Introduction
The human genome has three distinct genes encoding
members of the kinesin-13 family, known as Kif2a (chromosome 5q12),
Kif2b (chromosome 17q22) and MCAK/Kif2c (chromosome 1p34). Kif2a is
essential for bipolar spindle assembly and chromosome movement
(1). It localizes to spindle poles
in human cells and cells lacking Kif2a form monopolar spindles
instead of bipolar spindles in mitosis (2), leading to arrest of cell cycle
progression. A previous study demonstrated that the overexpression
of Kif2a is involved in the progression, invasion and metastasis of
squamous cell carcinoma of the oral tongue (SCCOT) (3). However, as yet, very little
information is available on the correlation between Kif2a and
apoptosis of tumor cells. Additionally, the signaling pathways that
Kif2a is involved in remain to be determined.
Phosphatidylinositol-3-kinase (PI3K) is a lipid
kinase that generates second messengers involved in the regulation
of many cell functions including proliferation, survival and
invasion (4). One of its major
effectors is protein kinase B (Akt). Results of previous studies
have shown that the PI3K/Akt pathway is frequently activated in
many types of human cancer, including head and neck squamous cell
carcinoma (5–8), and that phospho-Akt and Akt2 were
overexpressed in oral squamous cell carcinoma (9). Therefore, the evidence suggests that
the PI3K/Akt signaling pathway is likely associated with the cell
proliferation, apoptosis and migration of SCCOT. However, the
mechanism responsible for PI3K/Akt activation in SCCOT has yet to
be determined.
SCCOT is a common tumor of the head and neck
(10). Since Kif2a overexpression
and PI3K/Akt activation are frequent events in oral squamous cell
carcinoma and are associated with tumor progression of SCCOT, we
investigated whether there is any correlation between Kif2a and
PI3K/Akt signaling. We examined the effects of downregulated Kif2a
expression on the PI3K/Akt pathway and the pro-apoptotic role of
silencing Kif2a in SCCOT cells. In addition, we studied the effects
of a pharmaceutical inhibitor and agonist of PI3K on the
anti-apoptotic function of Kif2a in SCCOT cells. The results
suggest that silencing Kif2a inhibits the PI3K/Akt signaling
pathway, leading to cell apoptosis.
Materials and methods
Cell culture
Human tongue squamous cell carcinoma cell lines
(Tca8113 cells) were purchased from the Culture Collection of
Chinese Academy of Science (Shanghai, China) and routinely cultured
in Dulbecco’s modified Eagle’s medium (Gibco-BRL, Carlsbad, CA,
USA) containing 10% fetal bovine serum (Gibco-BRL), penicillin (100
U/ml) and streptomycin (100 μg/ml) at 37°C in a humidified air
atmosphere containing 5% CO2.
Silencing Kif2a gene using small
interfering RNA (si-RNA)
si-RNA expression vector was constructed by
introducing synthetic double-stranded oligonucleotides (Kif2a:
5′-CACCGGCAAAGAGATTGACCTGGTTCAAGAGACC AGGTCAATCTCTTTGCCTTTT TTG-3′;
nonsense control: 5′-CACCGTTCTCCGAACGTGTCACGTCAAGAGATTAC
GTGACACGTTCGGAGAATTTTTTG-3′) synthesized by GeneChem (Shanghai,
China) and inserted into the pGPU/GFP/Neo vector (Shanghai
GenePharma, Ltd., Shanghai, China). Authenticity of the constructs
was confirmed by sequencing Tca8113 cells that were planted in
six-well plates in Opti-MEM (Invitrogen Life Technologies,
Carlsbad, CA, USA) until the cells reached 40% confluence. The
cells were transfected with the kif2a si-RNA (si-Kif2a) or nonsense
si-RNA (si-NC) (100 nmol/l) premixed with the Lipofectamine™ 2000
(Invitrogen Life Technologies) in Opti-MEM. Mock transfection with
Lipofectamine™ 2000 only was also included as a control. Six hours
after transfection, the cells were placed in fresh complete medium
without penicillin and streptomycin. Human insulin-like growth
factor 1 (IGF-1) was purchased from InvivoGen (San Diego, CA, USA)
and Peprotech, Inc. (Rocky Hill, NJ, USA).
Western blot analysis
Western blot analysis was performed as previously
described (3). In brief, cells
were collected and lysed in 1% NP-40 lysis buffer. Then cell
extract was resolved on 10% polyacrylamide gels using minigel
apparatus and transferred to a polyvinylidene fluoride membrane.
The membrane was blotted with rabbit anti-Kif2a polyclonal
antibodies at dilution of 1:10,000 (Abcam, Cambridge, UK) for 2 h
at room temperature. Blots were then exposed to horseradish
peroxidase-conjugated goat anti-rabbit lgG (dilution 1:10,000),
followed by development using an electrochemiluminescence
reagent.
Real-time reverse
transcription-polymerase chain reaction (RT-PCR) analysis
After Tca8113 cells were collected, total RNA was
extracted using the TRIzol (Invitrogen Life Technologies) method as
recommended by the manufacturer. RNA was reverse transcribed by the
First Strand cDNA Synthesis kit (Fermentas, Burlington, Canada).
According to the protocol recommended by Takara SYBR Premix Ex Taq™
(Takara Bio, Inc., Shiga, Japan), real-time quantitative PCR
analysis was performed using a LightCycler 2.0 (Roche Diagnostics,
Basel, Switzerland). Thermal cycling parameters were as follows: an
initial incubation of 95°C for 30 sec and then 40 cycles of 95°C
for 5 sec, 55°C for 20 sec and 72°C for 15 sec. Forward and reverse
primer sequences for PI3K, Akt, B-cell lymphoma 2 (Bcl-2),
Bcl-2-associated X protein (Bax) and β-actin are shown in Table I.
 | Table IPrimer sequences of genes. |
Table I
Primer sequences of genes.
| Gene | Primer sequence
(5′-3′) | Product size
(bp) |
|---|
| β-actin | F:
CGTTGACATCCGTAAAGACC
R: TAGAGCCACCAA TCCACAC | 176 |
| PI3K | F:
CATCACTTCCTCCTGCTCTAT
R: CAGTTGTTGGCAATCTTCTTC | 377 |
| Akt | F:
TGCATTGCCGAGTCCAGAA
R: GCATCCGAGAAA CAAAACATCA | 139 |
| Bcl-2 | F:
GCAGAGATGTCCAGTCAG
R: CCCACCGAACTCAAAGAAGG | 129 |
| Bax | F:
ATGGGCTGGACACTGGACTTC
R: GAGCGAGGCGGTGAGGAC | 146 |
Annexin V-fluorescein isothiocyanate
(FITC) assay
To assess the degree of apoptosis of different
groups of Tca8113 cells, the extent of Annexin V-FITC/propidium
iodide (PI) staining was determined by flow cytometry using the
Annexin V/PI staining kit from Bender MedSystems (Vienna, Austria).
Samples were read on an Epics XL-MCL flow cytometer (Beckman
Coulter, Brea, FL, USA) and were analyzed by WinMDI 2.8
software.
GeneChip hybridization and data
analysis
GeneChip hybridization and data analysis were
performed by KangChen Bio-tech (Shanghai, China). Purified and
fragmented cRNA probes were hybridized onto Affymetrix Human
Genome-U133A 2.0 GeneChips (Affymetrix, Inc., Santa Clara, CA,
USA). Each RNA pool was hybridized to an individual chip and
hybridization was performed in the presence of herring sperm DNA
(0.1 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) for 16 h at 45°C.
Chips were then washed, stained with streptavidin- phycoerythrin
and scanned with a confocal microscope scanner (GeneArray Scanner
2500; Hewlett-Packard, Palo Alto, CA, USA) according to Affymetrix
guidelines. The standard Affymetrix analysis software algorithms
(Microarray Suite 5.0) were used for data capturing, which selects
the spots representative of a transcript and subtracts the
background from the significant signals. The images from the
scanned chip were processed on GeneSpring 7.2 software (Silicon
Genetics, Redwood City, CA, USA). Gene expression data for each
replicate experiment were normalized using the ‘per chip
normalization’ and ‘per gene normalization’ algorithms implemented
in the GeneSpring program. Genes with expression levels altered by
≥1.5-fold in Kif2a silenced relative to si-NC samples were
considered to be differentially expressed. The significance of gene
expression differences between the two experimental conditions was
calculated using a one-way analysis of variance. Biological theme
analyses were conducted by Expression Analysis Systematic Explorer
and the Database for Annotation, Visualization and Integrated
Discovery (DAVID) 2.0 program.
Statistical analysis
Statistical analysis was performed using the SPSS
16.0 software package for Windows. Values are presented as the
means ± standard deviation (SD). The Student’s t-test was used for
paired data that were normally distributed. P<0.05 was
considered to indicate a statistically significant difference.
Results
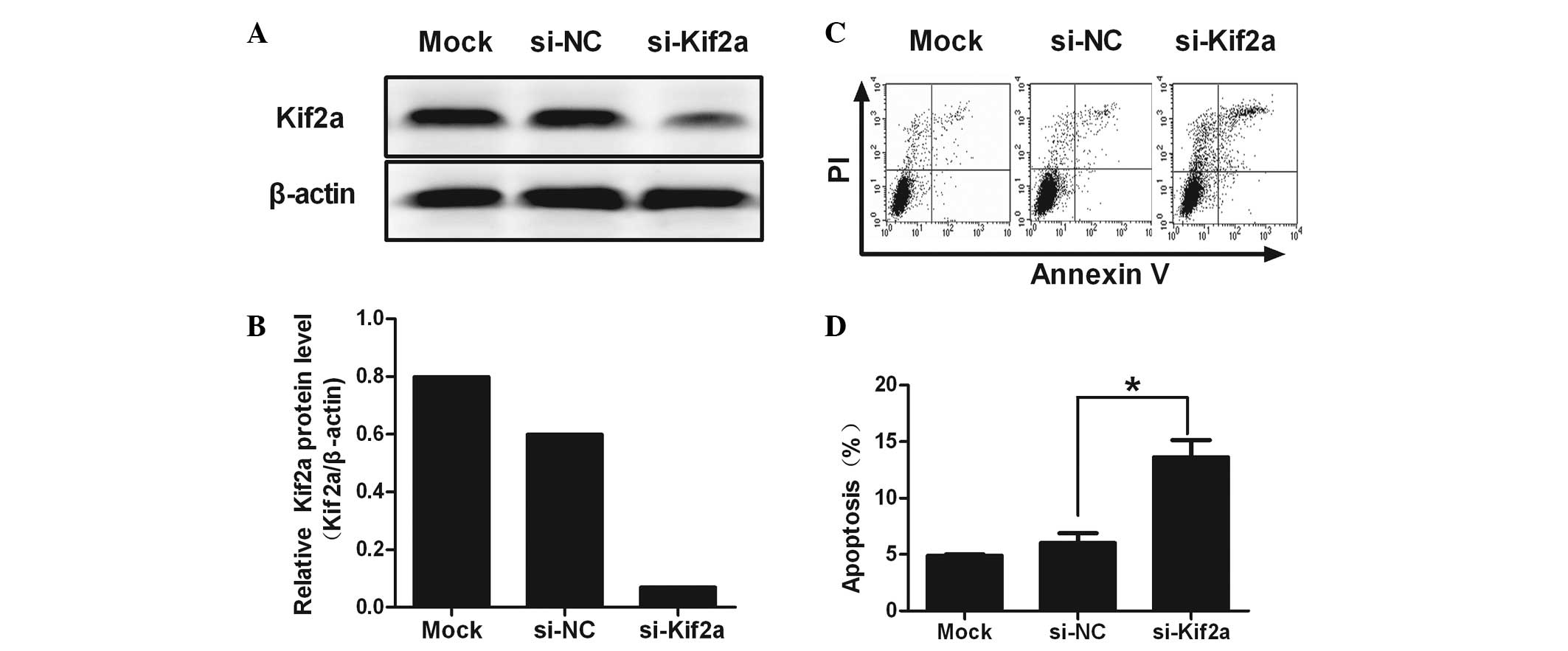
Kif2a expression is suppressed by
si-Kif2a
RNA interference targeting Kif2a was determined by
western blot analysis. Reduction of Kif2a protein expression by
~80.67% was observed in si-Kif2a-transfected cells compared to
si-NC and lipofectamine-only treated cells (Fig. 1A and B; P=0.022). These results
indicated that si-Kif2a had a gene-silencing effect targeting Kif2a
at the protein level.
Apoptosis is induced by si-Kif2a in
Tca8113 cells
Flow cytometric analysis revealed a significant
increase in the percentage of apoptotic cells in Tca8113-Kif2a as
compared to Tca8113-nonsense (NC) and Tca8113 cells (28.39±2.89 vs.
5.28±0.50 and 3.73±0.57%; P<0.01). No significant difference was
observed between Tca8113-NC and Tca8113. It was revealed that
silencing Kif2a induced significant apoptosis in Tca8113 cells
(Fig. 1C and D).
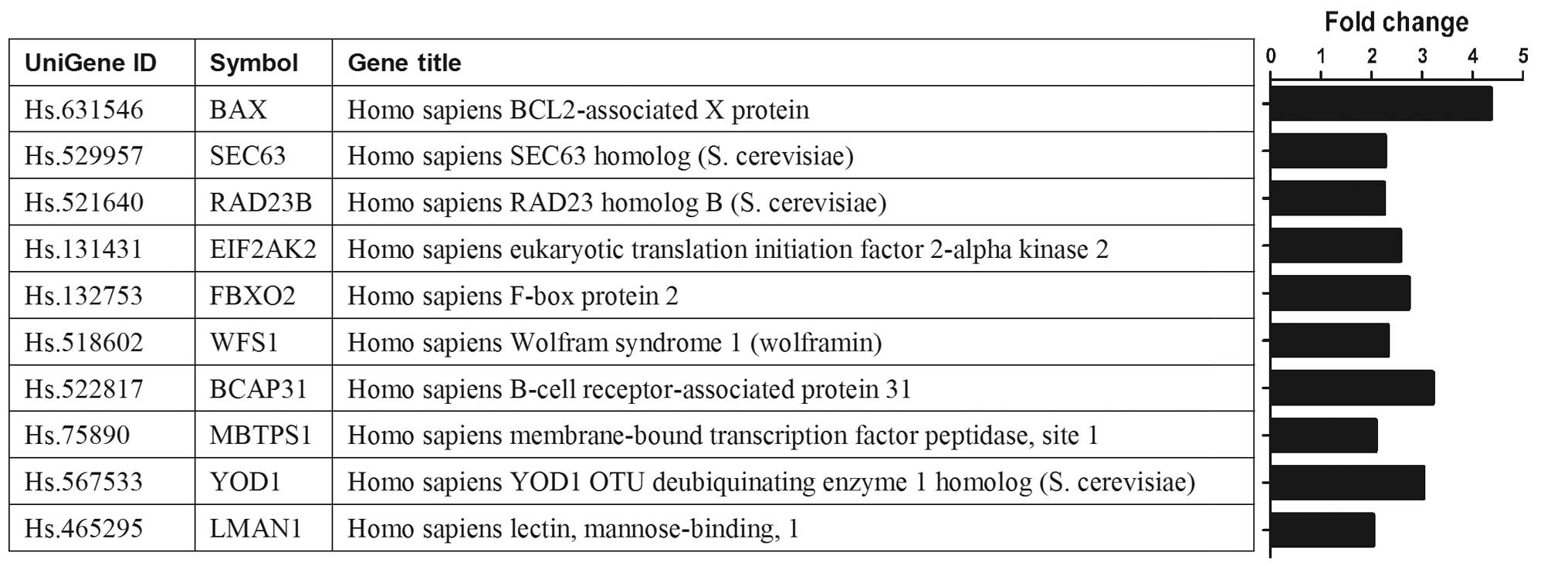
Gene expression patterns in
si-Kif2a-transfected cells
Analysis of the gene expression profiles of the
cells transfected with si-Kif2a and si-NC revealed that 744 genes
were upregulated and 1,282 genes were downregulated. The DAVID
database indicated multiple biological pathways that appear to be
enriched in the two cell groups. Most notably, the enrichment score
for the protein processing in endoplasmic reticulum was
significantly elevated. The enriched genes involved in the protein
processing in endoplasmic reticulum included multiple members, such
as BAX, SEC63, RAD23B, EIF2AK2, FBXO2, WFS1, BCAP31, MBTPS1, YOD1
and LMAN1. Of these genes, the expression of BAX gene has a
significant change showing >4-fold upregulation in Tca8113-Kif2a
cells (Fig. 2).
Expression of PI3K, Akt, Bcl-2 and Bax in
different groups of Tca8113 cells
The mRNA level of PI3K, Akt, Bcl-2 and Bax were
analyzed by real-time PCR. Data showed that PI3K, Akt and Bcl-2 in
Tca8113-Kif2a cells decreased significantly compared to Tca8113-NC
or Tca8113 cells (Fig. 3A–C).
However, the level of Bax in Tca8113-Kif2a cells was the highest
among the three groups (Fig. 3D).
From the data, we hypothesize that silencing of Kif2a may be
closely associated with the survival of Tca8113 cells by the
PI3K/Akt signal pathway, particularly by regulating the ratio of
Bcl-2/Bax.
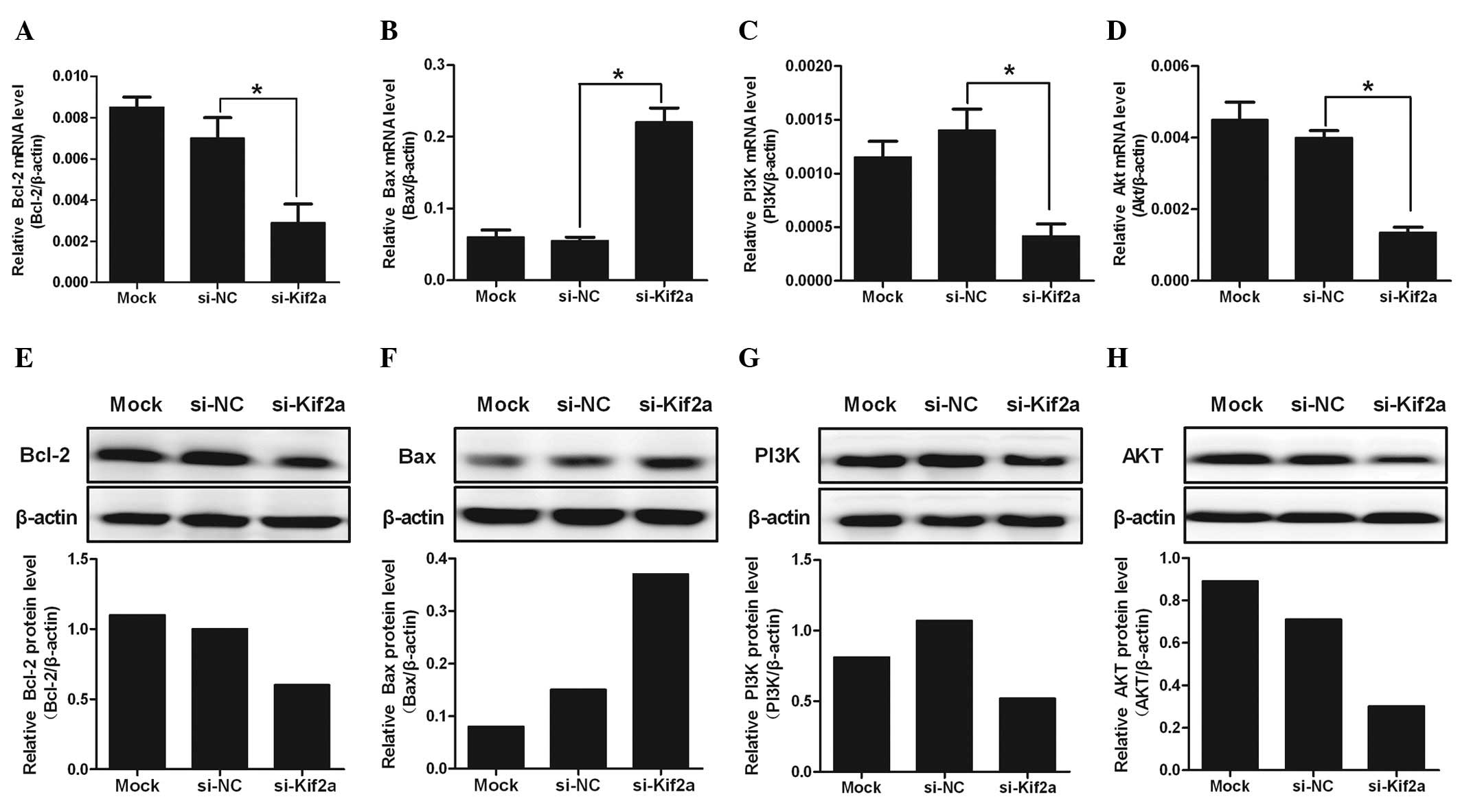 | Figure 3The expression of PI3K, Akt, Bcl-2 and
Bax in different groups of Tca8113 cells at the mRNA level. The
mRNA levels of (A) PI3K, (B) Akt, (C) Bcl-2 and (D) Bax were
analyzed quantitatively by real-time polymerase chain reaction. The
protein expression levels of (E) PI3K, (F) Akt, (G) Bcl-2 and (H)
Bax were analyzed quantitatively by western blot analysis. The
level of PI3K, Akt and Bcl-2 mRNA in Tca8113-Kif2a decreased more
clearly than Tca8113-nonsense or Tca8113 cells. However, the level
of Bax in Tca8113-Kif2a cells was highest among the three groups
(*P<0.05). The data are presented as the means ± SD.
Experiments were repeated ≥3 times. Mock, lipofectamine only;
si-NC, nonsense si-RNA; si-Kif2a, kif2a si-RNA. |
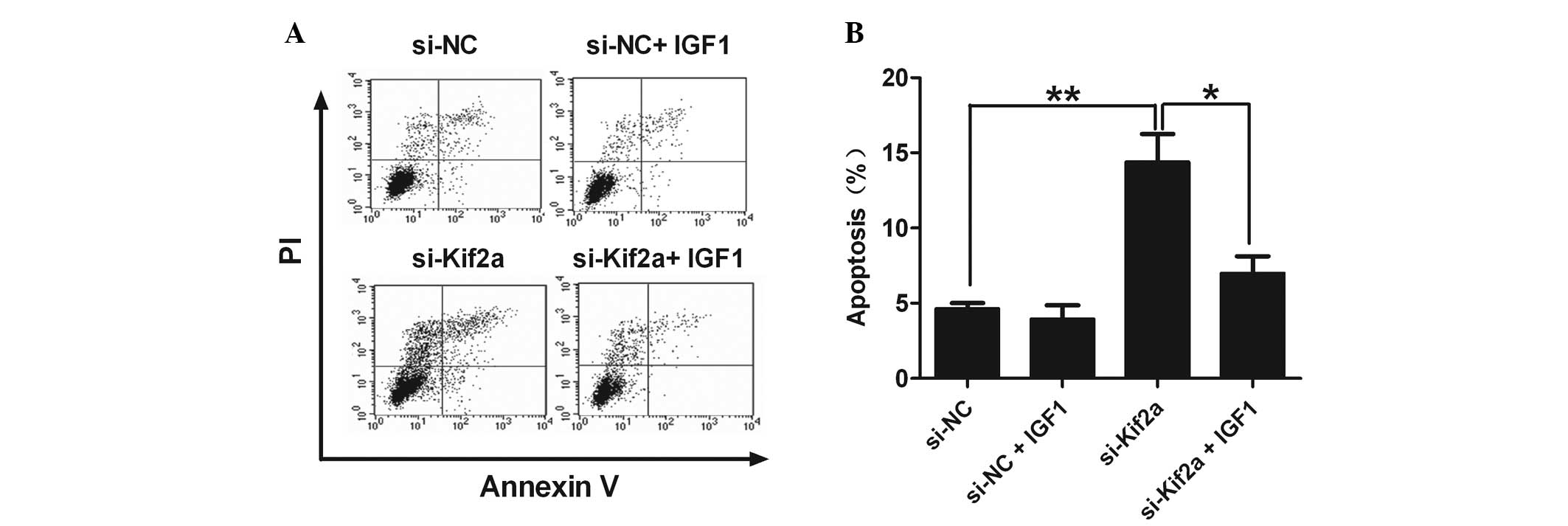
IGF-1 abrogates the upregulation of
apoptosis of Tca8113-Kif2a cells
IGF-1 is the specific agonist of PI3K/Akt. When
comparing the percentage of apoptotic cells in the si-Kif2a only
group with the si-Kif2a and IGF-1 group, we found that IGF-1
effectively eliminated the upregulation of apoptosis in
Tca8113-Kif2a cells (Fig. 4). It
was revealed that silencing Kif2a significantly induced Tca8113
cell apoptosis through the PI3K/Akt pathway.
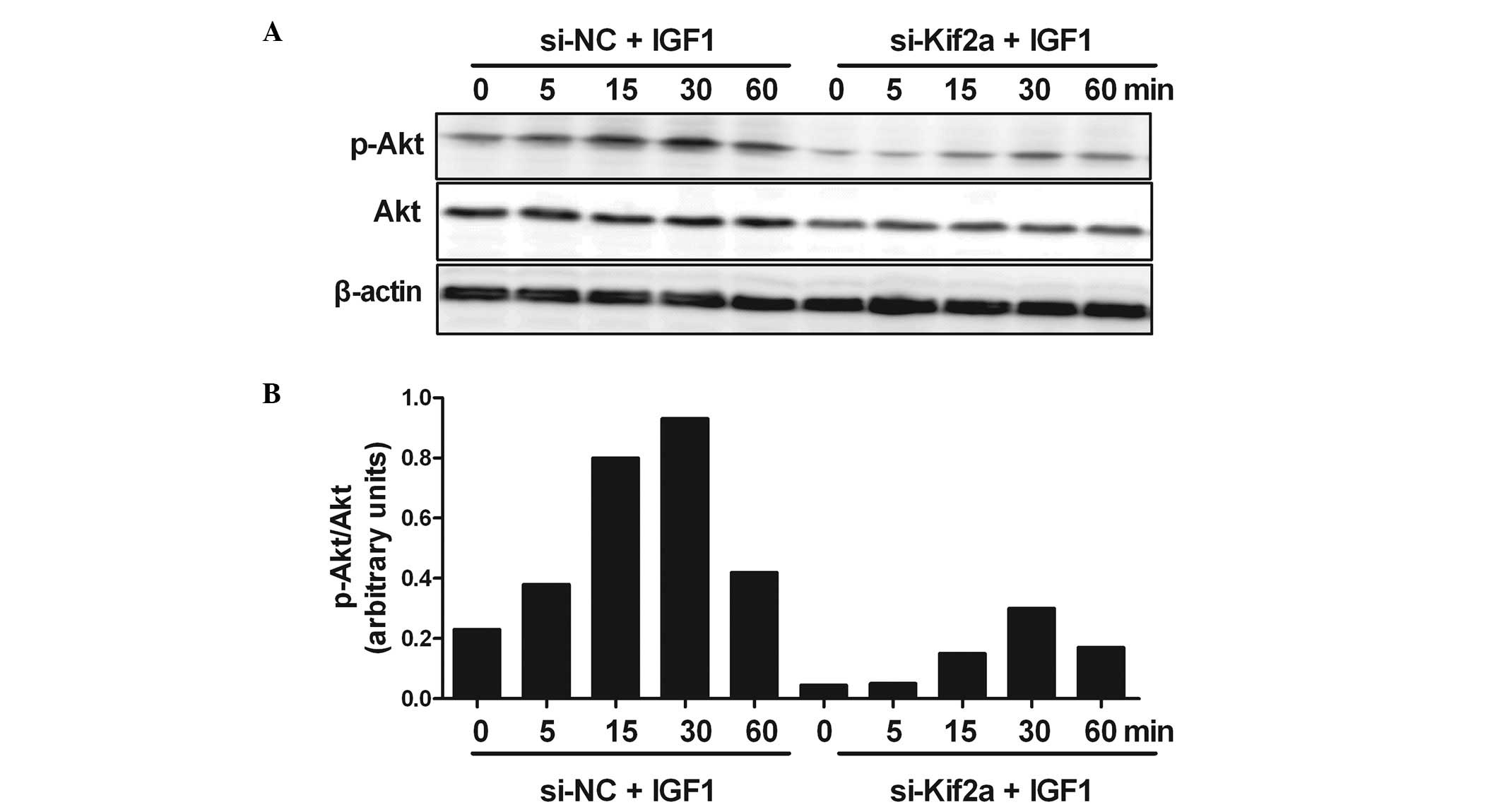
Silencing Kif2a induces apoptosis of
Tca8113 cells by inhibiting the PI3K/Akt signaling pathway
In order to understand the molecular mechanisms of
the apoptosis of Tca8113 cells by silencing Kif2a, we investigated
PI3K/Akt signaling. The result showed strong phosphorylation of Akt
at 30 min after adding IGF-1 in the si-NC group. In the
experimental group, the Akt response was reduced by silencing Kif2a
at all the indicated time points. However, the phosphorylation of
Akt at 30 min after the addition of IGF-1 the si-Kif2a group also
reached a peak (Fig. 5).
Discussion
In this study, to the best of our knowledge, the
results demonstrate for the first time that silencing Kif2a leads
to apoptosis of SCCOT cells and inhibits the PI3K/Akt signaling
pathway, which may be one of the mechanisms responsible for
inducing SCCOT cell apoptosis.
Several novel points arise from this study. First,
we reported that silencing of Kif2a induces apoptosis of SCCOT
cells. Previous studies on Kif2a were usually focused on its
function and its mechanisms in depolymerizing microtubules. There
has been no report on its role in regulating the apoptosis of tumor
cells. Our data showed that silencing Kif2a increased the
percentage of apoptotic cells.
Second, we provided evidence that Kif2a is an
upstream regulator of the PI3K/Akt pathway. Our finding that
silencing Kif2a inhibits PI3K/Akt is significant since the
overexpression and activation of PI3K/Akt is a frequent event in
many types of human cancer (5,11),
although the mechanisms underlying its activation are not fully
understood. Previously, it was reported that the EVI1 oncogene
increases the sensitivity of colon cancer cells to taxol-mediated
apoptosis through activation of PI3K/Akt (12) and ID-1 protects esophageal cancer
cells from tumor necrosis factor-α-induced apoptosis through
activation of the PI3K/Akt signaling pathway (13). Whether a similar association exists
between the PI3K/Akt pathway and other known oncogenes requires
further investigation.
Third, the results showed that silencing Kif2a by
si-RNA suppressed the PI3K/Akt signaling pathway and induced SCCOT
cells to apoptosis, suggesting a potentially novel therapeutic
strategy for SCCOT. Although PI3K/Akt is not the sole mediator of
Kif2a-dependent cell survival, its functions in regulating cell
cycle control, driving tumorigenesis and imparting chemoresistance
to anticancer treatment make it an attractive target for cancer
therapy (14). A number of
candidate drugs targeting this pathway, such as inhibitors of PI3K,
epidermal growth factor receptor, platelet-derived growth factor
receptor and mammalian target of rapamycin (mTOR), as well as
monoclonal HER2 antibody, have been studied. Rapamycin, an
inhibitor of the Akt downstream mTOR, also has poor aqueous
solubility and chemical stability, although it has significant
anti-proliferative activity in several murine tumor systems
(15). A rapamycin analog,
CCI-779, with improved pharmaceutical properties and comparable
efficacy, was approved in phase I and II of clinical studies; phase
III trials are in progress (16).
However, inhibitors of mTOR may not block all the functions of the
PI3K/Akt pathway as they only affect one of the many downstream
pathways of PI3K/Akt signaling. While new reagents targeting this
pathway are being developed and tested, consideration should be
given to targeting Kif2a as an alternative strategy in cancer
therapy, since our previous study indicates that Kif2a has multiple
effects on tumor progression including tumor growth, invasion and
metastasis. From a therapeutic standpoint, since Kif2a is
overexpressed in SCCOT, but occurs at very low levels in normal
tissues (3), inhibition of Kif2a
should have very few side-effects on normal tissues. Furthermore,
with the development of improved delivery systems in RNA
interference technology and the recent success in the application
of therapeutic si-RNA in non-human primates (17), RNAi-based therapeutic reagents
targeting Kif2a may be a promising alternative or adjunct to
cytotoxic chemotherapy for SCCOT.
In conclusion, we confirmed that Kif2a is an
important molecule associated with the survival and progression of
SCCOT cells. Thus, we hypothesize that Kif2a is a prognostic
marker, or even a molecular target, for SCCOT therapy.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant nos. 30772269, 81072202,
81271105 and 81202142) and Shandong Natural Science Foundation (no.
Y2007C128).
References
|
1
|
Ganem NJ and Compton DA: The KinI kinesin
Kif2a is required for bipolar spindle assembly through a functional
relationship with MCAK. J Cell Biol. 166:473–478. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ganem NJ, Upton K and Compton DA:
Efficient mitosis in human cells lacking poleward microtubule flux.
Curr Biol. 15:1827–1832. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang CQ, Qu X, Zhang XY, et al:
Overexpression of Kif2a promotes the progression and metastasis of
squamous cell carcinoma of the oral tongue. Oral Oncol. 46:65–69.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bader AG, Kang S, Zhao L and Vogt PK:
Oncogenic PI3K deregulates transcription and translation. Nat Rev
Cancer. 5:921–929. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brader S and Eccles SA: Phosphoinositide
3-kinase signalling pathways in tumor progression, invasion and
angiogenesis. Tumori. 90:2–8. 2004.PubMed/NCBI
|
|
6
|
Yen CC, Chen YJ, Lu KH, et al: Genotypic
analysis of esophageal squamous cell carcinoma by molecular
cytogenetics and real-time quantitative polymerase chain reaction.
Int J Oncol. 23:871–881. 2003.PubMed/NCBI
|
|
7
|
Zhang G, Zhou X, Xue L, et al:
Accumulation of cytoplasmic beta-catenin correlates with reduced
expression of E-cadherin, but not with phosphorylated Akt in
esophageal squamous cell carcinoma: immunohistochemical study.
Pathol Int. 55:310–317. 2005. View Article : Google Scholar
|
|
8
|
Bian Y, Terse A, Du J, et al: Progressive
tumor formation in mice with conditional deletion of TGF-beta
signaling in head and neck epithelia is associated with activation
of the PI3K/Akt pathway. Cancer Res. 69:5918–5926. 2009. View Article : Google Scholar
|
|
9
|
Iamaroon A and Krisanaprakornkit S:
Overexpression and activation of Akt2 protein in oral squamous cell
carcinoma. Oral Oncol. 45:e175–e179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Greenlee RT, Hill-Harmon MB, Murray T and
Thun M: Cancer statistics, 2001. CA Cancer J Clin. 51:15–36. 2001.
View Article : Google Scholar
|
|
11
|
Aggarwal BB: Nuclear factor-kappaB: the
enemy within. Cancer Cell. 6:203–208. 2004.PubMed/NCBI
|
|
12
|
Liu Y, Chen L, Ko TC, Fields AP and
Thompson EA: Evi1 is a survival factor which conveys resistance to
both TGFbeta- and taxol-mediated cell death via PI3K/AKT. Oncogene.
25:3565–3575. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li B, Cheung PY, Wang X, Tsao SW, Ling MT,
Wong YC and Cheung AL: Id-1 activation of PI3K/Akt/NFκB signaling
pathway and its significance in promoting survival of esophageal
cancer cells. Carcinogenesis. 28:2313–2320. 2007.PubMed/NCBI
|
|
14
|
Xin M and Deng X: Nicotine inactivation of
the proapoptotic function of Bax through phosphorylation. J Biol
Chem. 280:1078l–10789. 2005.PubMed/NCBI
|
|
15
|
Hennessy BT, Smith DL, Ram PT, Lu Y and
Mills GB: Exploiting the PI3K/AKT pathway for cancer drug
discovery. Nat Rev Drug Discov. 4:988–1004. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Morgensztern D and McLeod HL:
PI3K/Akt/mTOR pathway as a target for cancer therapy. Anticancer
Drugs. 16:797–803. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zimmermann TS, Lee AC, Akinc A, et al:
RNAi-mediated gene silencing in non-human primates. Nature.
441:111–114. 2006. View Article : Google Scholar : PubMed/NCBI
|