Introduction
Cervical carcinoma is the second most prevalent type
of malignancy and the fifth most common cause of cancer-related
mortality in females worldwide. Invasion and metastasis are major
causes of cancer-associated mortality (1). Persistent infection with high-risk
types of the human papillomavirus (HPV) is known to cause cervical
cancer, however, additional genetic and epigenetic alterations are
required for progression from precancerous disease to invasive
cancer. DNA methylation is an early and frequent molecular
alteration in cervical carcinogenesis. Dysregulated activation of
numerous genes, including CD44 and SOX9, has been implicated in
cervical cancer; however, the mechanism of regulation in human
cervical cancer cells remains to be elucidated (2–4). It
has been demonstrated that the inactivation of tumor suppressor
genes and activation of oncogenes caused by genetic and epigenetic
alterations is important in carcinogenesis. miRNAs are closely
associated with the occurrence and regulation of cervical cancer
(5). Emerging studies have
evaluated the association of miRNA single nucleotide polymorphisms
with cancer risk; however, the results remain inconclusive
(6). In addition, the etiology of
cervical carcinoma remains poorly understood.
CD44 refers to a multifunctional family of type I
transmembrane proteins. The CD44 gene contains at least 21 exons,
11 of which can be variably spliced and produce a variety of
heavily glycosylated cell surface proteins, termed CD44 variant
isoforms. These proteins have been implicated in several biological
processes, including cell adhesion, cell to cell interactions for
example in lymphocyte homing hemopoiesis, cell migration and
metastasis. These abilities are important in chronic inflammation
and in cancer. In cancer, deregulation of the adhesion mechanisms
increases the ability of tumor cells to metastasize. CD44 may
function in certain cells through interactions with type I receptor
tyrosine kinases, including erbB2 (2,7–8).
Published data have demonstrated that CD44 mediates constitutive
type I receptor signaling in cervical carcinoma cells (2). The assessment of CD44 isoform
expression may be of clinical value in deciding upon adjuvant
therapy, resulting in a more individualized management of therapy
(9). In oral tongue squamous cell
carcinoma, reduced expression of CD44 may be an indicator of high
tumor invasiveness by increasing cervical lymph node metastasis
(10). One common confounder for
the analysis of clinical tumor specimens is the cellular
heterogeneity of CD44+/CD44− cells. Flow
cytometry sorting technology is able to overcome this problem and
obtain pure CD44+ or CD44− cells for
mechanistic study (11–14). Furthermore, FOS-like antigen 1
(Fra-1) is a proto-oncogene, located on chromosome 11q13, encoding
a 1.7 kb mature mRNA. It is a negative inhibitor of activator
protein-1 activity and has transforming activity.
The present study used flow cytometric analysis to
examine the expression levels of ATP-binding cassette sub-family G
member 2 (ABCG2), CD24, CD44, CD133, cytokeratin (CK) 14 and CK19
between cervical cancer tissues and corresponding normal non-tumor
tissue. In addition, the CD44+ or CD44− cells
from cervical cancer tissues were sorted.
Materials and methods
Tumor samples
In total, 12 participants were recruited at The
Third Xiangya Hospital, Central South University (Changsha, China).
Consent forms were obtained from individual patients and
experimental protocols were approved by the Institutional Review
Board of The Third Xiangya Hospital. The 12 participants were
females with histologically-confirmed cervical cancer (Table I). All subjects enrolled in the
present study were Chinese. Cervical cancer tissues and
corresponding non-tumor normal tissues were collected, and each
biopsy sample was divided into two sections, one was submitted for
routine histological diagnosis, and the remaining section was
subjected to flow cytometric analysis and cell sorting.
 | Table ICharacteristics of cervical cancer
for flow cytometric analysis. |
Table I
Characteristics of cervical cancer
for flow cytometric analysis.
| Sample | Gender | Age (years) | Diagnosis | HPV type | Peasent or not |
|---|
| 1 | Female | 43 | Squamous cell
cancer | 33,58 | Yes |
| 2 | Female | 57 | Squamous cell
cancer | 16 | No |
| 3 | Female | 43 | Squamous cell
cancer | 18,35 | No |
| 4 | Female | 43 | Squamous cell
cancer | 18,35 | No |
| 5 | Female | 29 | Squamous cell
cancer | 16 | No |
| 6 | Female | 43 | Squamous cell
cancer | 52 | No |
| 7 | Female | 53 | Squamous cell
cancer | 16 | Yes |
| 8 | Female | 65 | Squamous cell
cancer | 16 | Yes |
| 9 | Female | 70 | Squamous cell
cancer | 16 | No |
| 10 | Female | 37 | Squamous cell
cancer | 16,58 | Yes |
| 11 | Female | 43 | Squamous cell
cancer | 16 | Yes |
| 12 | Female | 46 | Squamous cell
cancer | 52 | Yes |
Flow cytometric analysis of ABCG2, CD24,
CD44, CD133, CK14 and CK19
Single-cell suspensions of cervical cancer tissue or
corresponding normal non-tumor tissue were prepared as follows: The
cancer tissue or non-tumor normal tissue was sliced into
1-mm3 sections and digested in serum-free Dulbecco’s
modified Eagle’s medium containing 1 mg/ml collagenase IV
(Invitrogen Life Technologies, Carlsbad, CA, USA), 1% hyaluronidase
(Sigma-Aldrich, St. Louis, MO, USA) and 0.25% DNase I (Merck &
Co., White House Station, NJ, USA). Enzymatic digestion was
incubated at 37°C until fully with oscillation every 10–15 min
prior to the substrate being passed through a 70-μm cell strainer.
The resulting cell suspension was centrifuged at 500 × g for 10 min
and resuspended in saline.
Single-cell suspensions were stained and incubated
at 4°C for 30 min with the following antibodies, respectively:
Fluorescein isothiocyanate (FITC)-conjugated CD44,
phycoerythrin-conjugated ABCG2 and FITC-conjugated Ck14. Isotype
controls were performed with FITC-conjugated rabbit anti-human IgG
(negative control) All antibodies were purchased from Beckman
Coulter (Miami, FL, USA) and used according to the manufacturer’s
instructions. The cells were washed twice and examined by
fluorescence-activated cell sorting (FACS) using a MoFlo™ XDP
High-Performance Cell sorter (Beckman Coulter). Data were acquired
and analyzed using Summit v5.2 software (Beckman Coulter, Inc.,
Fullerton, CA, USA).
CD44+/− cell sorting by
FACS
The expression levels of ABCG2, CD24, CD44, CD133,
CK14 and CK19 were examined as described above. At the same time,
the CD44+/CD44− cells were sorted for all
cervical cancer samples. The parameters of sorting were conducted
according to the manufacturer’s instructions.
CD44+/CD44− cells were pooled from every four
samples. The sorted cells were immediately stored in TRIzol
reagents or liquid nitrogen.
RNA extraction and quantitative PCR
(qPCR)
Total RNA was extracted from the sorted cells using
an RNeasy® kit (Qiagen, Carlsbad, CA, USA) according to
the manufacturer’s instructions. The total RNA samples (1 μg) were
used to generate cDNA. Reverse transcription was performed as
previously described (12).
Following the RT reaction, the PCR reaction was preceded by 94°C
for 5 min, then 30 cycles for lactotransferrin (LTF) of 94°C for 45
sec, 55°C for 45 sec and 72°C for 1 min followed by 72°C for 7 min.
All reverse transcription-PCR reactions were repeated at least
three times at different numbers of the extension cycle to avoid
false results of the PCR. GAPDH was used as an endogenous control
for normalization. The sequences of the primers used for reverse
transcription-PCR were synthesized (Table II). The expression of mRNA was
assessed by evaluated threshold cycle (CT) values. The CT values
were normalized with the expression levels of GAPDH and the
relative quantity of mRNA specific to each of the target genes was
calculated using the 2−ΔΔCT method (15,16).
 | Table IISequences of the primers used for
qPCR in the present study. |
Table II
Sequences of the primers used for
qPCR in the present study.
| Gene | Forward | Reverse | Annealing
temperature (°C) |
|---|
| SOX2 |
5′-AACCCCAAGATGCACAACTC-3′ |
5′-GCTTAGCCTCGTCGATGAAC-3′ | 60 |
| Bmi1 |
5′-CCAGGGCTTTTCAAAAATGA-3′ |
5′-CCGATCCAATCTGTTCTGGT-3′ | 60 |
| CD133 |
5′-TTGTGGCAAATCACCAGGTA-3′ |
5′-TCAGATCTGTGAACGCCTTG-3′ | 60 |
| ck5 |
5′-GGTTGATGCACTGATGGATG-3′ |
5′-TCATACTGGGCCTTGACCTC-3′ | 60 |
| Nanog |
5′-CAGAAGGCCTCAGCACCTAC-3′ |
5′-ATTGTTCCAGGTCTGGTTGC-3′ | 60 |
| Nestin |
5′-TCCAGGAACGGAAAATCAAG-3′ |
5′-GCCTCCTCATCCCCTACTTC-3′ | 60 |
| END_1 |
5′-CCACTCCTTCCACCAACACT-3′ |
5′-GCTGTCATTGGAGCACTTGA-3′ | 60 |
| NR4A2 |
5′-AGTCTGATCAGTGCCCTCGT-3′ |
5′-TATGCTGGGTGTCATCTCCA-3′ | 60 |
| OCT4 |
5′-AGTGAGAGGCAACCTGGAGA-3′ |
5′-ACACTCGGACCACATCCTTC-3′ | 60 |
| ABCG2 |
5′-AGCTGCAAGGAAAGATCCAA-3′ |
5′-TGCCCATCACAACATCATCT-3′ | 60 |
| TP53 |
5′-GTGGAAGGAAATTTGCGTGT-3′ |
5′-CCAGTGTGATGATGGTGAGG-3′ | 60 |
| RKIP |
5′-ATAGACCCACCAGCATTTCG-3′ |
5′-ACTGTGCCACTGCTGATGTC-3′ | 60 |
| LTF |
5′-TGAGAATGCTGGAGACGTTG-3′ |
5′-TCTGCCAGCTTCAAATCCTT-3′ | 60 |
| UBAP1 |
5′-GGTCAACATGGGCTACTCGT-3′ |
5′-GCCCTTCTCACAAAGCTGTC-3′ | 60 |
| Iaspp |
5′-GAAAGCCTGGAACGAGTCTG-3′ |
5′-GCGCTAGTGAGGTTGTCCTT-3′ | 60 |
| MTDH |
5′-CCAACTGGGAAATCCAAAAA-3′ |
5′-CCTGTTTTGGACGGGTTTTA-3′ | 60 |
| mta2 |
5′-GCCAAACCCTAACCAGATCA-3′ |
5′-CAGGCATACCACTGAGCAGA-3′ | 60 |
| SPLUNC1 |
5′-AGGTCTTCTGGACAGCCTCA-3′ |
5′-CTGTAGTCCGTGGATCAGCA-3′ | 60 |
| BRD7 |
5′-CAAATTTTGGCGTTCCAGTT-3′ |
5′-GGGACCCAAGAGACAGATCA-3′ | 60 |
| Fra-1 |
5′-CGAAGGCCTTGTGAACAGAT-3′ |
5′-CTTCTGCTTCTGCAGCTCCT-3′ | 60 |
| P63 |
5′-CACCTCCGTATCCCACAGAT-3′ |
5′-GTCTCACTGGAGCCCACACT-3′ | 60 |
| GADPH |
5′-CGACCACTTTGTCAAGCTCA-3′ |
5′-ACTGAGTGTGGCAGGGACTC-3′ | 60 |
Western blot analysis
The proteins of the sorted cells were prepared using
lysis buffer. The protein concentrations were determined using the
bicinchoninic acid (Pierce Chemical, Rockford, IL, USA) protein
assay method. Extracts containing 50 μg of proteins were separated
in 10% SDS-PAGE gels and electroblotted onto nitrocellulose
membranes (HyClone Laboratories, Logan, UT, USA). The membranes
were inhibited using Tris-buffered saline/Tween-20 (25 mM Tris-HCl,
150 mM NaCl, pH 7.5 and 0.05% Tween-20) containing 5% non-fat milk
followed by overnight incubation at 4°C with primary antibodies
(Rabbit anti-Fra-1 antibody; Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA; 1:500). Following three washes, the membranes were
incubated with horseradish peroxidase-conjugated second antibodies
(Santa Cruz Biotechnology, Inc.) and the specific signals were
visualized using an ECL detection system. Anti-β-actin antibody
(Santa Cruz Biotechnology, Inc; 1:3,000) was used as a loading
control.
Intracellular protein level detection by
FACS
Following washing in Dulbecco’s phosphate-buffered
saline (D-PBS), the sorted cells were permeabilized with detergents
(Triton X-100). The cells were washed twice with D-PBS and then the
single-cell suspensions were stained and incubated at 4°C for 30
min with FITC conjugated Fra-1 (Biorbyt, Cambridge, UK),
respectively. Isotype controls were performed with FITC-conjugated
rabbit anti-human IgG (negative control; Biorbyt). All the
antibodies were used according to the manufacturer’s instructions.
The cells were washed twice and examined by FACS using a MoFlo™ XDP
High-Performance Cell sorter (Beckman Coulter). Data were acquired
and analyzed using summit v5.2 software.
Expression analysis of miR-19a and
miR-19b in cervical cancer
As described above, total RNA was extracted from the
sorted cells using an RNeasy® kit (Qiagen, Carlsbad, CA,
USA) according to the manufacturer’s instructions. cDNA was
synthesized from 2 mg of total RNA with M-MLV Reverse Transcriptase
(Promega Corporation, Madison, WI, USA) in a 25 ml volume
containing 2 mg total RNA, 400 mM reverse transcription primer
[oligo(dT)18 for random primers for U6 rRNA and miR-19a and miR-19b
specific primers; Bulge-Loop™ miRNA qPCR primers purchased from
Guangzhou RiboBio, Co., Ltd. (Guangzhou, China); for miRNA] 4 U/ml
M-MLV, 1 U/ml inhibitor and 0.4 mM dNTP mix. qPCR was performed
using the reagents of SYBR-green I mix (Takara, Dalian, China) in a
20 ml reaction volume (10 ml SYBR-green I mix, 200 mM forward and
reverse primer, and 2 ml cDNA template) on an MJ Opticon Monitor
chromo4 instrument (Bio-Rad, Hercules, CA, USA) using the following
protocol: 95°C for 20 sec, 40 cycles of 95°C for 10 sec, 60°C for
20 sec, 70°C for 1 sec. Data analysis was performed using the
2−ΔΔCT method (15–16).
Statistical analysis
Differences of nonparametric variables were analyzed
by the Fisher’s exact test using the EPI software (EPI Info,
version 3.2.2; www.CDC.gov/epiinfo/). Differences of
the quantitative variables between groups were analyzed by
Student’s t-test using SPSS 11.0 (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
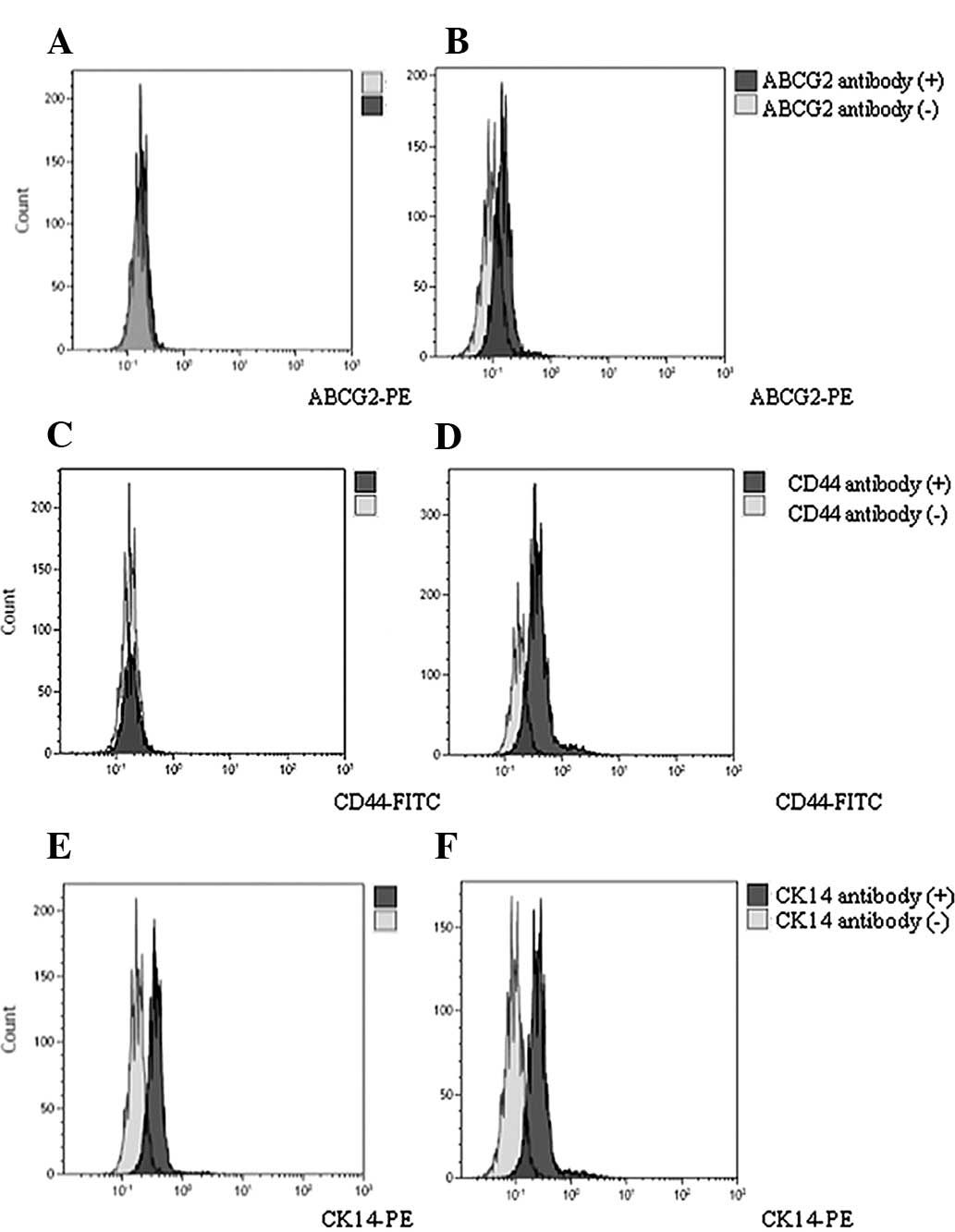
Expression of ABCG2, CD24, CD44, CD133,
CK14 and CK19 in cervical cancer and corresponding non-tumor normal
tissues
In the present study, all 12 cervical cancer tissue
samples were diagnosed as squamous cell cancer. There was a 75%
(9/12) infection rate of HPV 16 or 18. Other HPV types included HPV
33, 35, 52 and 58. In addition, there were 50% peasants
(subsistence farmers and the farm labourers; 6/12; Table I).
In total, 12 pairs of cervical cancer and
corresponding non-tumor normal tissues were analyzed by FACS. The
positive rate (%) of the marker CD44 in cervical cancer tissues and
corresponding non-tumor normal tissues was 6.74 and 1.29%,
respectively. The expression level of CD44 in cervical cancer
tissues was higher than in the corresponding non-tumor normal
tissues (t=3.12; P=0.0102). The positive rate (%) of the marker
ABCG2 in cervical cancer tissues and corresponding normal non-tumor
tissues was 2.82 and 1.08%, respectively. No significant
differences between the two groups were identified (t=1.38;
P=0.20>0.05). CD24, CD133, CK14 and CK19 had the same
distribution tendency as ABCG2. The positive rates between cervical
cancer tissues and corresponding non-tumor normal tissues were 8.27
and 8.39%, 0.89 and 1.02%, 1.01 and 1.15%, and 13.6 and 8.00% for
CD24, CD133, CK14 and CK19, respectively (P>0.05; Table III; Fig. 1).
 | Table IIIExpression of ABCG2, CD24, CD44,
CD133, CK14 and CK19 in cervical cancer by flow cytometry. |
Table III
Expression of ABCG2, CD24, CD44,
CD133, CK14 and CK19 in cervical cancer by flow cytometry.
| Mark | N | Expression level in
cervical cancer tissuesa | Expression level in
corresponding normal non-tumor tissuesb | t-value | P-value |
|---|
| ABCG2 | 12 | 2.82±1.33 | 1.08±0.23 | 1.38 | 0.20 |
| CD24 | 12 | 8.27±2.17 | 8.39±3.20 | −0.046 | 0.96 |
| CD44 | 12 | 6.74±1.24 | 1.29±0.72 | 3.12 | 0.01 |
| CD133 | 12 | 0.89±0.43 | 1.02±0.52 | −0.233 | 0.86 |
| CK14 | 12 | 1.01±0.45 | 1.15±0.28 | −0.37 | 0.72 |
| CK19 | 12 | 13.68±4.40 | 8.00±2.46 | 1.06 | 0.32 |
mRNA expression levels of genes
associated with cancer stem cells in CD44+ and
CD44− cells sorted from cervical cancer tissues
The results demonstrated that SOX2, BMI1 polycomb
ring finger oncogene (Bmi1), CD133, cytokeratin 5 (ck5), Nanog,
nestin, END_1, nuclear receptor subfamily 4, group A, member 2
(NR4A2), OCT4 and ABCG2 genes are associated with cancer stem
cells. These genes are important in various types of tumor. To
investigate the expression levels of these genes in
CD44+ cervical cancer cells and the effects between
these genes and CD44, CD44+ and CD44− cells
were sorted using a MoFlo™ XDP High-Performance Cell sorter
(Beckman Coulter). Then, RNA extraction and qPCR were performed
with the sorted CD44+/CD44− cells. Compared
with the CD44− cells, nestin, NR4A2 and OCT4 genes were
highly expressed in CD44+ cells. The fold changes were
3.55, 2.46 and 2.87, respectively (P<0.05). However, Bmi1 and
ck5 genes had low expression levels in CD44+ cells. The
fold changes were 0.39 and 0.23, respectively (P<0.05). No
significant differences between other genes, including SOX2, CD133
and Nanog, were identified in CD44+ and CD44−
cells (Fig. 2).
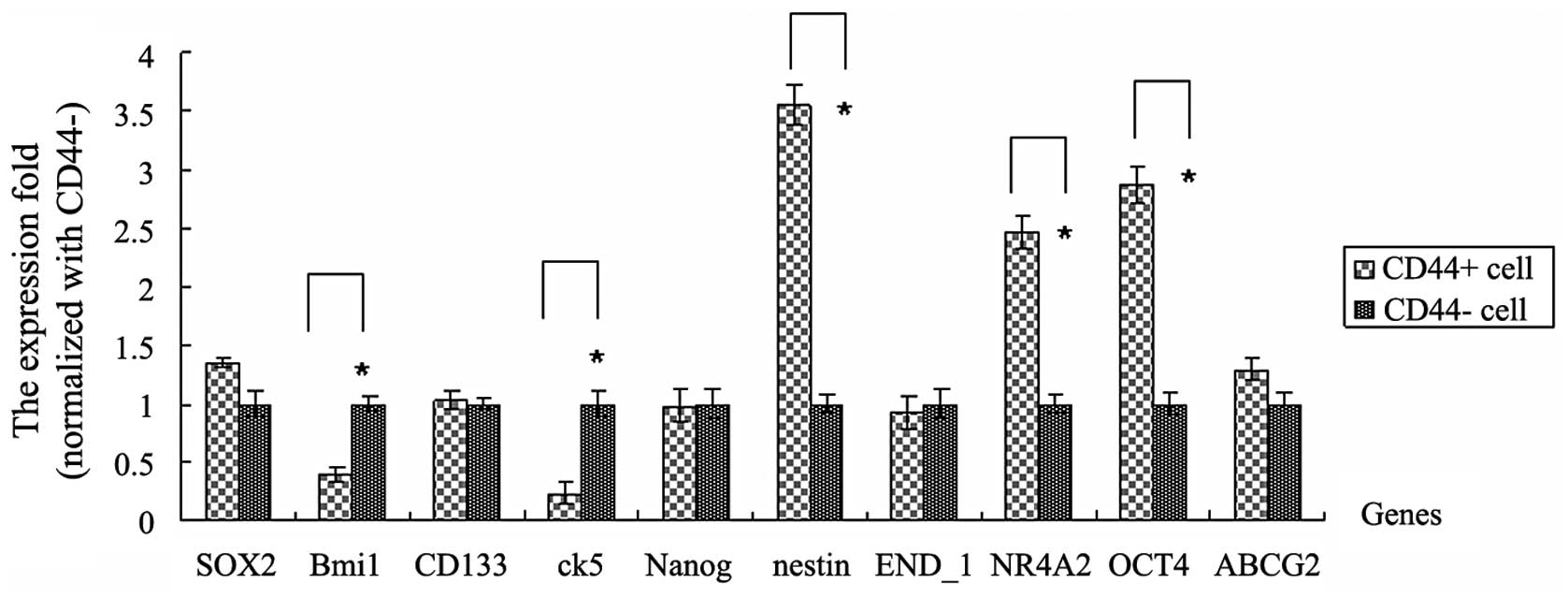 | Figure 2Expression levels of genes associated
with cancer stem cells between CD44+ and
CD44− cells. The mRNA expression levels were analyzed by
qPCR for SOX2, Bmi1, CD133, ck5, Nanog, nestin, END_1, NR4A2, OCT4
and ABCG2 genes. CD44+ and CD44− cells were
sorted from cervical cancer tissues. The mean fold change in
expression of the target gene was calculated using the following
formula: ΔΔCT = (CT,Target −
CT,GAPDH)CD44+ − (CT,Target −
CT,GAPDH)CD44-. At least three replicates of
each reaction were performed. Sample spreadsheet of data analysis
using the 2−ΔΔCT method. Bmi1, B lymphoma Mo-MLV
insertion region 1 homolog; ck5, cytokeratin 5; NR4A2, nuclear
receptor subfamily 4, group A, member 2; ABCG2, ATP-binding
cassette sub-family G member 2. |
mRNA expression levels of other
tumor-associated genes in CD44+ and CD44−
cells sorted from cervical cancer tissues
Fra-1, tumor protein p53 (TP53), Raf kinase
inhibitor protein (RKIP), LTF, ubiquitin associated protein 1,
(UBAP1), protein phosphatase 1, regulatory subunit 13 like (Iaspp),
metadherin (MTDH), metastasis associated 1 family, member 2 (mta2),
short palate, lung, and nasal epithelial clone-1 (Splunc1),
bromodomain containing 7 (BRD7) and p63 are all genes that are
associated with several types of tumor. To investigate these genes
in CD44+ cervical cancer cells and the effect between
these genes and CD44, CD44+ and CD44− cells
were sorted using a MoFlo™ XDP High-Performance Cell sorter
(Beckman Coulter). Then, RNA extraction and qPCR were conducted
with the CD44+/CD44− sorted cells.
The results demonstrated that the TP53 and LTF genes
exhibited a low expression in CD44+ cells compared with
CD44− cells. The fold changes were 0.36 and 0.42 to TP53
and LTF genes, respectively (P<0.05). However, Fra-1 and p63
genes were highly expressed in CD44+ cells. There was
3.55 fold to Fra-1 gene and 2.56 fold to P63 gene fold in
CD44+ cells compared with CD44− cells,
respectively (P<0.05). No significant differences were
identified in RKIP, UBAP1, Iaspp, MTDH, mta2, Splunc1 and BRD7
genes between CD44+ and CD44− cells (Fig. 3).
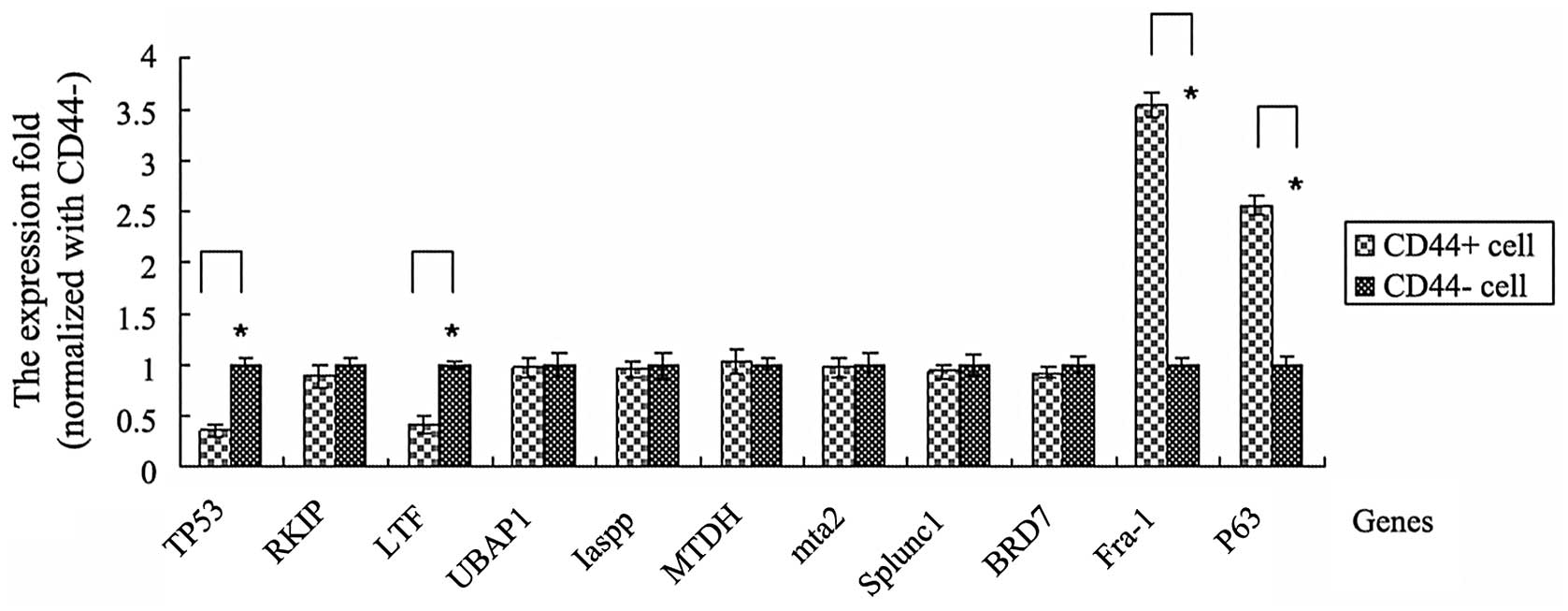 | Figure 3Expression levels of other
tumor-associated genes between CD44+ and
CD44− cells. The mRNA expression levels were analyzed by
qPCR for Fra-1, TP53, RKIP, LTF, UBAP1, Iaspp, MTDH, mta2, Splunc1,
BRD7 and P63 genes. CD44+ and CD44− cells
were sorted from cervical cancer tissues. The mean fold change in
the expression of the target gene was calculated using the
following formula: ΔΔCT = (CT,Target −
CT,GAPDH)CD44+ − (CT,Target −
CT,GAPDH)CD44-. At least three replicates of
each reaction were performed. Sample spreadsheet of data analysis
using the 2−ΔΔCT method. Fra-1, FOS-like antigen 1;
TP53, tumor protein p53; RKIP, Raf kinase inhibitor protein; LTF,
lactotransferrin; UBAP1, ubiquitin associated protein 1; Iaspp,
protein phosphatase 1, regulatory subunit 13 like; MTDH,
metadherin; mta2, metastasis associated 1 family, member 2;
SPLUNC1; short palate, lung, and nasal epithelial clone-1; BRD7,
bromodomain containing 7. |
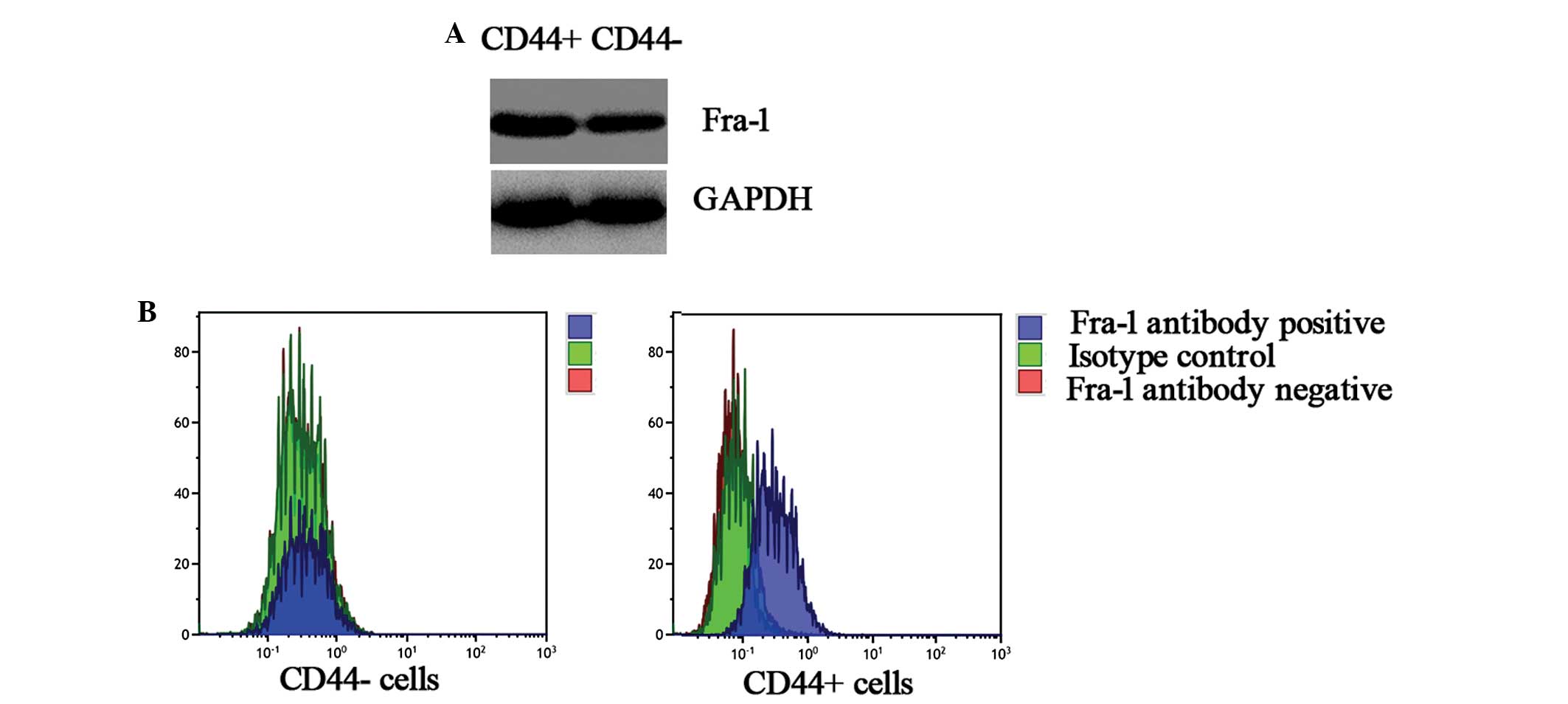
Analysis of protein expression levels of
Fra-1 between CD44+ and CD44− cells sorted
from cervical cancer tissues using western blot analysis
To verify whether the Fra-1 gene had a higher
expression level of CD44+ cells than the
CD44− sorted cells from cervical cancer tissues, its
protein expression levels were further examined (Fig. 4A). In comparison to the
CD44− cells, the expression level was high in the
CD44+ cells. This corresponded with the results of qPCR
and confirmed that Fra-1 was highly expressed in CD44+
cells sorted from cervical cancer tissues.
Analysis of protein expression levels of
Fra-1 in CD44+ and CD44− cells sorted from
cervical cancer tissues by FACS
To confirm that Fra-1 was expressed highly in
CD44+ cells sorted from cervical cancer tissues, the
protein expression levels of Fra-1 were further examined by FACS
(Fig. 4B). In comparison to the
CD44− cells, the expression level was high in the
CD44+ cells. This corresponded with the results of qPCR
and confirmed that Fra-1 was expressed highly in CD44+
cells.
Expression of miR-19a and miR-19b are
downregulated in CD44+ cells in cervical cancer
tissues
The open access program TargetScan (http://www.targetscan.org/) was used to predict the
targets of miR-19a and miR-19b. Fra-1 was found to be a potential
target of miR-19a and miR-19b. The endogenous expression of miR-19a
and miR-19b was compared in CD44+/CD44− cells
sorted from cervical cancer tissues by qPCR. As shown in Table IV, the expression of miR-19a and
miR-19b was downregulated by ~50% in CD44+ cells
compared with CD44− cells (Table IV). These results confirmed that
Fra-1 was highly expressed in CD44+ cells.
 | Table IVIdentification of the expression of
miR-19a and miR-19b in CD44+/CD44− cells
sorted from cervical cancer tissues. |
Table IV
Identification of the expression of
miR-19a and miR-19b in CD44+/CD44− cells
sorted from cervical cancer tissues.
| miRNA | Sample | U6 CT (mean ±
SD) | miRNA CT (mean ±
SD) | ΔCT (mean ±
SD) | ΔΔCT (mean ±
SD) | Folda |
|---|
| miR-19a |
CD44+ | 19.84±0.83 | 29.77±1.05 | 9.93±0.99 | 0.88 | 0.54 |
|
CD44− | 19.97±0.92 | 29.02±1.11 | 9.05±1.04 | | |
| miR-19b |
CD44+ | 19.91±0.89 | 31.45±1.06 | 11.54±1.01 | 0.93 | 0.52 |
|
CD44− | 20.03±0.96 | 30.64±1.15 | 10.61±1.07 | | |
Discussion
Cervical cancer that has been verified to be
associated with the human papillomavirus (HPV), is the second most
common type of cancer in females worldwide and is a leading cause
of cancer-related mortality in females in developing countries
(17,18). The most common high-risk HPV types
in cervical cancer are HPV 16 and 18, and the most common low-risk
types causing genital warts are HPV 6 and HPV 11 (19–22).
In the present study, there was a 75% (9/12) infection rate of HPV
16 or 18. Other HPV types included HPV 33, 35, 52 and 58. This
corresponded with previous studies (17,19).
CD44 has been implicated in numerous biological
processes, including cell adhesion and cell to cell interactions
for example lymphocyte homing hemopoiesis, cell migration and
metastasis. Published data have demonstrated that CD44 mediates
constitutive type I receptor signaling in cervical carcinoma cells
(2). The assessment of CD44
isoform expression may be of clinical value in deciding upon
adjuvant therapy, resulting in a more individualized management of
therapy. CD44 may function in certain cells through interactions
with type I receptor tyrosine kinases, including erbB2 (2,7–8).
However, one common confounder for the analysis of clinical tumor
specimens is the cellular heterogeneity. Sorted pure
CD44+/CD44− cells were used to reveal the
mechanisms of cervical cancer. To the best of our knowledge, there
are no studies in which the cell sorting of
CD44+/− technology was used to study the
mechanisms of cervical cancer.
In the present study, the CD44+ or
CD44− cells of cervical cancer tissues were sorted by
flow cytometry. Then, the mRNA expression levels of 21 genes were
analyzed and the expression levels of miR-19a and miR-19b were
detected by qPCR. The results demonstrated that the expression
level of CD44 in cervical cancer tissues was higher than in the
corresponding non-tumor normal tissues. Compared with the
CD44− cells, the Fra-1, nestin, NR4A2, OCT4 and P63
genes were highly expressed in CD44+ cells. The fold
changes were 3.55, 3.55, 2.46, 2.87 and 2.56, respectively
(P<0.05). Bmi1, ck5, TP53 and LTF genes were expressed at low
levels in CD44+ cells. Bourguignon et al
(23) confirmed that
hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302
expression leading to self-renewal, clonal formation and cisplatin
resistance in cancer stem cells from head and neck squamous cell
carcinoma. A study by Dhingra et al (24) demonstrated that nestin and CD44 are
significantly expressed in a subset of gastric adenocarcinoma,
particularly co-expression of nestin and CD44, and are highly
expressed in Lauren intestinal histological subtypes.
Overexpression of chromatin assembly factor-1 p60, poly
(ADP-ribose) polymerase 1 and nestin predicts metastasizing
behavior of oral cancer (25).
CD44 is a key tumor-promoting agent in transformed tumor cells
lacking p53. It was also suggested that the derepression of CD44
resulting from inactivation of p53 may potentially aid the survival
of immortalized, premalignant cells (26). Immunostaining of p53 family members
may aid the diagnosis and monitoring of high-risk pre-malignant
lesions of the oral epithelium. The combination of staining
patterns of p63, p73α and CD44v6 enabled us to isolate phenotypic
undifferentiated or transient amplifying areas, reflecting the
immaturity of the tumour cell lineage (27). The results from the present study
and other studies suggested that CD44 affected the expression of
important genes, including TP53 and nestin.
Fra-1 was highly expressed in CD44+
cells. Fra-1 was a potential target of miR-19a and miR-19b. The
expression of miR-19a and miR-19b was downregulated by ~50% in
CD44+ cells compared with CD44− cells. The
results suggested that CD44 dysregulated the activation of numerous
genes with important functions. Ramos-Nino et al revealed
that Fra-1 is associated with cell migration in human malignant
mesothelioma (MMs) and that Fra-1 modulation of CD44 may govern the
migration of selected MMs (28).
The results from the study by Kajanne et al demonstrated
that Fra-1 was an important molecule in prostate cancer (29). Thus, interaction of Fra-1 and CD44
may be important in cervical carcinoma.
The findings of the present study suggested that
CD44 dysregulated the activation of the Fra-1 gene and may exhibit
an important role.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant no. 81272975), the Key Project
of Hunan Provincial Natural Science Foundation (grant no.
12JJ2044), the Key Planned Science and Technology Project of Hunan
Province (grant no. 2012FJ2014), the Planned Science and Technology
Project of Hunan Province (grant no. 2011FJ3153), the Planned
Project of Development and Reform Commission of Hunan Province
(grant no. 2012-1493-1), the Planned Project of the Department of
Health of Hunan Province (grant nos. B2011-030 and B2012-029) and
the Planned Project of Key Subject Construction of The Third
Xiangya Hospital, Central South University.
References
|
1
|
Parish SL, Swaine JG, Son E and Luken K:
Determinants of cervical cancer screening among women with
intellectual disabilities: evidence from medical records. Public
Health Rep. 128:519–526. 2013.PubMed/NCBI
|
|
2
|
Wobus M, Kuns R, Wolf C, Horn LC, Köhler
U, Sheyn I, Werness BA and Sherman LS: CD44 mediates constitutive
type I receptor signaling in cervical carcinoma cells. Gynecol
Oncol. 83:227–234. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu JH, Liang XA, Wu YM, Li FS and Dai YM:
Identification of DNA methylation of SOX9 in cervical cancer using
methylated-CpG island recovery assay. Oncol Rep. 29:125–132.
2013.PubMed/NCBI
|
|
4
|
Dobo C, Stavale JN, de Lima FO, Ribeiro
DA, Arias V, Gomes TS and Oshima CT: HSP27 is commonly expressed in
cervical intraepithelial lesions of brazilian women. Asian Pac J
Cancer Prev. 14:5007–5010. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saavedra KP, Brebi PM and Roa JC:
Epigenetic alterations in preneoplastic and neoplastic lesions of
the cervix. Clin Epigenetics. 4:132012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Wang Q, Liu H, Shao N, Tan B,
Zhang G, Wang K, Jia Y, Ma W, Wang N and Cheng Y: The association
of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with
cancer risk: a meta-analysis of 32 studies. Mutagenesis.
27:779–788. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Makrydimas G, Zagorianakou N, Zagorianakou
P and Agnantis NJ: CD44 family and gynaecological cancer. In Vivo.
17:633–640. 2003.PubMed/NCBI
|
|
8
|
Ibrahim EM, Stewart RL, Corke K, Blackett
AD, Tidy JA and Wells M: Upregulation of CD44 expression by
interleukins 1, 4, and 13, transforming growth factor-beta1,
estrogen, and progestogen in human cervical adenocarcinoma cell
lines. Int J Gynecol Cancer. 16:1631–1642. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Speiser P, Wanner C, Tempfer C, Mittelböck
M, Hanzal E, Bancher-Todesca D, Gitsch G, Reinthaller A and Kainz
C: CD44 is an independent prognostic factor in early-stage cervical
cancer. Int J Cancer. 74:185–188. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mostaan LV, Khorsandi MT, Sharifian SM,
Shandiz FH, Mirashrafi F, Sabzari H, Badiee R, Borghei H and
Yazdani N: Correlation between E-cadherin and CD44 adhesion
molecules expression and cervical lymph node metastasis in oral
tongue SCC: Predictive significance or not. Pathol Res Pract.
207:448–451. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oberprieler NG and Taskén K: Analysing
phosphorylation-based signalling networks by phospho flow
cytometry. Cell Signal. 23:14–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou Y, Zeng Z, Zhang W, Xiong W, Wu M,
Tan Y, Yi W, Xiao L, Li X, Huang C, et al: Lactotransferrin: a
candidate tumor suppressor-deficient expression in human
nasopharyngeal carcinoma and inhibition of NPC cell proliferation
by modulating the mitogen-activated protein kinase pathway. Int J
Cancer. 123:2065–2072. 2008. View Article : Google Scholar
|
|
13
|
Krutzik PO, Irish JM, Nolan GP and Perez
OD: Analysis of protein phosphorylation and cellular signaling
events by flow cytometry: techniques and clinical applications.
Clin Immunol. 110:206–221. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, Tang A, Zhou Y, Tang J, Luo Z,
Jiang C, Li X, Xiang J and Li G: Tumor-conditioned mesenchymal stem
cells display hematopoietic differentiation and diminished influx
of Ca2+. Stem Cells Dev. 21:1418–1428. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou Y, Wang W, Zheng D, Peng S, Xiong W,
Ma J, Zeng Z, Wu M, Zhou M, Xiang J, et al: Risk of nasopharyngeal
carcinoma associated with polymorphic lactotransferrin haplotypes.
Med Oncol. 29:1456–1462. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salehi M, Taheri T, Mohit E, Zahedifard F,
Seyed N, Taslimi Y, Sattari M, Bolhassani A and Rafati S:
Recombinant Leishmania tarentolae encoding the HPV type 16
E7 gene in tumor mice model. Immunotherapy. 4:1107–1120. 2012.
|
|
18
|
Sahiner F, Gümral R, Sener K, Yiğit N,
Dede M, Yapar M and Kubar A: Investigation of HPV-DNA in cervical
smear samples by two different methods: MY09/11 consensus PCR and
type-specific real-time PCR. Mikrobiyol Bul. 46:624–636. 2012.(In
Turkish).
|
|
19
|
Balbi G, Napolitano A, Giordano F, Capuano
S, Manganaro MA, Di Martino L, Fusco D, Grauso F and Seguino E:
Role of the association of high-risk HPV identified by real-time
PCR in cervical preneoplastic lesions. Eur J Gynaecol Oncol.
33:467–471. 2012.PubMed/NCBI
|
|
20
|
Raychaudhuri S and Mandal S: Current
status of knowledge, attitude and practice (KAP) and screening for
cervical cancer in countries at different levels of development.
Asian Pac J Cancer Prev. 13:4221–4227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gadducci A, Guerrieri ME and Greco C:
Tissue biomarkers as prognostic variables of cervical cancer. Crit
Rev Oncol Hematol. 86:104–129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mocarska A, Starosławska E,
Zelazowska-Cieślińska I, Łosicki M, Stasiewicz D, Kieszko D and
Burdan F: Epidemiology and risk factors of the cervical squamous
cell carcinoma. Pol Merkur Lekarski. 33:101–106. 2012.(In
Polish).
|
|
23
|
Bourguignon LY, Wong G, Earle C and Chen
L: Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes
miR-302 expression leading to self-renewal, clonal formation, and
cisplatin resistance in cancer stem cells from head and neck
squamous cell carcinoma. J Biol Chem. 287:32800–32824. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dhingra S, Feng W, Brown RE, Zhou Z,
Khoury T, Zhang R and Tan D: Clinicopathologic significance of
putative stem cell markers, CD44 and nestin, in gastric
adenocarcinoma. Int J Clin Exp Pathol. 4:733–741. 2011.PubMed/NCBI
|
|
25
|
Mascolo M, Ilardi G, Romano MF, Celetti A,
Siano M, Romano S, Luise C, Merolla F, Rocco A, Vecchione ML, De
Rosa G and Staibano S: Overexpression of chromatin assembly
factor-1 p60, poly(ADP-ribose) polymerase 1 and nestin predicts
metastasizing behaviour of oral cancer. Histopathology.
61:1089–1105. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Godar S, Ince TA, Bell GW, Feldser D,
Donaher JL, Bergh J, Liu A, Miu K, Watnick RS, Reinhardt F, et al:
Growth-inhibitory and tumor-suppressive functions of p53 depend on
its repression of CD44 expression. Cell. 134:62–73. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bidaud P, Chasle J, Sichel F, Rousseau S,
Petit P, Pottier D, Picquenot JM, Louis MY and Lechevrel M:
Expression of p53 family members and CD44 in oral squamous cell
carcinoma (OSCC) in relation to tumorigenesis. Histol Histopathol.
25:331–339. 2010.PubMed/NCBI
|
|
28
|
Ramos-Nino ME, Blumen SR, Pass H and
Mossman BT: Fra-1 governs cell migration via modulation of CD44
expression in human mesotheliomas. Mol Cancer. 6:812007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kajanne R, Miettinen P, Tenhunen M and
Leppä S: Transcription factor AP-1 promotes growth and
radioresistance in prostate cancer cells. Int J Oncol.
35:1175–1182. 2009.PubMed/NCBI
|