Introduction
Rheumatoid arthritis (RA) is an autoimmune disease
that affects ~1% of the adult population (1). RA is considered to be a complex
disease and its etiology and pathogenesis remain unclear (2). Genetic and environmental factors, and
their interactions, are involved in the development of RA (3). The strongest genetic associations are
with genes located in the human leukocyte antigen (HLA) region,
particularly the HLA-DRB1 gene (4,5).
Rheumatoid factor (RF) production in RA patients is correlated with
smoking. Mattey et al(6)
demonstrated that patients that were HLA-DRB1*0401 positive and had
previously smoked were likely to be RF positive. Various genes,
including the protein tyrosine phosphatase non-receptor 22 (PTPN22)
gene, are also known to be associated with RA (7). Karlson et al(8) demonstrated that HLA-DRB1 shared
epitope (HLA-SE) and PTPN22 were correlated with presence of cyclic
citrullinated peptide (CCP) and RF. However, the strength of the
association between these genes and RA vary geographically. For
example, PTPN22 is associated with RA in populations of European
origin, but not in non-European populations (9,10).
In contrast, peptidyl arginine deiminase type IV, solute carrier
family 22 (organic cation/ergothioneine transporter) member 4 and
Fc receptor-like 3 are associated with RA in the Japanese
population and in other Asian groups (11–13);
however, yields weak or negative results in European populations
(14,15). These divergent results suggest the
genetic heterogeneity of RA across human populations.
Forkhead proteins are a large family of functionally
diverse transcription factors that have been implicated in a
variety of cellular processes (16). Forkhead box J3 (FOXJ3) is a member
of the FOX gene family, which encodes transcription factors
composed of a conserved DNA binding motif termed the forkhead
domain (17). Based on
interpretations of the mouse and human genome sequences, human FOX
genes have ~43 members, which have been classified into 17
subclasses: FOXA to FOXQ (18).
Numerous members of this family are crucial in the regulation of
immune responses and it has been demonstrated that FOXP3 is
important in autoimmune diseases (19). Li et al(20) showed that a FOXJ1 polymorphism
(rs3192453, 3375G>C) was associated with allergic rhinitis and
that another FOXJ1 polymorphism was related to systemic lupus
erythematosus and RA in the Korean population (21). However, no genetic study of FOXJ3
has currently been conducted.
In the present study, it was investigated whether
FOXJ3 polymorphisms were correlated with susceptibility to RA.
Patients and methods
Patients
A case-control study was conducted to investigate
the correlation between RA and FOXJ3 polymorphisms. Unrelated
patients (n=307) with RA (59 males and 248 females) were recruited
from rheumatology centers in the Republic of Korea (Kyung Hee
University Hospital, Seoul, Korea). Each patient was diagnosed by a
rheumatologist based on the ACR 1987 rheumatoid arthritis
diagnostic criteria (22). For the
control group, 476 subjects (207 males and 269 females) were
recruited from participants undergoing a general health check-up
program, following the confirmation of no clinical implication of
rheumatic disease or any other disorder. All procedures were
conducted according to the Declaration of Helsinki guidelines.
Written informed consent was obtained from each subject. This study
was approved by the Ethics Review Committee of the Medical Research
Institute, School of Medicine, Kyung Hee University (Seoul,
Korea).
Clinical data, such as gender, age at disease onset
and the presence of bone erosion were obtained by reviewing medical
records at the time of enrollment. Laboratory data included
erythrocyte sedimentation rate (ESR), C-reactive protein (CRP),
anti-cyclic citrullinated peptide (anti-CCP) and RF values.
SNP selection and genotyping
All tagging SNPs of FOXJ3 analyzed in this study
were selected from our unpublished results of a 100K genome-wide
association analysis (Illumina, Inc., San Diego, CA, USA) in RA.
From the International HapMap Project data set (http://www.hapmap.org), the SNPs with an optimal minor
allele frequency (≥5%), heterozygosity >0.1 and the best
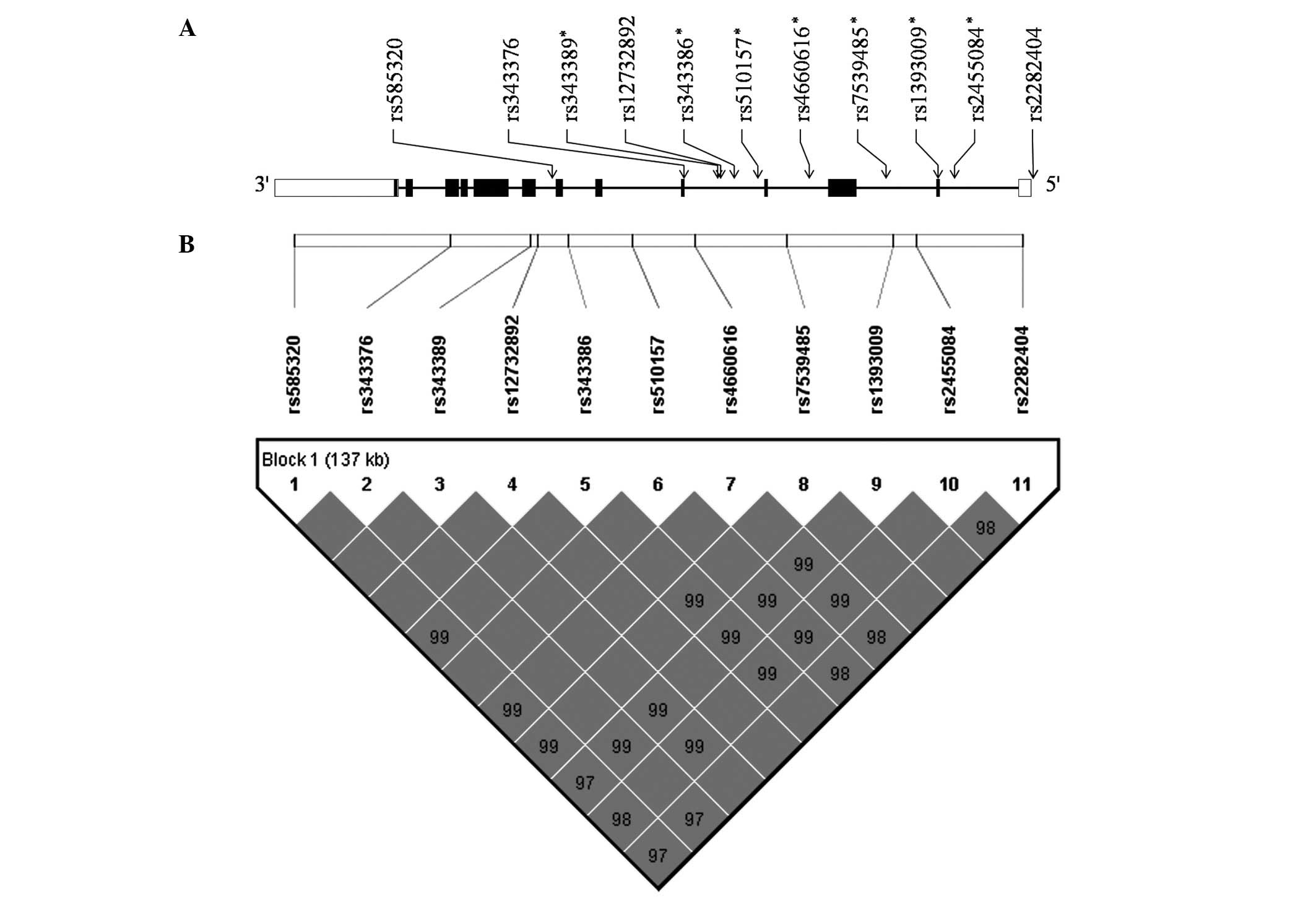
coverage to serve as tag SNPs were genotyped. A total of 11 SNPs
[rs2282404 (5′-near gene), rs2455084 (intron 1), rs1393009 (intron
1), rs7539485 (intron 2), rs4660616 (intron 3), rs510157 (intron
4), rs343386 (intron 4), rs12732892 (intron 4), rs343389 (intron
4), rs343376 (exon 5, Ala161Val) and rs585320 (intron 7)] were
selected. The locations of the 11 selected SNPs in the region of
FOXJ3 are shown in Fig. 1A.
Genomic DNA was extracted using a commercially available Qiagen DNA
extraction kit (Qiagen, Tokyo, Japan) from blood samples collected
in EDTA. SNP genotyping was performed using direct sequencing with
genomic DNA that was amplified using primers for each SNP. The
sequences of the primers are shown in Table I. The samples were sequenced using
an ABI Prism 377 automatic sequencer (Applied Biosystems, Inc.,
Foster City, CA, USA). Sequence data were analyzed using the
SeqManII software (DNAStar, Inc., Madison, WI, USA).
 | Table IPrimer sequences for FOXJ3
polymorphisms. |
Table I
Primer sequences for FOXJ3
polymorphisms.
| SNP | Primer | Size (bp) |
|---|
| rs2282404 | Sense:
5′-CCATAACCTGGGTAGCTACCTTGAG-3′
Antisense: 5′-TTCCGAGCGACTTGGGGTCCTTGT-3′ | 441 |
| rs2455084 | Sense:
5′-TGCGATGGCTGCTTCCTAAATGTGG-3′
Antisense: 5′-AAATGACGTCCTGAACCCTAGCCTT-3′ | 488 |
| rs1393009 | Sense:
5′-GAACCTTGCATGCTTAACTCTGTGG-3′
Antisense: 5′-ACAAGCCTGTCCATACAAACCCATC-3′ | 452 |
| rs7539485 | Sense:
5′-CATGATCATAGCTCACTGCAGCCT-3′
Antisense: 5′-CATCAGACAAGTCAGTGGGGAAAGG-3′ | 603 |
| rs4660616 | Sense:
5′-CTGGCTGACTCAGTATGAGAAGTGA-3′
Antisense: 5′-GGAAGATCTGCCATGTGCCCTAAA-3′ | 492 |
| rs510157 | Sense:
5′-CCTGGCCACAAAGCGAGGTTTAAA-3′
Antisense: 5′-TGTAATTCCAGGTGATTTGGGGGGT-3′ | 557 |
| rs343386 | Sense:
5′-GAGCTTCCCTTTGGTCCTTCTTGTG-3′
Antisense: 5′-AGACAGCTACATCATTCGGGAGCT-3′ | 757 |
| rs12732892 | Sense:
5′-GATTAGTGTGGCTGTCAGTGTATTG-3′
Antisense: 5′-CATACCAAAACCTATGGAGTATAGC-3′ | 512 |
| rs343389 | Sense:
5′-CCATTCAGGTAGTCTGTACCTTT-3′
Antisense: 5′-GCAGAAACCTTACAGACCAGGAGAG-3′ | 481 |
| rs343376 | Sense:
5′-CCATGTAGATTTGTGAGGTCCTGGG-3′
Antisense: 5′-CCTAAACTGTCCCTCAGAGTTAAG-3′ | 661 |
| rs585320 | Sense:
5′-GTGTCAACATGGTTTCCAGCTGTAG-3′
Antisense: 5′-CTGCTACGTTTTTCTTTCCCGTGTG-3′ | 433 |
Statistical analysis
Hardy-Weinberg equilibrium (HWE) for 11 SNPs was
assessed using SNPStats (http://bioinfo.iconcologia.net/SNPstats). A linkage
disequilibrium (LD) block of polymorphisms was tested using
Haploview version 4.2 (Broad Institute of MIT and Harvard,
Cambridge, MA, USA). The haplotypes and their frequencies were
calculated by the expectation-maximization algorithm. Multiple
logistic regression analyses were conducted to calculate the odds
ratios, 95% confidence intervals and corresponding P-values.
SNPStats, HapAnalyzer, Helixtree software (Golden Helix, Inc.,
Bozeman, MT, USA) and SNPAnalyzer (ISTECH, Inc., Goyang, Korea)
were used to evaluate genetic data. Logistic regression analysis in
all three analysis models (log-additive, dominant and recessive
models for rare alleles) was utilized to demonstrate alternative
effects of the variants (23).
P<0.05 was considered to indicate a statistically significant
difference.
Results
The clinical characteristics of the 307 RA patients
(248 females and 59 males) and 476 control subjects are shown in
Table II. The mean age of the RA
patients was 45.56±12.91 years and the mean age of the control
subjects was 44.45±12.70 years. The mean disease duration was
4.68±4.53 years. The mean level of ESR in RA patients was
41.00±29.29 mm/h and CRP was 2.26±4.84 mg/dl. Among the 307 RA
patients, 268 patients were positive for RF (87.3%) and 143
patients exhibited bone erosion (46.6%).
 | Table IIGeneral characteristics of study
subjects. |
Table II
General characteristics of study
subjects.
| Characteristic | Patients with RA
(n=307) | Controls (n=476) |
|---|
| Age (years, mean ±
SD) | 46.56±12.91 | 44.45±12.70 |
| Gender
(male/female) | 59/248 | 207/269 |
| Disease duration
(years, mean ± SD) | 4.68±4.53 | |
| Erythrocyte
sedimentation rate (mm/h, mean ± SD) | 41.00±29.29 | |
| C-reactive protein
(mg/dl, mean ± SD) | 2.26±4.84 | |
| Patients with bone
erosion (n) | 143 | |
| RF positive (n) | 268 | |
| Anti-CCP positive
(n) | 90 | |
In the control group, the genotype distributions for
all SNPs were in HWE (data not shown). The genotype frequencies of
the polymorphisms were compared between the RA and the normal
control groups using logistic regression models with adjustment for
age and gender. The genotype distributions of FOXJ3 gene
polymorphisms in the RA and control groups are shown in Table III. Of the 11 SNPs, seven
(rs2455084, rs1393009, rs7539485, rs4660616, rs510157, rs343386 and
rs343389) were significantly associated with the risk of RA. The
rare alleles of rs2455084 (rare allele, A), rs1393009 (rare allele,
C), rs7539485 (rare allele, T), rs4660616 (rare allele, G),
rs510157 (rare allele, T), rs343386 (rare allele, C) and rs343389
(rare allele, T) were associated with a decreased risk of RA. The
rest of the SNPs (rs2282404, rs12732892, rs343376 and rs585320)
were not associated with RA (Table
III).
 | Table IIIGenotype frequencies of FOXJ3
polymorphisms in patients with rheumatoid arthritis and control
subjects. |
Table III
Genotype frequencies of FOXJ3
polymorphisms in patients with rheumatoid arthritis and control
subjects.
| SNP | | RA | Control | Log-additive | Dominant | Recessive |
|---|
| |
|
|
|
|
|
|---|
| (Locus) | Genotype | n (%) | n (%) | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| rs2282404 | C/C | 147 (50.0) | 246 (55.7) | 0.82 (0.65–1.04) | 0.098 | 0.80 (0.59–1.07) | 0.130 | 0.73 (0.42–1.27) | 0.270 |
| (promoter) | T/C | 122 (41.5) | 168 (38.0) | | | | | | |
| T/T | 25 (8.5) | 28 (6.3) |
| rs2455084 | T/T | 184 (61.3) | 252 (53.5) | 1.31
(1.04–1.65) | 0.020 | 1.38
(1.03–1.85) | 0.032 | 1.55
(0.90–2.68) | 0.110 |
| (intron 1) | A/T | 96 (32.0) | 172 (36.5) | | | | | | |
| A/A | 20 (6.7) | 47 (10.0) |
| rs1393009 | T/T | 183 (62.9) | 252 (53.4) | 1.37
(1.09–1.73) | 0.007 | 1.48
(1.10–1.99) | 0.010 | 1.58
(0.91–2.76) | 0.096 |
| (intron 1) | T/C | 89 (30.6) | 173 (36.6) | | | | | | |
| C/C | 19 (6.5) | 47 (10.0) |
| rs7539485 | C/C | 183 (61.8) | 239 (54.6) | 1.28
(1.01–1.61) | 0.037 | 1.35
(1.00–1.82) | 0.051 | 1.46
(0.85–2.51) | 0.160 |
| (intron 2) | T/C | 92 (31.1) | 155 (35.4) | | | | | | |
| T/T | 21 (7.1) | 44 (10.1) |
| rs4660616 | T/T | 182 (60.1) | 251 (53.3) | 1.28
(1.02–1.60) | 0.033 | 1.32
(0.98–1.77) | 0.063 | 1.56
(0.92–2.66) | 0.095 |
| (intron 3) | T/G | 100 (33) | 171 (36.3) | | | | | | |
| G/G | 21 (6.9) | 49 (10.4) |
| rs510157 | G/G | 184 (62.0) | 239 (53.5) | 1.33
(1.06–1.68) | 0.015 | 1.42
(1.05–1.91) | 0.022 | 1.55
(0.90–2.68) | 0.110 |
| (intron 4) | T/G | 93 (31.3) | 163 (36.5) | | | | | | |
| T/T | 20 (6.7) | 45 (10.1) |
| rs343386 | T/T | 182 (60.3) | 249 (53.1) | 1.28
(1.02–1.60) | 0.032 | 1.34
(1.00–1.80) | 0.050 | 1.49
(0.87–2.55) | 0.140 |
| (intron 4) | T/C | 99 (32.8) | 173 (36.9) | | | | | | |
| C/C | 21 (7.0) | 47 (10.0) |
| rs12732892 | T/T | 217 (72.6) | 320 (68.1) | 1.17
(0.90–1.52) | 0.240 | 1.24
(0.90–1.71) | 0.180 | 1.08
(0.54–2.18) | 0.830 |
| (intron 4) | T/C | 69 (23.1) | 128 (27.2) | | | | | | |
| C/C | 13 (4.3) | 22 (4.7) |
| rs343389 | C/C | 183 (61.0) | 252 (53.5) | 1.29
(1.02–1.61) | 0.029 | 1.36
(1.01–1.82) | 0.040 | 1.47
(0.86–2.52) | 0.150 |
| (intron 4) | T/C | 96 (32.0) | 172 (36.5) | | | | | | |
| T/T | 21 (7.0) | 47 (10.0) |
| rs343376 | C/C | 219 (72.5) | 321 (68.3) | 1.16
(0.90–1.51) | 0.250 | 1.22
(0.89–1.68) | 0.210 | 1.14
(0.57–2.29) | 0.700 |
| (exon 5, Ala161
Val) | T/C | 70 (23.2) | 126 (26.8) | | | | | | |
| T/T | 13 (4.3) | 23 (4.9) |
| rs585320 | T/T | 217 (72.1) | 320 (67.7) | 1.14
(0.88–1.48) | 0.300 | 1.24
(0.90–1.70) | 0.190 | 0.97
(0.50–1.90) | 0.940 |
| (intron 7) | T/C | 69 (22.9) | 130 (27.5) | | | | | | |
| C/C | 15 (5.0) | 23 (4.9) |
FOXJ3, located on chromosome 1p34.2, consists of 622
amino acids and has a molecular mass of 68,928 Da. The missense
polymorphism rs343376 (Ala162Val) is the only SNP with
heterozygosity >0.1 among the coding SNPs located in the FOXJ3
gene region (http://www.ncbi.nlm.nih.gov/SNP). CC, CT and TT
genotype frequencies are 0.467, 0.417 and 0.117 in European; 0.822,
0.178 and 0.000 in Chinese; 0.750, 0.227 and 0.023 in Japanese and
0.467, 0.433 and 0.100 in Sub-Saharan African populations
(http://www.ncbi.nlm.nih.gov/SNP). The
CC, CT and TT genotype frequencies in the population of the present
study were 0.680, 0.270 and 0.040, which are similar to those in
the Japanese population. However, the rs343376 polymorphism was not
associated with RA in the study population (Table III). The SNP genotype frequencies
obtained from the study population are similar to frequencies
observed in the Japanese population, according to the NCBI
website.
A strong LD block, including all SNPs, was
constructed using the Gabriel method (Fig. 1B). Four haplotypes in the block
exhibited frequencies >0.05 and all were used for haplotype
association analysis. The frequency of the TCCTTGTCTTC haplotype
was 0.465, TCCTTGTCTTT 0.271, CTTCCTGTCAC 0.175 and TCTTCTGTCAC
0.086, respectively. Two haplotypes (TCCTTGTCTTT and TCTTCTGTCAC)
were significantly associated with RA (Table IV).
 | Table IVHaplotype association of FOXJ3 in
patients with rheumatoid arthritis and control subjects. |
Table IV
Haplotype association of FOXJ3 in
patients with rheumatoid arthritis and control subjects.
| | RA | Control | Log-additive | Dominant | Recessive |
|---|
| |
|
|
|
|
|
|---|
| Haplotype | Genotype | n (%) | n (%) | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| TCCTTGTCTTC | H/H | 77 (25.1) | 105 (22.1) | 1.00
(0.82–1.22) | 0.995 | 0.86
(0.62–1.18) | 0.350 | 1.18
(0.84–1.66) | 0.329 |
| H/− | 140 (45.6) | 246 (51.7) |
| −/− | 90 (29.3) | 125 (26.3) |
| TCCTTGTCTTT | H/H | 25 (8.1) | 28 (5.9) | 1.28
(1.02–1.61) | 0.035 | 1.35
(1.01–1.80) | 0.043 | 1.42
(0.81–2.48) | 0.221 |
| H/− | 122 (39.7) | 165 (34.7) |
| −/− | 160 (52.1) | 283 (59.5) |
| CTTCCTGTCAC | H/H | 13 (4.2) | 22 (4.6) | 0.86
(0.67–1.12) | 0.264 | 0.82
(0.60–1.12) | 0.212 | 0.91
(0.45–1.84) | 0.798 |
| H/− | 74 (24.1) | 133 (27.9) |
| −/− | 220 (71.7) | 321 (67.4) |
| TCTTCTGTCAC | H/H | 3 (1.0) | 4 (0.8) | 0.66
(0.45–0.96) | 0.032 | 0.60
(0.40–0.91) | 0.017 | 1.16
(0.26–5.24) | 0.843 |
| H/− | 34 (11.1) | 84 (17.6) |
| −/− | 270 (87.9) | 388 (81.5) |
To determine whether FOXJ3 SNPs influence the
clinical features of RA, the RA group was divided according to
clinical characteristics such as ESR (≥30 or <30 mm/h), CRP
(≥0.5 or <0.5 mg/dl), bone erosion (positive or not), anti-CCP
(positive and negative) and RF (positive or negative). We observed
an association between anti-CCP and SNP (rs585320). The SNP
rs585320 was significant in the recessive model (OR=0.12, 95%
CI=0.01–0.99, P-value=0.015; Table
V). However, no association was observed between FOXJ3
polymorphisms and the clinical characteristics (ESR, CRP, bone
erosion and RF).
 | Table VGenotype frequencies of FOXJ3
polymorphisms in rheumatoid arthritis patients with anti-CCP
(negative) and anti-CCP (positive). |
Table V
Genotype frequencies of FOXJ3
polymorphisms in rheumatoid arthritis patients with anti-CCP
(negative) and anti-CCP (positive).
| SNP | | anti-CCP (−) | anti-CCP (+) | Log-additive | Dominant | Recessive |
|---|
| |
|
|
|
|
|
|---|
| (Locus) | Genotype | n (%) | n (%) | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| rs2282404 | C/C | 36 (43.4) | 43 (48.3) | 0.89
(0.56–1.40) | 0.60 | 0.83
(0.45–1.52) | 0.54 | 0.94
(0.35–2.50) | 0.89 |
| (promoter) | T/C | 38 (45.8) | 37 (41.6) | | | | | | |
| T/T | 9 (10.8) | 9 (10.1) |
| rs2455084 | T/T | 45 (52.3) | 58 (64.4) | 0.64
(0.39–1.05) | 0.08 | 0.62
(0.33–1.14) | 0.12 | 0.44
(0.13–1.53) | 0.18 |
| (intron 1) | A/T | 33 (38.4) | 28 (31.1) | | | | | | |
| A/A | 8 (9.3) | 4 (4.4) |
| rs1393009 | T/T | 44 (55.0) | 58 (64.4) | 0.69
(0.42–1.14) | 0.15 | 0.68
(0.36–1.26) | 0.22 | 0.48
(0.13–1.70) | 0.24 |
| (intron 1) | T/C | 29 (36.2) | 28 (31.1) | | | | | | |
| C/C | 7 (8.8) | 4 (4.4) |
| rs7539485 | C/C | 44 (54.3) | 58 (65.2) | 0.66
(0.40–1.09) | 0.10 | 0.65
(0.35–1.21) | 0.17 | 0.42
(0.12–1.46) | 0.16 |
| (intron 2) | T/C | 29 (35.8) | 27 (30.3) | | | | | | |
| T/T | 8 (9.9) | 4 (4.5) |
| rs4660616 | T/T | 44 (51.2) | 58 (64.4) | 0.63
(0.38–1.03) | 0.06 | 0.59
(0.32–1.09) | 0.100 | 0.44
(0.13–1.52) | 0.18 |
| (intron 3) | T/G | 34 (39.5) | 28 (31.1) | | | | | | |
| G/G | 8 (9.3) | 4 (4.4) |
| rs510157 | G/G | 45 (55.6) | 58 (65.2) | 0.68
(0.41–1.11) | 0.12 | 0.67
(0.36–1.24) | 0.20 | 0.42
(0.12–1.47) | 0.16 |
| (intron 4) | T/G | 28 (34.6) | 27 (30.3) | | | | | | |
| T/T | 8 (9.9) | 4 (4.5) |
| rs343386 | T/T | 45 (52.3) | 58 (64.4) | 0.64
(0.39–1.05) | 0.08 | 0.62
(0.33–1.13) | 0.12 | 0.44
(0.13–1.53) | 0.18 |
| (intron 4) | T/C | 33 (38.4) | 28 (31.1) | | | | | | |
| C/C | 8 (9.3) | 4 (4.4) |
| rs12732892 | T/T | 58 (69.0) | 66 (73.3) | 0.70
(0.40–1.21) | 0.20 | 0.81
(0.42–1.57) | 0.53 | 0.13
(0.02–1.12) | 0.03 |
| (intron 4) | T/C | 20 (23.8) | 23 (25.6) | | | | | | |
| C/C | 6 (7.1) | 1 (1.1) |
| rs343389 | C/C | 45 (54.2) | 58 (64.4) | 0.67
(0.41–1.09) | 0.11 | 0.66
(0.36–1.22) | 0.18 | 0.43
(0.12–1.48) | 0.17 |
| (intron 4) | T/C | 30 (36.1) | 28 (31.1) | | | | | | |
| T/T | 8 (9.6) | 4 (4.4) |
| rs343376 | C/C | 58 (69.0) | 66 (74.2) | 0.67
(0.38–1.17) | 0.16 | 0.76
(0.39–1.49) | 0.43 | 0.13
(0.02–1.14) | 0.03 |
| (exon 5, Ala161
Val) | T/C | 20 (23.8) | 22 (24.7) | | | | | | |
| T/T | 6 (7.1) | 1 (1.1) |
| rs585320 | T/T | 58 (67.4) | 66 (73.3) | 0.65
(0.38–1.12) | 0.12 | 0.75
(0.39–1.44) | 0.39 | 0.12
(0.01–0.99) | 0.015 |
| (intron 7) | T/C | 21 (24.4) | 23 (25.6) | | | | | | |
| C/C | 7 (8.1) | 1 (1.1) |
Discussion
RA is a common autoimmune disease with a complex
etiology involving genetic and environmental factors. The
association between FOXJ3 polymorphisms and susceptibility to RA
was investigated in the present study. The results suggested that
certain SNPs in the FOXJ3 gene contribute to RA. The case-control
analysis demonstrated that of 11 SNPs analyzed in FOXJ3, seven
(rs2455084, rs1393009, rs7539485, rs4660616, rs510157, rs343386 and
rs343389) are significantly associated with a susceptibility to RA.
Moreover, two haplotypes (TCCTTGTCTTT and TCTTCTGTCAC) are
significantly associated with RA. To the best of our knowledge,
this is the first study to demonstrate that FOXJ3 polymorphisms are
associated with RA.
It was also analyzed whether FOXJ3 polymorphisms are
related to diagnostic markers reflecting RA disease activity, such
as the levels of CRP, ESR, anti-CCP and RF, and the presence of
bone erosion. Based on a value for ESR of 30 mm/h, CRP of 0.5
mg/dl, the existence of bone erosion and the presence of anti-CCP
(positive or negative) and RF (positive and negative); the RA
patient sample was divided into two clinical subgroups and the
correlation between clinical data and FOXJ3 polymorphisms was
investigated. The results demonstrated that anti-CCP was associated
with FOXJ3 SNP.
Forkhead proteins are a large family of functionally
diverse transcription factors that have been implicated in a
variety of cellular processes. Recently, several studies have
demonstrated that this family is involved in the regulation of
immune responses. Nik Tavakoli et al(24) reported that FOXP3 is critical in
the regulation and development of Treg cells. FOXN1
modulates the thymic stroma and regulates thymopoiesis, and FOXOs
are also involved in the maintainance of quiescence and tolerance
of T and B cells (25). Moreover,
it has been suggested that FOXJ1 was associated with autoimmune
diseases, such as allergic rhinitis and systemic lupus
erythematosus (20,21). However, studies concerning the
function of FOXJ3 remain to be conducted. Thus far it has only been
suggested that FOXJ3 may be involved in the development of skeletal
muscle, and the peripheral and central nervous system (17). Therefore, further studies are
required to investigate the function of FOXJ3 (rs585320).
In the present study, seven significant SNPs were
located in introns and did not result in an amino acid change.
According to the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) and the HapMap
database (http://www.hapmap.org/), seven SNPs in
exons of the FOXJ3 gene have been identified; however, with the
exception of rs343376, these SNPs remain unknown or <0.1 about
heterozygosity of them. Since low heterozygosity means little
genetic variability, these SNPs were excluded from the present
study and only one coding SNP (rs343376) was analyzed. This SNP
(rs343376) was not associated with RA in the study population. It
was suggested that significant polymorphisms in introns may be
markers rather than direct contributors to genetic functions. The
effects of other genetic variations linked to these polymorphisms
may exhibit functional significance. However, there remains a
possibility that these intronic polymorphisms are involved in its
genetic function via the change resulting from alternative splicing
(26).
In conclusion, the results of the present study
suggest that FOXJ3 polymorphisms are associated with susceptibility
to RA in the study population. To the best of our knowledge, this
study is the first to analyze the association between FOXJ3 SNPs
and RA. Moreover, the polymorphism/association information
identified in this study may be useful for further genetic studies
of RA.
Acknowledgements
This work was supported by the Priority Research
Centers Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Education, Science and Technology
(2009-0093829) and by a grant from the Institute of Bio-Science and
Technology (IBST) at Dankook University in 2010.
References
|
1
|
Silman AJ and Pearson JE: Epidemiology and
genetics of rheumatoid arthritis. Arthritis Res. 4(Suppl 3):
S265–S272. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Firestein GS: Evolving concepts of
rheumatoid arthritis. Nature. 423:356–361. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Klareskog L, Padyukov L, Rönnelid J and
Alfredsson L: Genes, environment and immunity in the development of
rheumatoid arthritis. Curr Opin Immunol. 18:650–655. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hall FC, Weeks DE, Camilleri JP, et al:
Influence of the HLA-DRB1 locus on susceptibility and severity in
rheumatoid arthritis. QJM. 89:821–829. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kong KF, Yeap SS, Chow SK and Phipps ME:
HLA-DRB1 genes and susceptibility to rheumatoid arthritis in three
ethnic groups from Malaysia. Autoimmunity. 35:235–239. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mattey DL, Dawes PT, Clarke S, et al:
Relationship among the HLA-DRB1 shared epitope, smoking, and
rheumatoid factor production in rheumatoid arthritis. Arthritis
Rheum. 47:403–407. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Begovich AB, Carlton VE, Honigberg LA, et
al: A missense single-nucleotide polymorphism in a gene encoding a
protein tyrosine phosphatase (PTPN22) is associated with rheumatoid
arthritis. Am J Hum Genet. 75:330–337. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karlson EW, Chibnik LB, Cui J, et al:
Associations between human leukocyte antigen, PTPN22, CTLA4
genotypes and rheumatoid arthritis phenotypes of autoantibody
status, age at diagnosis and erosions in a large cohort study. Ann
Rheum Dis. 67:358–363. 2008. View Article : Google Scholar
|
|
9
|
Ikari K, Momohara S, Inoue E, et al:
Haplotype analysis revealed no association between the PTPN22 gene
and RA in a Japanese population. Rheumatology (Oxford).
45:1345–1348. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kawasaki E, Awata T, Ikegami H, et al:
Systematic search for single nucleotide polymorphisms in a lymphoid
tyrosine phosphatase gene (PTPN22): association between a promoter
polymorphism and type 1 diabetes in Asian populations. Am J Med
Genet A. 140:586–593. 2006. View Article : Google Scholar
|
|
11
|
Suzuki A, Yamada R, Chang X, et al:
Functional haplotypes of PADI4, encoding citrullinating enzyme
peptidylarginine deiminase 4, are associated with rheumatoid
arthritis. Nat Genet. 34:395–402. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tokuhiro S, Yamada R, Chang X, et al: An
intronic SNP in a RUNX1 binding site of SLC22A4, encoding an
organic cation transporter, is associated with rheumatoid
arthritis. Nat Genet. 35:341–348. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kochi Y, Yamada R, Suzuki A, et al: A
functional variant in FCRL3, encoding Fc receptor-like 3, is
associated with rheumatoid arthritis and several autoimmunities.
Nat Genet. 37:478–485. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Plenge RM, Padyukov L, Remmers EF, et al:
Replication of putative candidate-gene associations with rheumatoid
arthritis in >4,000 samples from North America and Sweden:
association of susceptibility with PTPN22, CTLA4, and PADI4. Am J
Hum Genet. 77:1044–1060. 2005. View
Article : Google Scholar
|
|
15
|
Barton A, Bowes J, Eyre S, et al: A
functional haplotype of the PADI4 gene associated with rheumatoid
arthritis in a Japanese population is not associated in a United
Kingdom population. Arthritis Rheum. 50:1117–1121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Carlsson P and Mahlapuu M: Forkhead
transcription factors: key players in development and metabolism.
Dev Biol. 250:1–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Landgren H and Carlsson P: FoxJ3, a novel
mammalian forkhead gene expressed in neuroectoderm, neural crest,
and myotome. Dev Dyn. 231:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kaestner KH, Knochel W and Martinez DE:
Unified nomenclature for the winged helix/forkhead transcription
factors. Genes Dev. 14:142–146. 2000.PubMed/NCBI
|
|
19
|
Dejaco C, Duftner C, Grubeck-Loebenstein B
and Schirmer M: Imbalance of regulatory T cells in human autoimmune
diseases. Immunology. 117:289–300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li CS, Chae SC, Lee JH, et al:
Identification of single nucleotide polymorphisms in FOXJ1 and
their association with allergic rhinitis. J Hum Genet. 51:292–297.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li CS, Zhang Q, Lim MK, et al: Association
of FOXJ1 polymorphisms with systemic lupus erythematosus and
rheumatoid arthritis in Korean population. Exp Mol Med. 39:805–811.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arnett FC, Edworthy SM, Bloch DA, et al:
The American Rheumatism Association 1987 revised criteria for the
classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lewis CM: Genetic association studies:
design, analysis and interpretation. Brief Bioinform. 3:146–153.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nik Tavakoli N, Hambly BD, Sullivan DR and
Bao S: Forkhead box protein 3: essential immune regulatory role.
Int J Biochem Cell Biol. 40:2369–2373. 2008.PubMed/NCBI
|
|
25
|
Coffer PJ and Burgering BM: Forkhead-box
transcription factors and their role in the immune system. Nat Rev
Immunol. 4:889–899. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ast G: How did alternative splicing
evolve? Nat Rev Genet. 5:773–782. 2004. View Article : Google Scholar : PubMed/NCBI
|