hum Pathol.
29:175–180. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Endo K, Yoon BI, Pairojkul C, Demetris AJ
and Sirica AE: ERBB-2 overexpression and cyclooxygenase-2
up-regulation in human cholangiocarcinoma and risk conditions.
Hepatology. 36:439–450. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hansel DE, Rahman A, Hidalgo M, et al:
Identification of novel cellular targets in biliary tract cancers
using global gene expression technology. Am J Pathol. 163:217–229.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yeh CN, Pang ST, Wu RC, Chen TW, Jan YY
and Chen MF: Prognostic value of MUC4 for mass-forming intrahepatic
cholangiocarcinoma after hepatectomy. Oncol Rep. 21:49–56.
2009.PubMed/NCBI
|
|
21
|
National Institutes of Health. Guide for
the Care and Use of Laboratory Animals. NIH publication no. 85–23,
revised 1996.
|
|
22
|
Pang ST, Dillner K, Wu X, Pousette A,
Norstedt G and Flores-Morales A: Gene expression profiling of
androgen deficiency predicts a pathway of prostate apoptosis that
involves genes related to oxidative stress. Endocrinology.
143:4897–906. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular datasets. Nucleic Acids Res. 40:D109–D114.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nature Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
26
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dasgupta A, Chatterjee R and Chowdhury JR:
Thioacetamide-induced hepatocarcinoma in rat. Oncology. 38:249–253.
1981. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fitzhugh OG and Nielson AA: Liver tumors
in rats fed thiourea or thioacetamide. Science. 108:626–628. 1948.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Al-Bader AA, Mathew TC, Abul H, Al-Mosawi
M, Panigrahi D and Dashti HM: Bacterial translocation in
thioacetamide induced liver cirrhosis. J R Coll Surg Edinb.
43:278–82. 1998.PubMed/NCBI
|
|
30
|
Lee JW, Shin KD, Lee M, et al: Role of
metabolism by flavin-containing monooxygenase in
thioacetamide-induced immunosuppression. Toxicol Lett. 136:163–172.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dashti H, Jeppsson B, Hagerstrand I, et
al: Early biochemical and histological changes in rats exposed to a
single injection of thioacetamide. Pharmacol Toxicol. 60:171–174.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gallagher CH, Gupta DN, Judah JD and Rees
KK: Biochemical changes in acute thioacetamide intoxication. J Path
Bact. 72:193–201. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gupta DN: Acute changes in the liver after
administration of thioacetamide. J Pathol Bacteriol. 72:183–192.
1956. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Muller D, Zimmerman SI and Schiller F:
Drug metabolism in rat liver injured by thioacetamide. Arch
Toxicol. 5:368–371. 1982. View Article : Google Scholar
|
|
35
|
Dashti HM, Mathew TC, Jadaon MM and
Ashkanani E: Zinc and liver cirrhosis: biochemical and
histopathologic assessment. Nutrition. 13:206–212. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sirica AE, Lai GH and Zhang Z: Biliary
cancer growth factor pathways, cyclo-oxygenase-2 and potential
therapeutic strategies. J Gastroenterol Hepatol. 16:363–372. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nelson DR, Zeldin DC, Hoffman SM, Maltais
LJ, Wain HM and Nebert DW: Comparison of cytochrome P450 (CYP)
genes from the mouse and human genomes, including nomenclature
recommendations for genes, pseudogenes and alternative-splice
variants. Pharmacogenetics. 14:1–18. 2004. View Article : Google Scholar
|
|
38
|
Henderson CJ, Russell AL, Allan JA and
Wolf CR: Sexual differentiation and regulation of cytochrome P-450
CYP2C7. Biochim Biophys Acta. 1118:99–106. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Arun KA and Bernard HS: Latent
overexpression of hepatic CYP2C7 in adult male and female rats
neonatally exposed to phenobarbital: a developmental profile of
gender-dependent P450s. J Pharmacol Exp Ther. 293:1027–1033.
2000.PubMed/NCBI
|
|
40
|
Hagenbuch B, Stieger B, Foguet M, Lübbert
H and Meier PJ: Functional expression, cloning and characterization
of the hepatocyte Na+/bile acid cotransport system. Proc
Natl Acad Sci USA. 88:10629–10633. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Trauner M, Meier PJ and Boyer JL:
Molecular regulation of hepatocellular transport systems in
cholestasis. J Hepatol. 31:165–178. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kjellén L and Lindahl U: Proteoglycans:
structures and interactions. Annu Rev Biochem. 60:443–475.
1991.
|
|
43
|
Santra M, Eichstetter I and Iozzo RV: An
anti-oncogenic role for decorin. Downregulation of ErbB2 leads to
growth suppression and cytodifferentiation of mammary carcinoma
cells. J Biol Chem. 275:35153–35161. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Z, Li XJ, Liu Y, Zhang X, Li YY and
Xu WS: Recombinant human decorin inhibits cell proliferation and
downregulates TGF-beta1 production in hypertrophic scar
fibroblasts. Burns. 33:634–641. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Abhijit GB, Indraneel B, William ML and
Jamboor KV: Aberrant expression and localization of decorin in
human oral dysplasia and squamous cell carcinoma. Cancer Res.
63:7769–7776. 2003.PubMed/NCBI
|
|
46
|
Pujuguet P, Simian M, Liaw J, Timpl R,
Werb Z and Bissell MJ: Nidogen-1 regulates laminin-1-dependent
mammary-specific gene expression. J Cell Sci. 113:849–858.
2000.PubMed/NCBI
|
|
47
|
Baxter RM, Crowell TP, George JA, Getman
ME and Gardner H: The plant pathogenesis related protein GLIPR-2 is
highly expressed in fibrotic kidney and promotes epithelial to
mesenchymal transition in vitro. Matrix Biol. 1:20–29. 2007.
View Article : Google Scholar
|