Introduction
Bone marrow mesenchymal stem cells (BMSCs) are
multipotent progenitor cells with the capacity of differentiating
into osteoblasts, chondrocytes, adipocytes and myoblasts (1–3).
Previous studies have shown that the multi-differentiation
processes of BMSCs are tightly regulated by various factors.
Recently, microRNAs (miRNAs or miRs) have been reported to have
crucial roles in the osteogenic differentiation of BMSCs. For
example, miR-204 and -211 inhibit osteogenesis of hBMSCs by acting
as endogenous negative regulators of Runt-related transcription
factor-2 (Runx-2) (4).
miR-138 suppresses osteogenic differentiation of hBMSCs by
repressing the expression of focal adhesion kinase (FAK), as well
as its downstream targets (5).
Furthermore, miR-637 significantly suppresses osteogenic
differentiation in hBMSCs through direct repression of osterix
(Osx) expression (6).
Previous studies have reported that miR-125b is a crucial
post-transcriptional regulator of genes that are involved in cell
proliferation or differentiation processes of various cell lineages
(7–10). However, the role of miR-125b in
hBMSCs and its downstream target genes have not been fully
elucidated.
Osteoporosis, a common disease of the skeletal
system, is characterized by the loss of bone mass due to an
imbalance in relative bone secretion versus bone resorption
(11). Disruptive bone metabolism
or levels of bone secretion which are disproportionate to
resorption result in decreased bone mass or osteoporosis. Excessive
accumulation of adipocytes is frequently observed in the bone
marrow of patients with senile osteoporosis, which may suggest an
imbalanced differentiation of BMSCs into osteoblasts or adipocytes
(12). The osteogenic
differentiation of BMSCs is a well-coordinated process and is
regulated by key transcription factors, including Runx-2 and
Osx (13,14). However, whether miR-125b has a
regulatory role in the proliferation and osteogenic differentiation
of hBMSCs remains to be elucidated. In this context, the present
study aimed to assess the impact of miR-125b and Osx
expression on the proliferation and osteogenic differentiation of
hBMSCs derived from senile osteoporotic patients.
Materials and methods
Bone marrow donors
Written informed consent was obtained from all
participating subjects and the present study was approved by the
ethics committee of the Fourth Military Medical University (Xi’an,
China). Bone marrow aspirates of 5–10 ml from the iliac crest were
obtained from five normal donors and four elderly osteoporotic
patients who had undergone an iliac bone graft procedure during
surgery. Normal donors had a T-score of >-1 standard deviation
(SD) and no history of bone fractures. A standardized clinical
evaluation was performed to exclude possible comorbidities.
Exclusion criteria included premature menopause, the presence of a
disease and the use of drugs that are able to affect bone or
calcium and phosphorus metabolism. Osteoporosis was diagnosed in
accordance with the World Health Organization parameters, with a
T-score of <-2.5 SD. Calcium, phosphate, parathyroid
hormone, carboxy-terminal telopeptide of type I collagen, alkaline
phosphatase (ALP), 25-hydroxyvitamin D levels and the urinary
calcium excretion rate were measured in the serum of all elderly
patients with osteoporotic fractures to exclude any secondary
causes of osteoporosis.
Isolation and culture of hBMSCs
hBMSCs were isolated and cultured as previously
described (15). Bone marrow
aspirates of 5–10 ml were placed in tubes containing heparin (100
U/l) and were mixed with isochoric phosphate-buffered saline (PBS).
The cell suspension was mixed with an equal volume (1.073 g/ml) of
Percoll solution (Pharmacia, Uppsala, Sweden) in 15 ml conical
tubes (Nunc A/S, Roskilde, Denmark) and centrifuged (L500
Centrifuge; Xiangyi Centrifuge Instrument Co., Ltd., Hunan, China)
at 644.8 × g for 30 min. Mononuclear cells were collected from the
middle layer and interface, diluted with two volumes of PBS and
collected by centrifugation at 161.2 × g. The cells were
resuspended in complete culture medium (low-glucose Dulbecco’s
modified Eagle medium with 10% fetal bovine serum, 100 U/ml
penicillin, 100 mg/ml streptomycin and 2 mM L-glutamine; Hyclone,
Logan, UT, USA). The cells were seeded at 4,500
cells/cm2 in 25 cm2 culture flasks (Nunc A/S)
and were incubated at 37°C in 5% CO2 with 95% humidity.
After three days of incubation, the cultured medium and
non-adherent cells were discarded, adherent cells were washed twice
with PBS and resuspended in fresh culture medium. After 10–14 days
of culture, the cells were harvested using 0.25% trypsin and 1 mM
EDTA (Gibco, Grand Island, NY, USA) and were replated at
104 cells/cm2 in 25 cm2 culture
flasks (Nunc A/S) to expand the cells through successive
passages.
For osteogenic differentiation, hBMSCs were seeded
at 104 cells/cm2 in 25 cm2 culture
flasks (for RNA isolation; Nunc A/S) or in six-well plates (for
staining). Cells were grown to 45–75% confluency over 24–48 h in
standard growth medium. On every third day, the medium was replaced
with osteogenic differentiation medium (10% fetal bovine serum,
Hyclone; 100 nM dexamethasone, 45 mM L-ascorbic acid and 10 mM
β-glycerophosphate; Sigma, St. Louis, MO, USA). Samples were
stained or harvested for RNA isolation on the 14th day of
differentiation.
Transfection of miR-125b mimic and
inhibitor
The miR-125b mimic, miR-125b inhibitor and a
non-specific negative control (NC) were designed and synthesized by
Invitrogen Life Technologies (Shanghai, China). The sequences
included: miR-125b mimic, 5′-UCCCUGAGACCCUAACUUGUGAA
CAAGUUAGGGUCUCAGGGAUU-3′; miR-125b inhibitor,
5′-UCACAAGUUAGGGUCUCAGGGA-3′ and NC, 5′-CAG
UACUUUUGUGUAGUACAA-3′.
The negative control, a random sequenced anti-miRNA
molecule based on the miRNAs in C. elegans, has been
extensively compared to the human, mouse and rat genome sequences
and miRNA sequences by the Basic Logarithmic Alignment Search Tool
for nucleotides (BLASTn; National Library of Medicine, Bethesda,
MD, USA) and validated to not produce identifiable effects on the
known miRNA functions.
Cells were transfected using Lipofectamine 2000
reagent (Invitrogen, Carlsbad, CA, USA). The hBMSCs were plated in
six-well plates (Nunc A/S) to 45% confluence. In addition, 20 μM
miR-125b mimic or a miR-125b inhibitor were mixed with
Lipofectamine 2000, which was prepared according to the
manufacturer’s instructions. The final concentrations of miR-125b
mimic or miR-125b inhibitor and the negative control were adjusted
to 45 nM. The culture conditions were conducive and the survival
rate of hBMSCs was up to 90% following transfection. After the
transfection (4 h), the medium was replaced with osteogenic
differentiation medium to induce osteoblast differentiation.
RNA extraction
Total RNA was isolated from cultured cells using
TRIzol® reagent (Invitrogen), according to the
manufacturer’s instructions. The integrity and purity of total RNA
was verified spectrophotometrically and by gel-electrophoresis on
formaldehyde denaturation gel.
Quantitative polymerase chain reaction
(qPCR)
Gene expression levels were measured by the iQ5
real-time PCR detection system (Bio-Rad, Hercules, CA, USA).
microRNAs were prepared with an All-in-One™ miRNA qRT-PCR detection
kit (AOMD-Q020; GeneCopoeia, Inc., Rockville, MD, USA) according to
the manufacturer’s instructions. Briefly, the extracted RNA was
reverse-transcribed in the presence of a poly-A polymerase with an
oligo-dT adaptor. qPCR was then performed with SYBR®
Green (Takara, Shiga, Japan) detection with a forward primer for
the mature miRNA sequence and a universal adaptor reverse primer.
RNA was reverse-transcribed using the All-in-One™ miRNA qPCR Primer
(GeneCopoeia, Inc.).
The sequence of the F-Primer for human snRNA U6 was:
TCGTGAAGCGTTCCATATTTTTAA. The sequence of the F-Primer for Homo
sapiens (hsa)-miR-125b was: CCCTGAGACCCTAACTTGTGAAA and the
sequence of the Mature1_Seq for hsa-miR-125b was:
UCCCUGAGACCCUAACUUGUGA. The expression of Runx-2,
ALP, Osteocalcin (OC), collagen type I α1
(COL1α1) and Osx were quantified by qPCR using
SYBR® Green assays (Takara) and GAPDH was used as
an internal control.
PCR primers included: Runx-2 forward,
CCACCACTCAC TACCACACCTA and reverse, CCTGACGAAGTGCC ATAGTAGA;
ALP forward, GCATAACATCAGGGACAT TGAC and reverse,
TGGCTTTCTCGTCACTCTCAT; OC forward, CTCACACTCCTCGCCCTATT and
reverse, CG CTGGGTCTCTTCACTAC; COL1α1 forward, AGGGCT
CCAACGAGATCGAGATCCG and reverse, TACAG GAAGCAGACAGGGCCAACG;
Osx forward, ACCTACCC ATCTGACTTTGCTC and reverse,
CTGCCCACTATTTCC CACTG; GAPDH forward, GGAGACAACCTGGTCCTCAG
and reverse, CTCTGTCTCTCTGACCTCACAG.
The Ct value obtained for the gene of interest was
normalized to a housekeeping gene, GAPDH, to obtain the ΔCt
values. The ΔΔCt values were then obtained by subtracting the ΔCt
values for each gene of interest from the ΔCt values for the
control sample. Results were calculated using the equation
RQ=2−ΔΔCt, where RQ is the relative
quantity, and expressed as the fold change, relative to the gene
expression levels in the control samples.
Cell proliferation analysis
MTT (Sigma) and colony-forming assays were used to
analyze cell proliferation. Cells were plated in 96-well plates at
3×103 cells per well and cultured for 2, 4, 6, 8 and 10
days. The cell growth and viability were assessed following
incubation in 0.1 mg/ml MTT at 37°C for 4 h. The cells were lysed
in dimethylsulfoxide (Cellgro, Herndon, VA, USA) at room
temperature for 10 min. Using a microplate reader (Thermo Fisher
Scientific, Inc., Waltham, MA, USA), the absorbance in each well
was measured at 492 nm. Colony-forming efficiency was determined by
plating the cells at 10 cells/cm2 in 60-mm dishes (Nunc
A/S). After 14 days of culture, dishes were stained with Giemsa and
the number of visible colonies was counted. Colony-forming
efficiency was calculated as the percentage of the number of cells
initially plated that gave rise to visible colonies (>45
cells).
Evaluation of osteogenic differentiation
in hBMSCs by ALP staining
ALP staining and analysis were performed after 14
days of incubation in differentiated culture medium. Cells were
induced for osteoblast differentiation, washed twice in PBS, fixed
in methanol/formalin (9:1; Ziyi, Shanghai, China) for 30 min and
stained with the
5-bromo-4-chloro-3′-indolyphosphate
p-toluidine salt/nitro-blue tetrazolium chloride
Alkaline Phosphatase Color Development kit (C3206; Beyotime,
Haimen, China) according to the manufacturer’s instructions. The
ALP activity in the monolayers was determined using the
p-Nitrophenyl Phosphate Liquid Substrate System (Sigma) according
to the manufacturer’s instructions. Using a Multiskan Ascent
microplate reader (Thermo Fisher Scientific, Inc.), the absorbance
was measured at 405 nm.
Mineralization assay in hBMSCs by
Alizarin Red staining (ARS)
Following being fixed in 4% paraformaldehyde (m/v)
(Jianglaibio, Shanghai, China) for 10 min, hBMSCs were evaluated by
ARS (ScienCell, San Diego, CA, USA) staining. Briefly, the cells
were stained with 2% ARS (pH 4.1) for 15 min and washed twice in
PBS. Orange and red staining indicated calcium nodules.
Statistical analysis
Statistical analysis of data was performed using the
independent t-test and results were expressed as the mean ± SD.
P<0.05 was considered to indicate a statistically significant
difference. All statistical calculations were performed using SPSS
software, version 17.0 (SPSS, Inc., Chicago, IL, USA).
Results
Upregulated expression of miR-125b in
osteoporotic hBMSCs
The hBMSCs were isolated from the iliac crest of
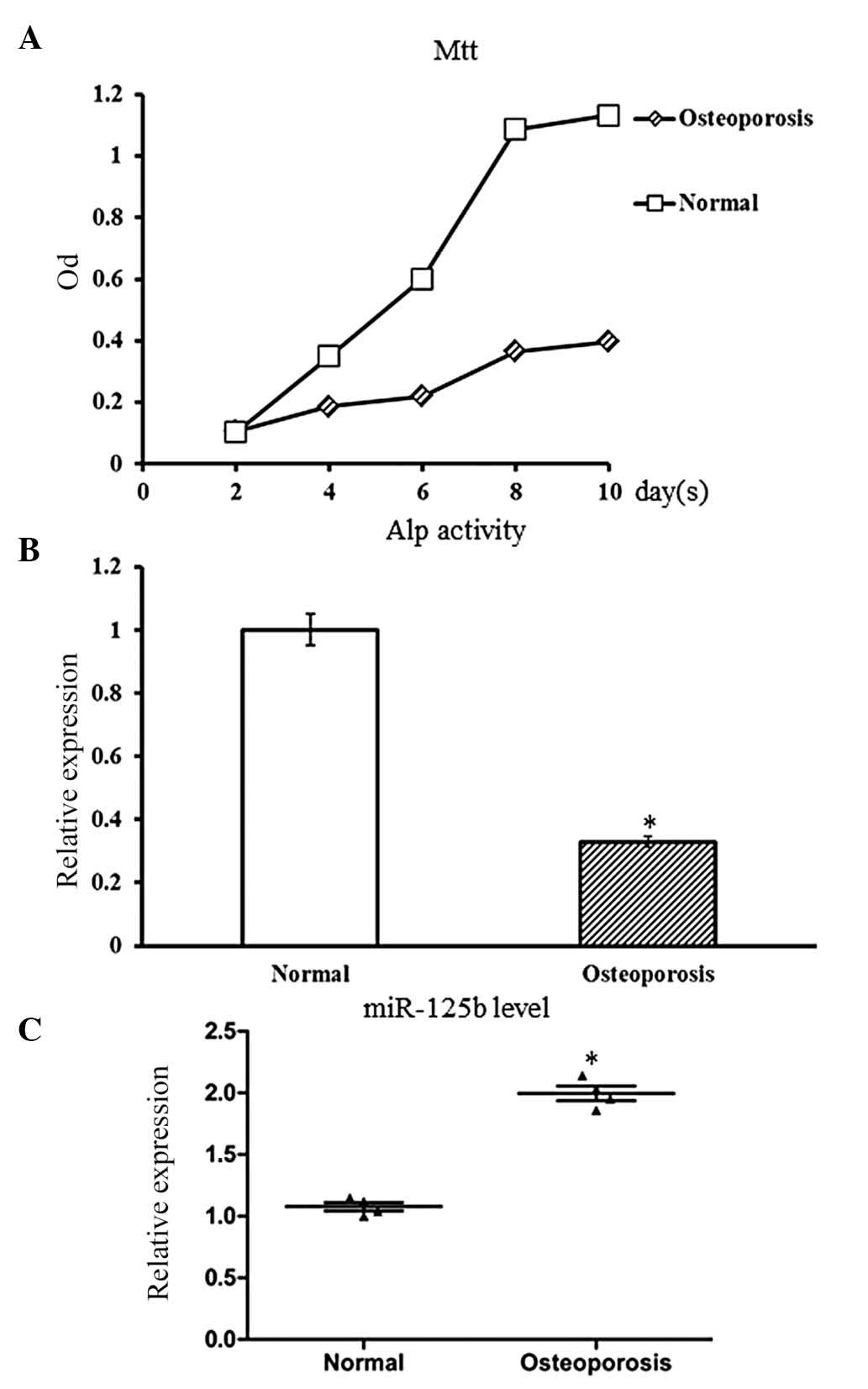
elderly patients with osteoporosis (n=4) and controls (n=5). The
clinical characteristics of each donor are summarized in Table I. Before comparing the expression
levels of miR-125b, the proliferation and osteogenic
differentiation activity of hBMSCs that were derived from
osteoporotic patients and control cases were estimated. The MTT
assay was performed on hBMSCs of passage three to estimate the cell
proliferation rate. Compared with hBMSCs derived from the control
subjects, the hBMSCs derived from osteoporotic patients showed a
slower growth rate with a low cell density (P<0.05; Fig. 1A). This result suggested a
decreased proliferation rate of osteoporotic hBMSCs.
 | Table IClinical characteristics of the
normal donors and patients with osteoporosis. |
Table I
Clinical characteristics of the
normal donors and patients with osteoporosis.
| Groups | Age (years) | Gender | BMD
t-score |
|---|
| Osteoporotic
patient | 76 | Female | −2.7 SD |
| Osteoporotic
patient | 82 | Male | −2.8 SD |
| Osteoporotic
patient | 82 | Female | −3.0 SD |
| Osteoporotic
patient | 88 | Female | −2.6 SD |
| Control | 19 | Female | 0.8 SD |
| Control | 28 | Male | 0.6 SD |
| Control | 40 | Female | −0.8 SD |
| Control | 44 | Male | −0.4 SD |
| Control | 38 | Male | 0.7 SD |
Upon osteogenic induction, hBMSCs were able to
undergo osteogenic differentiation into osteoblasts. The osteogenic
differentiation ability of hBMSCs was evaluated by ALP staining.
Compared with the control hBMSCs, the senile osteoporotic hBMSCs
showed decreased ALP activity (P<0.05; Fig. 1B), which indicated suppression of
osteogenic differentiation of osteoporotic hBMSCs.
qPCR was used to estimate the expression levels of
miR-125b in osteoporotic hBMSCs as well as the cells derived from
the control subjects. Compared to the control hBMSCs, miR-125b
expression levels in osteoporotic hBMSCs were significantly
upregulated (P<0.05; Fig.
1C).
In conclusion, it was identified that the
osteoporotic hBMSCs exhibited decreased cell proliferation and
osteogenic differentiation, which correlated with increased
expression levels of miR-125b. These results suggested that
miR-125b may have a negative impact on cell proliferation or
differentiation of hBMSCs; however, this requires validation and
further investigation.
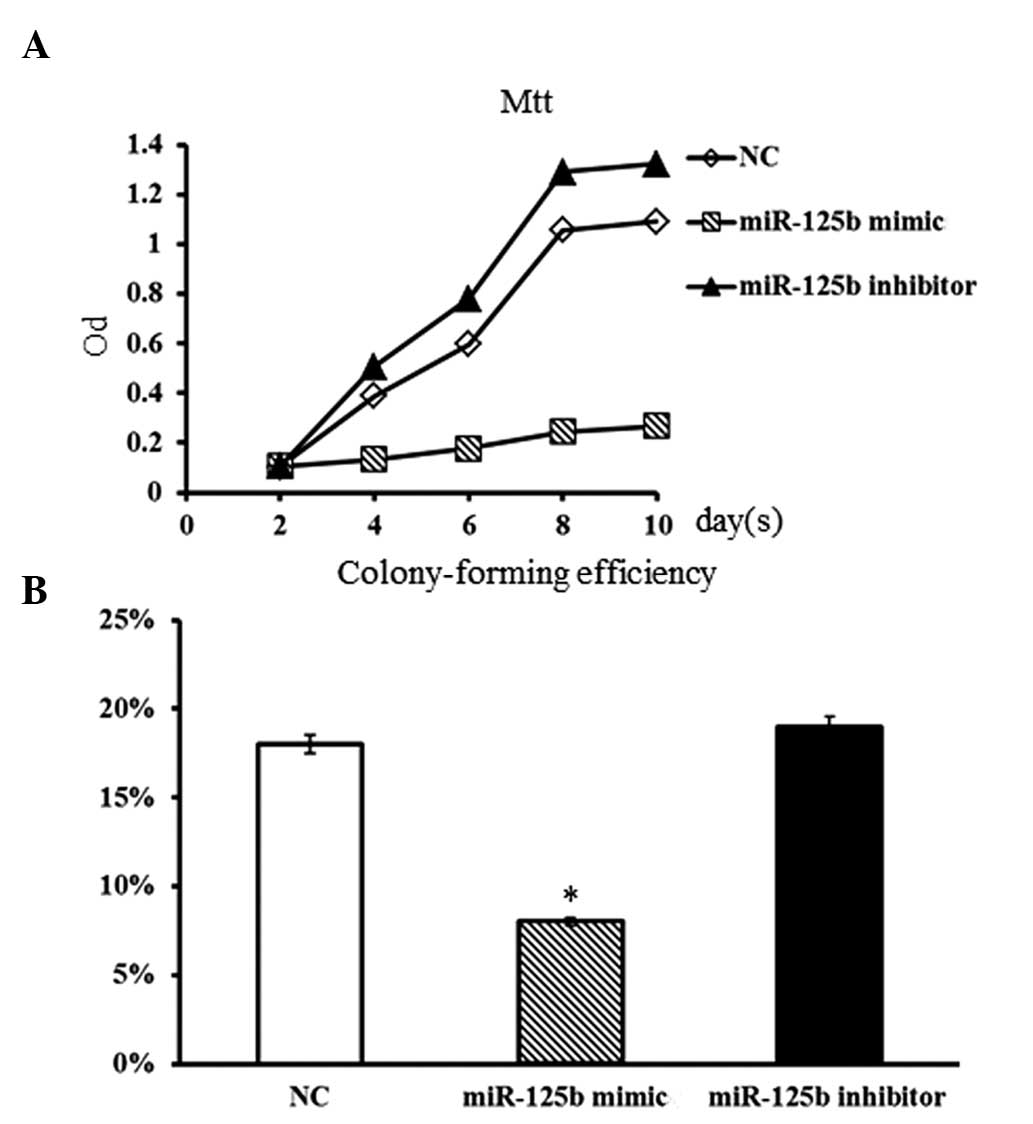
Impact of miR-125b on cell proliferation
in hBMSCs
To study the impact of miR-125b expression on cell
proliferation of hBMSCs, a miR-125b mimic and an miR-125b inhibitor
were respectively transfected into normal individual hBMSCs. The
miR-125b mimic, miR-125b inhibitor and non-specific negative
control were designed and synthesized according to the protocol by
Liu et al (9). The
proliferative rate of hBMSCs was evaluated by MTT and
colony-forming assays. Compared with the non-transfected (control)
group, hBMSCs transfected with miR-125b inhibitor showed a slightly
higher proliferation rate (Fig.
2A). However, the proliferative rate of hBMSCs was markedly
suppressed in the presence of the miR-125b mimic. Furthermore, the
colony-forming assay also showed consistent results. The
colony-forming ability of hBMSCs was obviously decreased in the
presence of the miR-125b mimic (P<0.05; Fig. 2B), while the inhibition of miR-125b
in hBMSCs resulted in a slight increase in the number of colonies
formed (P>0.05). These results suggested that the overexpression
of miR-125b in normal hBMSCs was able to efficiently suppress the
proliferative ability of hBMSCs.
Impact of miR-125b on the osteogenic
differentiation in hBMSCs
To study the role of miR-125b expression on
osteogenic differentiation in hBMSCs, the miR-125b mimic or the
miR-125b inhibitor were transfected into hBMSCs and cultured in
osteogenic differentiation medium. After 14 days of culture, the
expression of various bone-related genes in hBMSCs was determined
by qPCR. The expression of Runx-2, ALP, COL1α1
and OC were suppressed in hBMSCs transfected with the
miR-125b mimic (Fig. 3A). By
contrast, in the presence of miR-125b inhibitors, a significant
increase in the expression levels of Runx-2, ALP,
OC and COL1α1 was observed in hBMSCs (P<0.05)
(Fig. 3A).
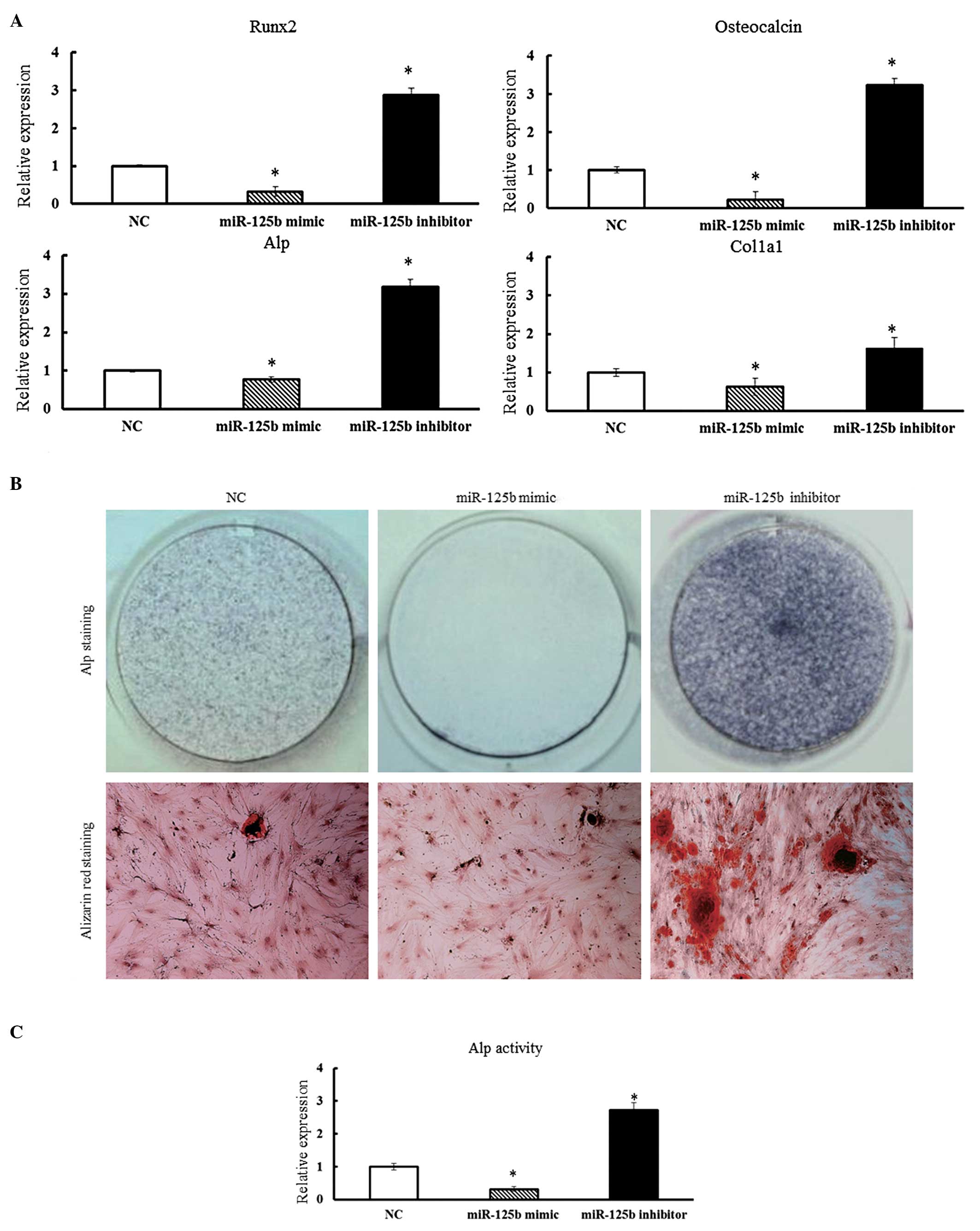 | Figure 3(A) Overexpression of miR-125b
suppressed the osteogenic differentiation of hBMSCs. Osteoblast
differentiation of hBMSCs was induced by osteogenic differentiation
medium and hBMSCs were collected 14 days after osteogenic
induction. qPCR analysis measured the expression of Runx-2,
ALP, OC and COL1α1 in hBMSCs. qPCR data were
subjected to Student’s t-tests. Error bars represent the
mean ± standard deviation of three independent experiments.
*P<0.05. (B) ALP staining was performed after 14 days
of culture to detect ALP activity and Alizarin Red staining was
performed after 21 days to evaluate mineralized bone matrix
formation. (C) ALP activity of hBMSCs was determined 14 days after
osteogenic induction by p-Nitrophenyl Phosphate Liquid
Substrate System and absorbance was measured at 405 nm. Data were
subjected to Student’s t-test. *P<0.05.
hBMSCs, human bone marrow-derived mesenchymal stem cells;
qPCR, quantitative polymerase chain reaction; Runx-2,
Runt-related transcription factor-2; ALP, alkaline
phosphatase; OC, osteocalcin; COL1α1, collagen type I α1; NC,
negative control; miR, micro RNA; OD, optical density. |
To further confirm these findings, transfected
hBMSCs from each group were subjected to ALP staining on day 14
after culturing in conditioned media. The hBMSCs transfected with
the miR-125b mimic showed poor ALP staining, while the hBMSCs
transfected with miR-125b inhibitor stained strongly positive for
ALP. This was a clear indication of the negative effects of
miR-125b expression on osteogenic differentiation of hBMSCs
(Fig. 3B and C). The degree of
matrix mineralization of hBMSCs was further evaluated by Alizarin
red staining on day 21 after transfection (Fig. 3B). Consistent with the results of
ALP staining, Alizarin red staining revealed that inhibition of the
function of miR-125b in hBMSCs obviously increased matrix
mineralization activity (Fig. 3C).
In conclusion, the results suggested that miR-125b has a negative
regulatory role in osteogenic differentiation processes and the
subsequent mineralization of hBMSCs.
Combined effects of Osx and miR-125b
expression on osteogenic differentiation of hBMSCs
The possible targets of miR-125b were predicted by
online software including (accession date: April 2012): TargetScan
(http://www.targetscan.org/) and PicTar
(http://pictar.mdc-berlin.de/).
Osx, one of the essential transcription factors for the
regulation of osteogenic differentiation, was identified as a
potential target of miR-125b.
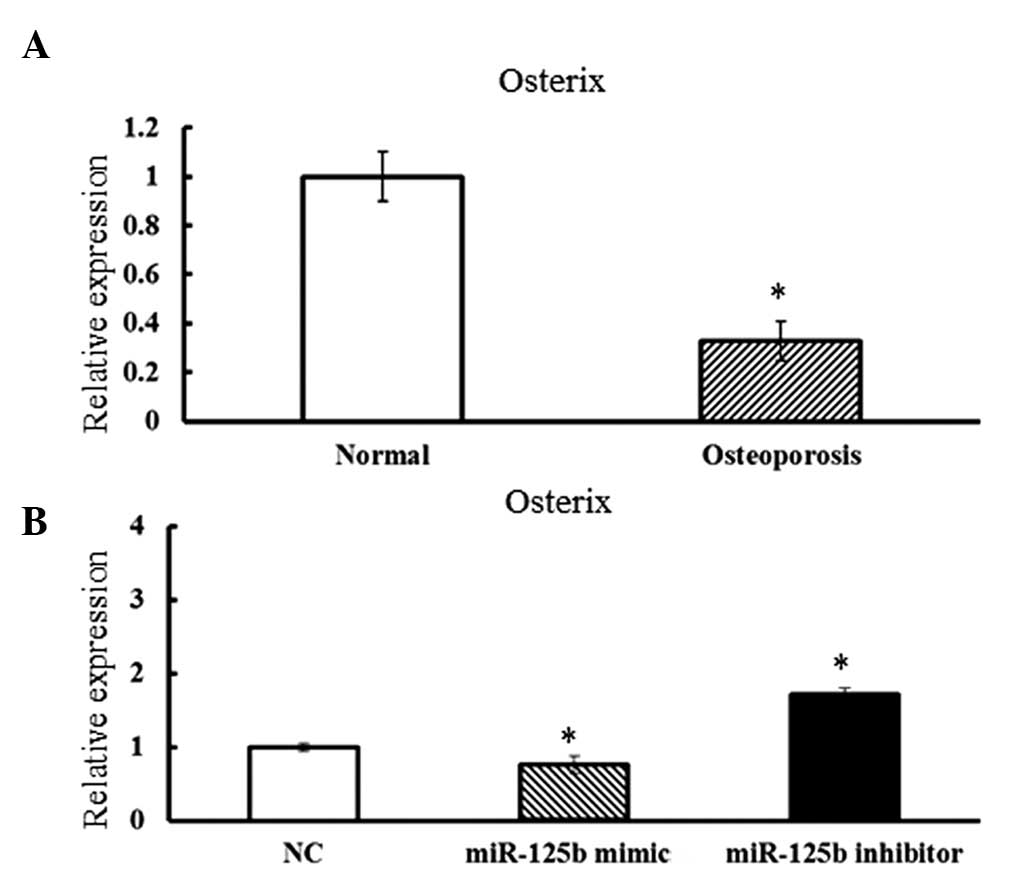
To assess whether Osx was functionally
associated with miR-125b, the expression levels of Osx in
osteoporotic hBMSCs were assessed. Compared with the cells derived
from normal adults, osteoporotic hBMSCs exhibited concurrently
lower expression levels of Osx and higher expression levels
of miR-125b (Fig. 4A).
Furthermore, when the miR-125b mimic was overexpressed in hBMSCs,
the expression levels of Osx were dramatically decreased.
Furthermore, significantly increased Osx expression levels
were observed in hBMSCs transfected with the miR-125b inhibitor
(Fig. 4B). The results strongly
suggested that Osx may be a target of miR-125b and may
mediate the osteogenic differentiation regulated by miR-125b in
hBMSCs.
Discussion
Osteoporosis is characterized by reduced bone mass,
which results from an imbalance in bone formation versus resorption
(16). The increased activity of
osteoclasts has been reported as the main factor involved in the
progression of osteoporosis; however, the defects of
BMSCs/osteoblasts in osteoporosis have not been fully elucidated.
BMSCs, as the main source of adult osteoprogenitors, were found to
exhibit age-associated changes in size, changes in proliferative
capacity and differentiation potential compared to other MSCs,
which suggests that BMSCs have an essential role in the
pathogenesis of bone loss (17).
While the regulation of the abnormal proliferation
and differentiation of BMSCs that are involved in the pathogenesis
of osteoporosis remains to be elucidated, recent studies have shown
that certain miRs may have a crucial role in the osteogenic
differentiation of BMSCs (5,18,19).
However, currently, there are few reports elucidating the roles of
miRs in the proliferation and osteogenic differentiation of hBMSCs
implicated in the process of senile osteoporosis.
The present study identified that miR-125b acted as
a key regulator in the progression of senile osteoporosis. The
expression levels of miR-125b in hBMSCs that were derived from
patients with osteoporosis and control subjects were assessed. The
expression levels of miR-125b were significantly higher in hBMSCs
derived from elderly patients compared with those from normal
younger subjects.
Furthermore, miR-125b inhibited hBMSC proliferation
and osteogenic differentiation. It was identified that Osx
was a potential target of miR-125b, contributing to the negative
regulatory function of miR-125b in osteogenic differentiation
processes in hBMSCs.
miR-125b has been reported to participate in
numerous cell activities. The overexpression of miR-125b in SKBR3
cells caused impaired anchorage-dependent growth and reduced
migration and invasion capacities by suppressing human epidermal
growth factor-2 and -3 signaling (8). miR-125b is also critical for the
suppression of human U251 glioma stem cell proliferation through
the cell cycle regulatory proteins CDK6 and CDC25A (20). Furthermore, miR-125b exhibited
suppressive effects on the proliferation and migration of
osteosarcoma cells by downregulating signal transducer and
activator of transcription 3 (9).
A recent study reported that miR-125b was downregulated during
osteogenic differentiation in human adipose-derived stem cells
(21). Moreover, miR-125b was also
shown to inhibit osteoblast differentiation of ST2 cells (22). However, in the present study, the
molecular mediator of miR-125b on osteogenic differentiation of ST2
cells was not elucidated. Bioinformatics projection and qPCR assays
identified that Osx (SP7) function may be a potential target
of miR-125b and may mediate osteogenic differentiation regulated by
miR-125b. Consistent with these results, Goettsch et al
(23) indicated that miR-125b
expression promoted osteogenic differentiation of human coronary
artery smooth muscle cells, targeted by Osx. Osx, which
functions downstream of Runx-2/core-binding factor α 1, was
required for osteoblast differentiation, bone formation (14) and upregulation in the osteogenic
differentiation of hBMSCs (24).
The present study demonstrated that the endogenous expression of
Osx decreased following hBMSC transfection with a miR-125b
mimic and increased following transfection with an miR-125b
inhibitor.
These results suggested that Osx may be
regulated by miR-125b; however, the mechanisms involved in the
suppressive effects of miR-125b on the proliferation of hBMSCs
requires elucidation. Recent studies have shown that miR-125b
expression directly repressed 20 novel targets in the p53 network
(25), which had a vital role in
the biology of MSCs (26) and
regulated osteoblast differentiation via the transcription factor
Osx (27). These studies
indicate that miR-125b-dependent regulation of the p53 pathway may
account for the negative effects on hBMSCs proliferation. Moreover,
there may be additional unknown signaling pathways involved in
senile osteoporosis, which may influence the high expression levels
of miR-125b.
In conclusion, to the best of our knowledge, the
present study was the first to report the roles of miRs in senile
osteoporosis. miR-125b may be involved in the progression of senile
osteoporosis by suppressing the proliferation and osteogenic
differentiation of hBMSCs. Further understanding of the abnormal
proliferation and osteogenic differentiation of osteoporotic hBMSCs
is required to elucidate the biological characteristics of hBMSCs
and also provide a novel clinical treatment for the prevention of
osteoporosis.
Acknowledgements
The present study was supported by the Ministry of
Science and Technology of China (no. 2011CB964703), the National
Natural Science Foundation of China (no. 30901504) and the China
Postdoctoral Science Foundation (nos. 20100480093 and
2012T50856).
References
|
1
|
Pittenger MF, Mackay AM, Beck SC, et al:
Multilineage potential of adult human mesenchymal stem cells.
Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tuli R, Tuli S, Nandi S, et al:
Characterization of multipotential mesenchymal progenitor cells
derived from human trabecular bone. Stem Cells. 21:681–693. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Le Blanc K and Pittenger M: Mesenchymal
stem cells: progress toward promise. Cytotherapy. 7:36–45.
2005.PubMed/NCBI
|
|
4
|
Huang J, Zhao L, Xing L and Chen D:
MicroRNA-204 regulates Runx2 protein expression and
mesenchymal progenitor cell differentiation. Stem Cells.
28:357–364. 2010.
|
|
5
|
Eskildsen T, Taipaleenmäki H, Stenvang J,
et al: MicroRNA-138 regulates osteogenic differentiation of human
stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA.
108:6139–6144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang JF, Fu WM, He ML, et al: MiR-637
maintains the balance between adipocytes and osteoblasts by
directly targeting Osterix. Mol Biol Cell. 22:3955–3961. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Willimott S and Wagner SD: MiR-125b
and miR-155 contribute to bcl2 repression and proliferation
in response to CD40 ligand (CD154) in human leukemic
B-cells. J Biol Chem. 287:2608–2617. 2012.PubMed/NCBI
|
|
8
|
Scott GK, Goga A, Bhaumik D, et al:
Coordinate suppression of ERBB2 and ERBB3 by enforced expression of
micro-RNA miR-125a or miR-125b. J Biol Chem. 282:1479–1486.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu LH, Li H, Li JP, et al:
miR-125b suppresses the proliferation and migration of
osteosarcoma cells through down-regulation of STAT3. Biochem
Biophys Res Commun. 416:31–38. 2011.
|
|
10
|
Lin KY, Zhang XJ, Feng DD, et al:
miR-125b, a target of CDX2, regulates cell differentiation
through repression of the core binding factor in hematopoietic
malignancies. J Biol Chem. 286:38253–38263. 2011.PubMed/NCBI
|
|
11
|
Manolagas SC and Jilka RL: Bone marrow,
cytokines, and bone remodeling. Emerging insights into the
pathophysiology of osteoporosis. N Engl J Med. 332:305–311. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Verma S, Rajaratnam JH, Denton J, et al:
Adipocytic proportion of bone marrow is inversely related to bone
formation in osteoporosis. J Clin Pathol. 55:693–698. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoshida CA, Yamamoto H, Fujita T, et al:
Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2
regulates limb growth through induction of Indian hedgehog. Genes
Dev. 18:952–963. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakashima K, Zhou X, Kunkel G, et al: The
novel zinc finger-containing transcription factor osterix is
required for osteoblast differentiation and bone formation. Cell.
108:17–29. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pittenger MF, Mackay AM, Beck SC, et al:
Multilineage potential of adult human mesenchymal stem cells.
Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gallagher JC and Sai AJ: Molecular biology
of bone remodeling: implications for new therapeutic targets for
osteoporosis. Maturitas. 65:301–307. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bellantuono I, Aldahmash A and Kassem M:
Aging of marrow stromal (skeletal) stem cells and their
contribution to age-related bone loss. Biochim Biophys Acta.
1792:364–370. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao J, Yang T, Han J, et al: MicroRNA
expression during osteogenic differentiation of human multipotent
mesenchymal stromal cells from bone marrow. J Cell Biochem.
112:1844–1856. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goff LA, Boucher S, Ricupero CL, et al:
Differentiating human multipotent mesenchymal stromal cells
regulate microRNAs: prediction of microRNA regulation by PDGF
during osteogenesis. Exp Hematol. 36:1354–1369. 2008. View Article : Google Scholar
|
|
20
|
Shi L, Zhang J, Pan T, et al:
MiR-125b is critical for the suppression of human U251
glioma stem cell proliferation. Brain Res. 1312:120–126.
2010.PubMed/NCBI
|
|
21
|
Zhang ZJ, Zhang H, Kang Y, et al: miRNA
expression profile during osteogenic differentiation of human
adipose-derived stem cells. J Cell Biochem. 113:888–898.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mizuno Y, Yagi K and Tokuzawa Y:
miR-125b inhibits osteoblastic differentiation by
down-regulation of cell proliferation. Biochem Biophys Res
Commun. 368:267–272. 2008.
|
|
23
|
Goettsch C, Rauner M, Pacyna N, et al:
miR-125b regulates calcification of vascular smooth muscle
cells. Am J Pathol. 179:1594–1600. 2011.PubMed/NCBI
|
|
24
|
Liu F, Akiyama Y, Tai S, et al: Changes in
the expression of CD106, osteogenic genes, and transcription
factors involved in the osteogenic differentiation of human bone
marrow mesenchymal stem cells. J Bone Miner Metab. 26:312–320.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Le MTN, Shyh-Chang N, Khaw SL, et al:
Conserved regulation of p53 network dosage by microRNA-125b
occurs through evolving miRNA-target gene pairs. PLoS Genet.
7:e10022422011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Armesilla-Diaz A, Elvira G and Silva A:
p53 regulates the proliferation, differentiation and spontaneous
transformation of mesenchymal stem cells. Exp Cell Res.
315:3598–3610. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu H and Li B: p53 control of bone
remodeling. J Cell Biochem. 111:529–534. 2010. View Article : Google Scholar
|