Introduction
Lung cancer, including small cell lung cancer and
non-small cell lung cancer, has become the most common cause of
cancer-related mortality worldwide (1). Therefore, understanding the
mechanisms of lung cancer for the development of effective
therapies is of great importance.
microRNAs (miRNAs), a class of noncoding RNAs,
between 19 and 30 nucleotides in length, post-transcriptionally
modulate gene expression by negatively regulating the stability or
translational efficiency of certain mRNAs (2,3).
Accumulating evidence has shown that the miRNAs have critical roles
in numerous processes, including developmental events, cell
differentiation and apoptosis, glucose and lipid metabolism and
cancer initiation and progression (4,5).
Furthermore, a number of recent studies have found that the
dysregulated expression of certain miRNAs is involved in lung
cancer and that this contributes to tumorigenesis (6–8).
miR-143, for example, has been found to be downregulated in lung
cancer tissues and cell lines. It has been demonstrated that, at
the molecular level, miR-143 significantly inhibits the migration
and invasion of lung cancer cells by targeting cluster of
differentiation 44v3 (9).
Therefore, the identification and understanding of the specific
roles of certain miRNAs may provide a novel and promising strategy
for the development of therapeutic treatments against cancer.
In the present study, miR-802 was investigated as a
positive regulator of proliferation in lung cancer cell lines
(A549, NCI-H358 and NCI-H1299). It was hypothesized that miR-143
may regulate proliferation by targeting the tumor suppressor
menin.
Material and methods
Tissue samples
A total of 40 pairs of tumor tissues and adjacent
normal tissues were collected from routine therapeutic surgery at
the Department of Thoracic Surgery, Qingdao Municipal Hospital
(Qingdao, China). All samples were obtained with informed consent
and approved by the Qingdao Municipal Hospital institutional review
board.
Cell culture
Three lung cancer cell lines (A549, NCI-H358 and
NCI-H1299) were obtained from the Chinese Academy of Sciences Cell
Bank (Shanghai, China). Cells were cultured in Dulbecco’s modified
Eagle’s medium (Gibco-BRL, Beijing, China) supplemented with 10%
fetal bovine serum (Gibco-BRL). Cultures were maintained at 37°C in
a humidified atmosphere with 5% CO2.
Analysis of miRNA expression using
TaqMan® quantitative polymerase chain reaction
(qPCR)
Total RNA from tissues or cell lines was harvested
using the miRNA Isolation kit (Ambion®, Austin, TX,
USA). The expression of mature miRNAs was assayed using the TaqMan
miRNA Assay (Applied Biosystems™, Foster City, CA, USA) specific
for hsa-miR-802. Briefly, 5 ng total RNA was reverse transcribed to
cDNA using specific stem-loop reverse transcription primers. qPCR
was performed using an Applied Biosystems 7500 Real-Time PCR System
and a TaqMan Universal PCR Master Mix. All the primers were
obtained from the TaqMan miRNA Assays. U6 small nuclear RNA
(Applied Biosystems) was used as an internal control.
BrdU assays
A cell proliferation ELISA (BrdU kit; Beyotime,
Haimen, China) was used to analyze the incorporation of BrdU during
DNA synthesis in accordance with the manufacturer’s instructions.
All experiments were performed in triplicate. Absorbance was
measured at 450 nm in the Spectra Max 190 ELISA reader (Molecular
Devices, Sunnyvale, CA, USA)
Plasmid construction and
transfection
For the miR-802 expression plasmid, human miR-802
precursor was cloned into pSilencer 4.1 (Ambion). The negative
control plasmid consisted of a scrambled sequence (Ambion). To
inhibit miR-802 function, an Ambion miRNA inhibitor of miR-802 was
used, along with the negative control. For transfection, a complex
of Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA) and 25 nM of the miRNAs mentioned above was prepared in
accordance with the manufacturer’s instructions.
miR-802 targets prediction
The putative targets of miR-802 were predicted using
the miRWalk software (10). The
algorithm produced a list of hundreds of target genes for miR-802
by searching for the presence of conserved heptamer and octamer
sites matching the seed region of miR-802.
Western blot analysis
Cells were harvested and lysed with ice-cold lysis
buffer (50 mM Tris-HCl, pH 6.8, 2 mM 2-mercaptoethanol, 2% w/v SDS
and 10% glycerol). The cells were centrifuged at 20,000 × g for 10
min at 4°C, and the proteins in the supernatant were quantified and
separated using 10% SDS PAGE and transferred to a nitrocellulose
membrane (Amersham Pharmacia Biotech, Amersham, UK). Following
blocking with 10% nonfat milk in phosphate-buffered saline,
membranes were immunoblotted with antibodies as indicated, followed
by horseradish peroxidase-linked secondary antibodies (Cell
Signaling Technology, Inc., Beverly, MA, USA). The signals were
detected using a SuperSignal™ West Pico Chemiluminescent Substrate
kit (Pierce, Rockford, IL, USA) in accordance with manufacturer’s
instructions. Anti-menin, -p18 and -p27 antibodies were purchased
from Abcam (Cambridge, MA, USA). Anti-β-catenin and -p65 antibodies
were purchased from Cell Signaling Technology, Inc. Protein levels
were normalized against GAPDH or Lamin B (Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA).
Luciferase reporter assay
Total cDNA from A549 cells was used to amplify the
3′-untranslated region (UTR) of menin by PCR. The menin 3′-UTR was
cloned into pMIR-REPORT™ (Ambion), resulting in pMIR-REPORT-menin.
Mutations were introduced into potential miR-802 binding sites
using the QuikChange® Site-Directed Mutagenesis kit
(Stratagene, La Jolla, CA, USA). Cells were transfected with the
pMIR-REPORT vectors containing the 3′-UTR variants, the miR-802
precursor and control plasmids for 36 h. The pRL-TK vector (Promega
Corporation, Madison, WI, USA) carrying the Renilla luciferase gene
was used as an internal control to normalize the transfection
efficiency. Luciferase values were determined using the
Dual-Luciferase Reporter Assay system (Promega Corporation).
Flow cytometric analysis
To assess the distribution of nuclear DNA content,
cells were collected, rinsed and fixed overnight in 75% cold
ethanol at −20°C. Subsequently, cells were treated with Tris-HCl
buffer (pH 7.4) supplemented with 100 μg/ml RNase A and stained
with 25 μg/ml propidium iodide (BD Biosciences, San Diego, CA,
USA). Cell cycle distribution was determined by flow cytometry
(Becton Dickinson, San Jose, CA, USA). In total, 10,000 cells were
acquired and analyzed for DNA content. All data were collected,
stored and analyzed by ModFit software (Becton Dickinson).
Statistical analysis
Data are expressed as the mean ± standard error of
the mean from at least three separate experiments. Differences
between groups were analyzed using a Student’s t-test analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-802 expression levels are increased
in lung carcinoma tissues
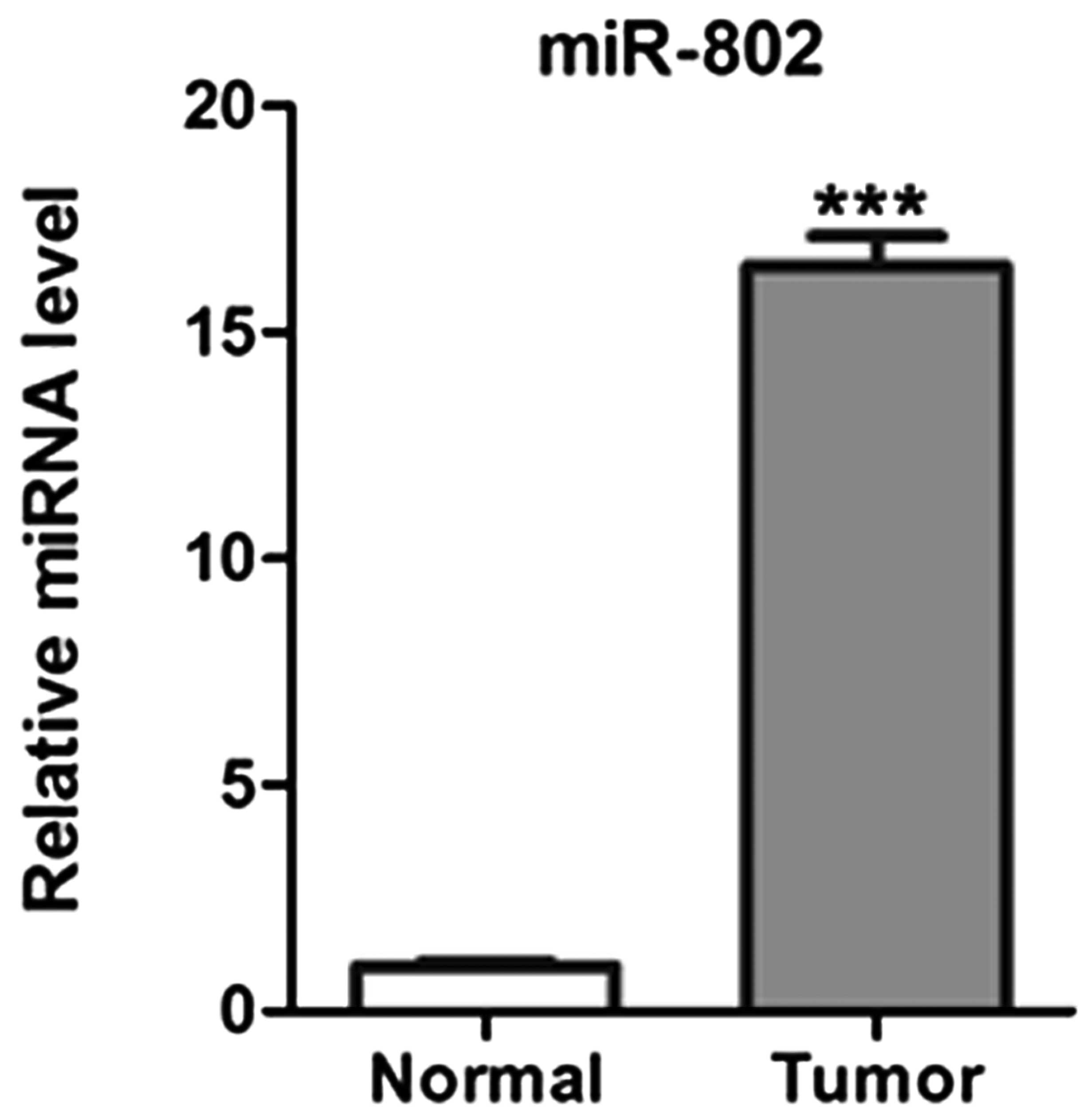
miR-802 expression levels were determined using
TaqMan qPCR in 40 pairs of human lung carcinoma tissues and
adjacent matched noncancerous tissues. As shown in Fig. 1, the expression levels of miR-802
were significantly upregulated in cancer tissues in comparison with
those in adjacent noncancerous tissues.
miR-802 overexpression promotes lung
cancer cell proliferation
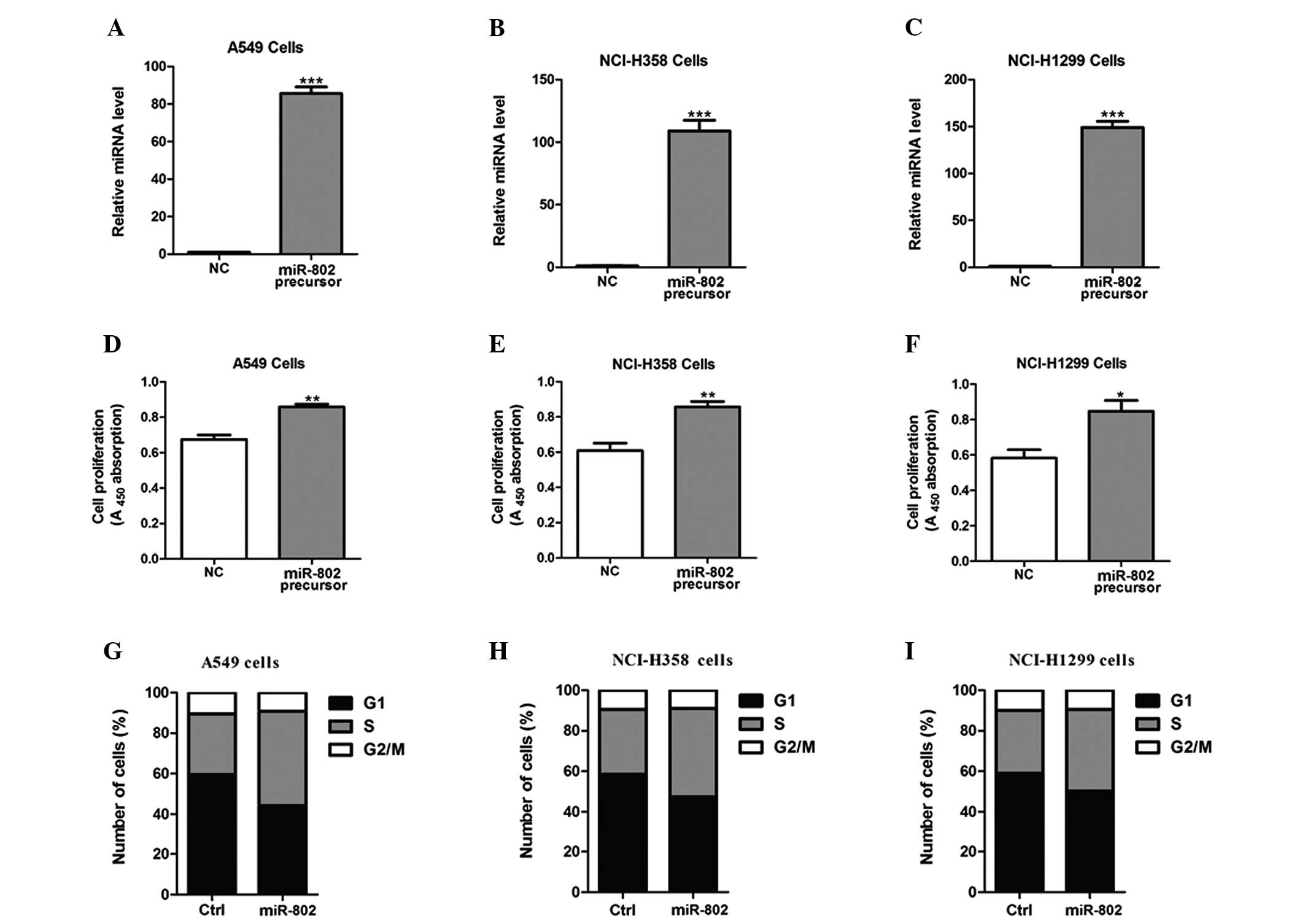
To determine the effects of miR-802 on lung
carcinoma cell growth, miR-802 precursor was transfected into A549,
NCI-H358 and NCI-H1299 cells (Fig.
2A–C). As a result of miR-802 overexpression, the proliferative
ability in these cells post-transfection was significantly enhanced
(Fig. 2D–F). Furthermore, cells
overexpressing miR-802 had a significantly lower percentage of
cells in the G1/G0 phase and increased percentage of cells in the S
phase, compared with negative control-transfected cells (Fig. 2G–I).
miR-802 antisense inhibits the
proliferation of lung cancer cells
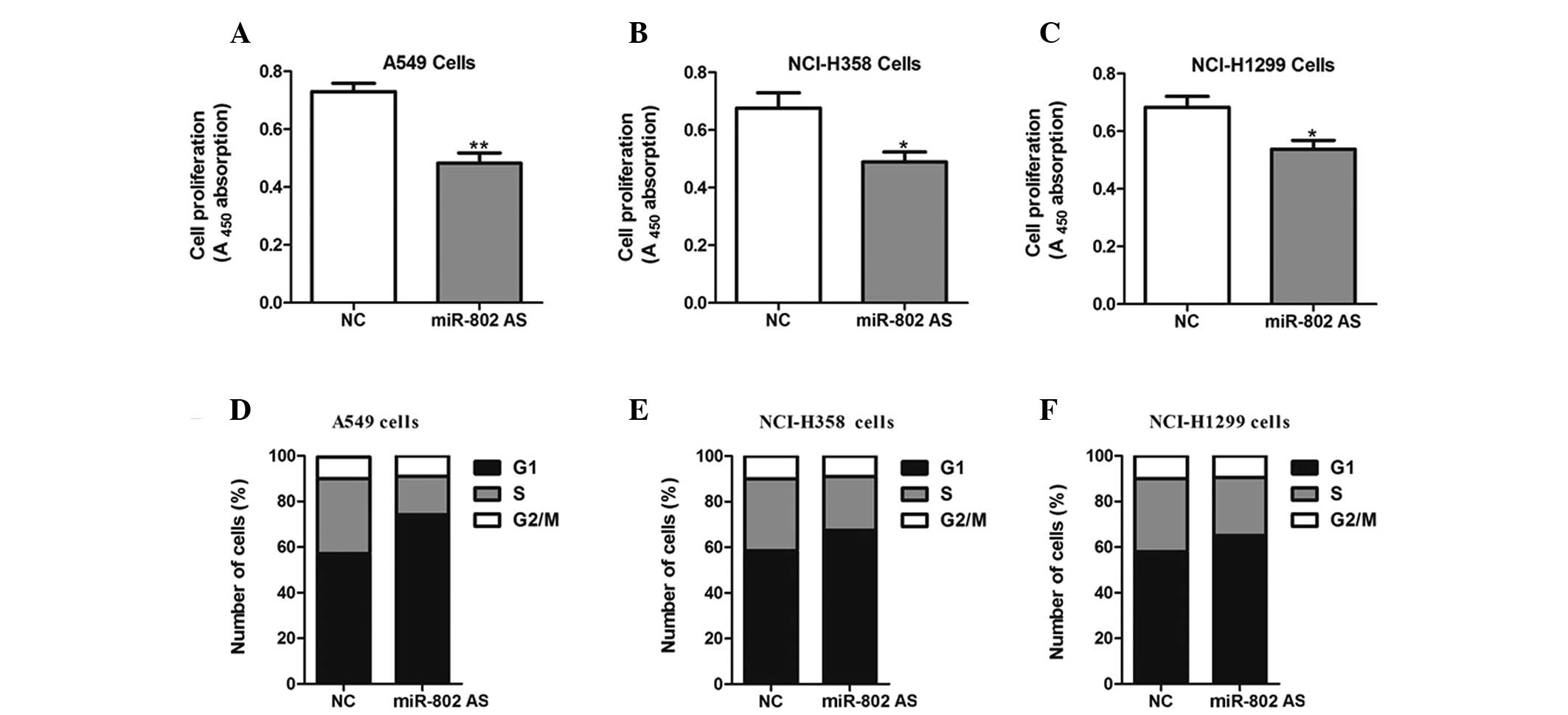
Cells were transfected with miR-802 antisense to
block the functions of endogenous miR-802. As a result of ectopic
expression of the hsa-miR-802 antisense, a reduction in
proliferative ability was observed in A549, NCI-H358 and NCI-H1299
cells compared with negative control-transfected cells (Fig. 3A–C). Additionally, inhibition of
miR-802 significantly increased the percentage of cells in the
G0/G1 phase and decreased the percentage of cells in the S phase
(Fig. 3D–F).
miR-802 targets the tumor suppressor
menin and downregulates its expression
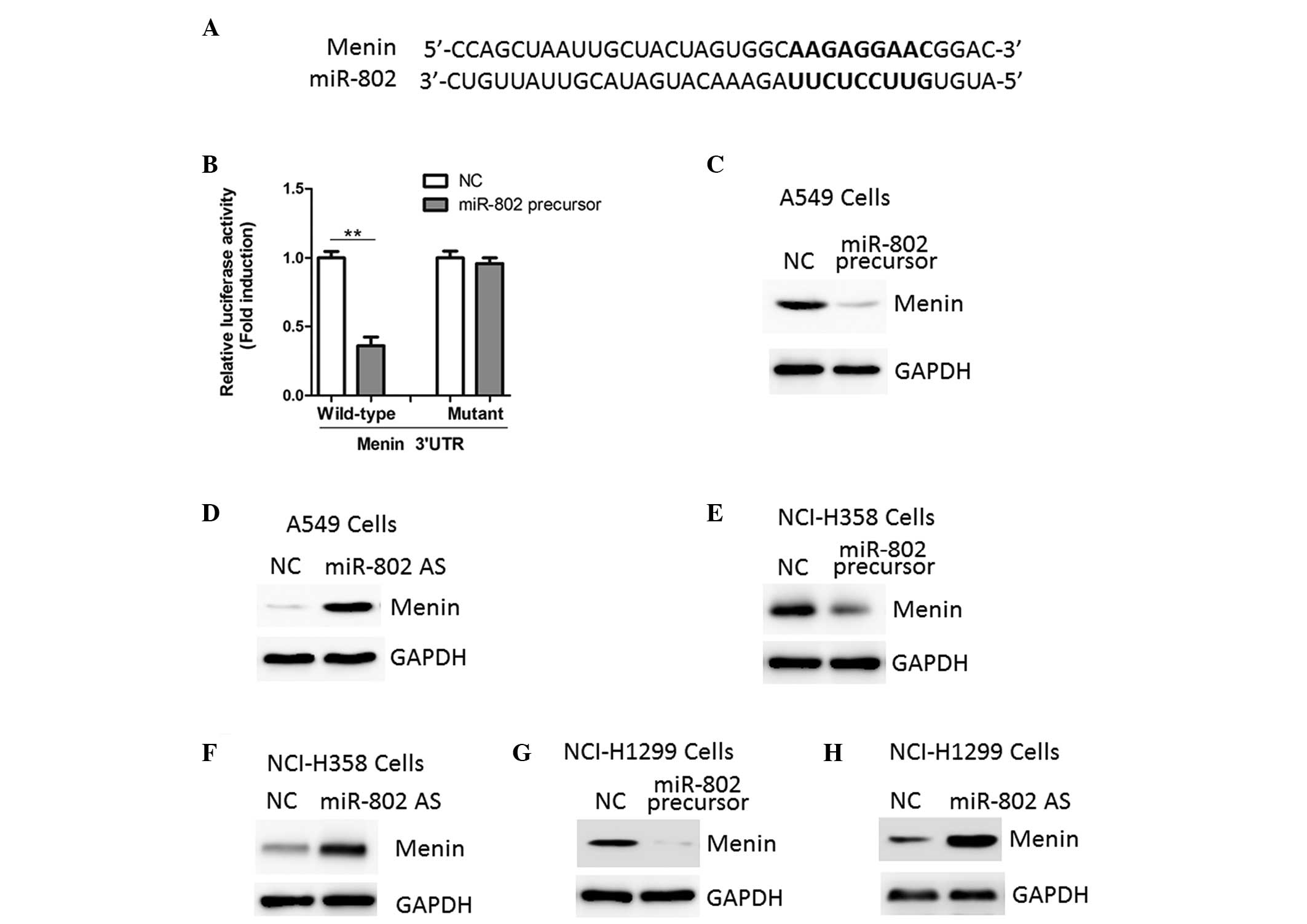
Using a stringent bioinformatics approach (miRWalk
software), several putative human miR-802 target genes were
identified (data not shown), including the tumor suppressor gene
menin (Fig. 4A). To investigate
whether miR-802 was able to directly bind to the 3′-UTR of menin
and repress its expression, luciferase report vectors containing
the putative miR-802 binding sites within the menin 3′-UTR were
constructed. As shown in Fig. 4B,
overexpression of miR-802 led to a reduction in luciferase activity
when the reporter construct contained the menin 3′-UTR in A549
cells. To confirm that menin was a direct target of miR-802,
further luciferase reporter assays were performed with vectors
containing a mutant menin 3′-UTR. The mutation in the miR-802
binding site from the menin 3′-UTR abolished this effect of
miR-802, indicating that miR-802 directly inhibits menin expression
by targeting its 3′-UTR (Fig.
4B).
The protein expression levels of menin showed an
inverse correlation with miR-802 expression in A549 cells. miR-802
precursor inhibited menin expression while miR-802 antisense
increased menin protein expression, as indicated by the results
from the western blot analysis (Fig.
4C and D). Similar results were also observed in NCI-H358 and
NCI-H1299 cells (Fig. 4E–H).
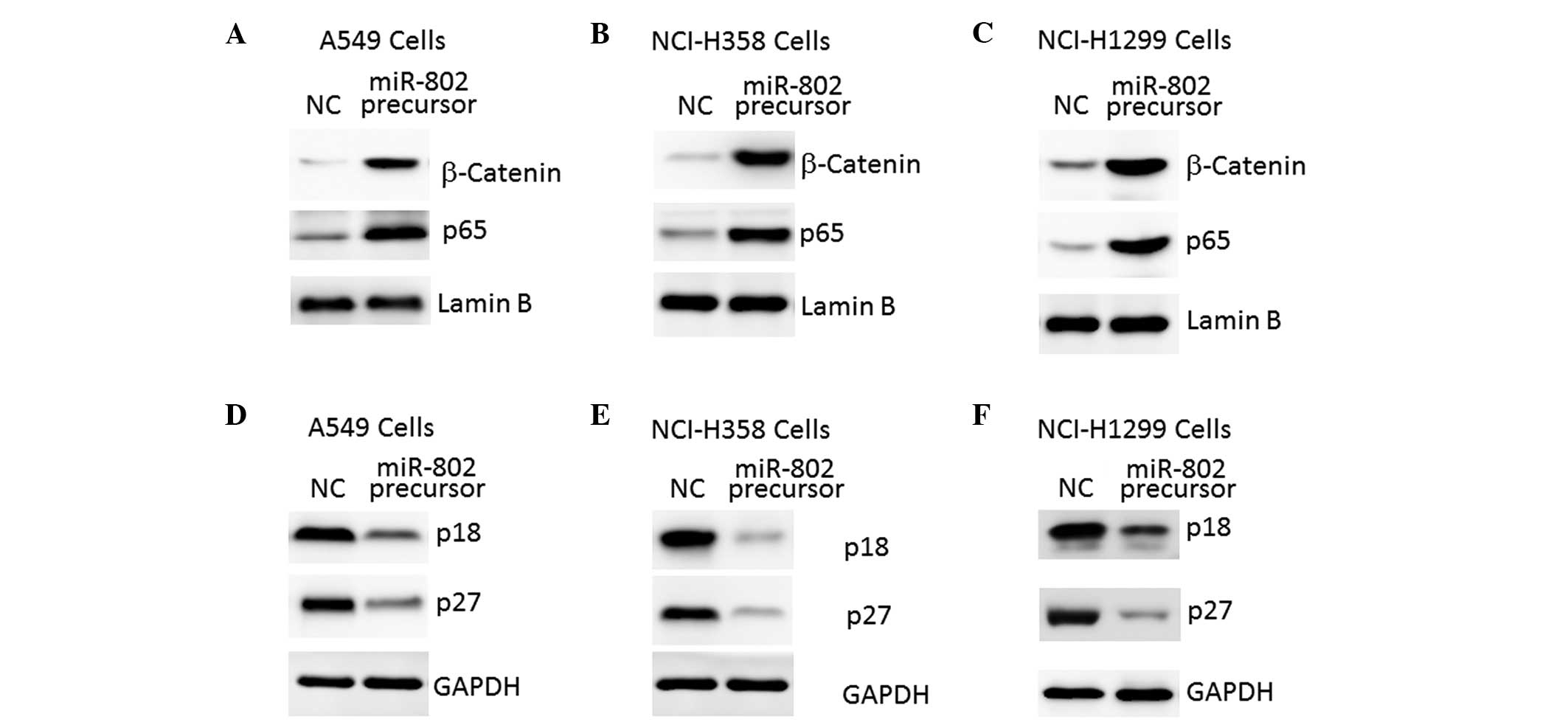
miR-802 regulates downstream signaling of
the tumor suppressor menin
A reduction in menin expression or activity has been
found to lead to increased Wnt/β-catenin and nuclear factor
κ-light-chain-enhancer of activated B cells (NF-κB)/p65 activation
in numerous types of tumor tissues and cells (11,12).
As shown in Fig. 5A and B,
overexpression of miR-802 led to activation of Wnt/β-catenin and
NF-κB/p65 signaling in lung carcinoma cells, as demonstrated by the
increased levels of nuclear β-catenin and p65 protein (Fig. 5A–C).
By contrast, menin has been found to upregulate
expression of p18 and p27, two negative regulators of cell cycle
progression, through recruitment of the histone methyltransferase
protein complex (13). In
accordance with this, reduced expression of p18 and p27 in lung
carcinoma cells overexpressing miR-802 was also observed in the
present study (Fig. 5D–F).
Therefore, these results further indicate that menin is an
important target gene of miR-802 in lung carcinoma cells.
Discussion
In the present study, miR-802 was characterized as
an miRNA that has an important role in the development of lung
carcinoma. A previous study demonstrated that miR-802 modulates the
biological efficacy of angiotensin II (Ang II) in the human
gastrointestinal tract through regulation of the Ang II type 1
receptor expression (14).
Furthermore, Kornfeld et al (15) demonstrated that the deregulated
expression of miR-802 has a critical role in the development of the
obesity-associated impairment of glucose metabolism through
targeting of hepatocyte nuclear factor-1β (14). However, the present study is the
first, to the best of our knowledge, to demonstrate that miR-802 is
expressed in lung cancer tissues, which suggests that miR-802 may
act as an oncogene in lung cancer.
To verify this hypothesis, the functional roles of
miR-802 in three lung cancer cell lines were investigated. Using
cell viability assays and cell cycle analysis it was demonstrated
that selective overexpression of miR-802 enhanced cell
proliferation, whilst inhibition of miR-802 repressed cell
proliferation. Furthermore, using the luciferase reporter system
assay in A549 cells, menin was identified as a novel direct target
of miR-802. In addition, western blot analysis confirmed that menin
and its downstream target genes were regulated by miR-802.
A previous study found that menin may be the
pathogenic gene for insulinoma in patients with multiple endocrine
neoplasia type 1 (16). Menin has
also been identified as a tumor suppressor in other types of
tumors, including pituitary and parathyroid tumors, as well as
adrenocortical and lung carcinoma (17). In addition, gene mutations in menin
have been found in sporadic carcinoid tumors of the lung (18,19).
At the molecular level, menin negatively regulates A549 cell
proliferation and invasion, which is mediated by the growth factor
pleiotrophin and its cell surface receptor, protein tyrosine
phosphatase β/ζ (20,21).
The present study confirms that miR-802 is
upregulated in lung cancer tissues. For the first time, to the best
of our knowledge, miR-802 has been demonstrated to significantly
promote the proliferation of lung cancer cells. Menin was also
identified as a novel target gene involved in miR-802-mediated cell
proliferation in lung cancer. These results indicate that miR-802
may have an important role in lung cancer, and suggest that
inhibition of miR-802 may be a potential therapeutic strategy for
the treatment of lung cancer.
References
|
1
|
Keith RL and Miller YE: Lung cancer
chemoprevention: current status and future prospects. Nat Rev Clin
Oncol. 10:334–343. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nat Rev Cancer. 11:644–656. 2011.PubMed/NCBI
|
|
5
|
Kasinski AL and Slack FJ: Epigenetics and
genetics. MicroRNAs en route to the clinic: progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar
|
|
6
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar
|
|
7
|
Sittka A and Schmeck B: MicroRNAs in the
lung. Adv Exp Med Biol. 774:121–134. 2013. View Article : Google Scholar
|
|
8
|
Redova M, Sana J and Slaby O: Circulating
miRNAs as new blood-based biomarkers for solid cancers. Future
Oncol. 9:387–402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma Q, Jiang Q, Pu Q, Zhang X, Yang W, et
al: MicroRNA-143 inhibits migration and invasion of human
non-small-cell lung cancer and its relative mechanism. Int J Biol
Sci. 9:680–692. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: prediction of possible miRNA binding sites by
‘walking’ the genes of 3 genomes. J Biomed Inform. 44:839–847.
2011.
|
|
11
|
Seigne C, Auret M, Treilleux I, Bonnavion
R, Assade F, et al: High incidence of mammary intraepithelial
neoplasia development in Men1-disrupted murine mammary glands. J
Pathol. 229:546–558. 2013. View Article : Google Scholar
|
|
12
|
Gang D, Hongwei H, Hedai L, Ming Z, Qian H
and Zhijun L: The tumor suppressor protein menin inhibits
NF-κB-mediated transactivation through recruitment of Sirt1 in
hepatocellular carcinoma. Mol Biol Rep. 40:2461–2466. 2013.
|
|
13
|
Veniaminova NA, Hayes MM, Varney JM and
Merchant JL: Conditional deletion of menin results in antral G cell
hyperplasia and hypergastrinemia. Am J Physiol Gastrointest Liver
Physiol. 303:G752–G764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sansom SE, Nuovo GJ, Martin MM, Kotha SR,
Parinandi NL and Elton TS: miR-802 regulates human angiotensin II
type 1 receptor expression in intestinal epithelial C2BBe1 cells.
Am J Physiol Gastrointest Liver Physiol. 299:G632–G642. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kornfeld JW, Baitzel C, Könner AC,
Nicholls HT, Vogt MC, et al: Obesity-induced overexpression of
miR-802 impairs glucose metabolism through silencing of Hnf1b.
Nature. 494:111–115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gaztambide S, Vazquez F and Castaño L:
Diagnosis and treatment of multiple endocrine neoplasia type 1
(MEN1). Minerva Endocrinol. 38:17–28. 2013.PubMed/NCBI
|
|
17
|
Matkar S, Thiel A and Hua X: Menin: a
scaffold protein that controls gene expression and cell signaling.
Trends Biochem Sci. 38:394–402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Debelenko LV, Brambilla E, Agarwal SK,
Swalwell JI, Kester MB, et al: Identification of MEN1 gene
mutations in sporadic carcinoid tumors of the lung. Hum Mol Genet.
6:2285–2290. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Haruki N, Yatabe Y, Travis WD, Nomoto S,
Osada H, et al: Characterization of high-grade neuroendocrine
tumors of the lung in relation to menin mutations. Jpn J Cancer
Res. 91:317–323. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao SB, Feng ZJ, Xu B, Wu Y, Yin P, et al:
Suppression of lung adenocarcinoma through menin and polycomb
gene-mediated repression of growth factor pleiotrophin. Oncogene.
28:4095–4104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feng ZJ, Gao SB, Wu Y, Xu XF, Hua X and
Jin GH: Lung cancer cell migration is regulated via repressing
growth factor PTN/RPTP β/ζ signaling by menin. Oncogene.
29:5416–5426. 2010.PubMed/NCBI
|