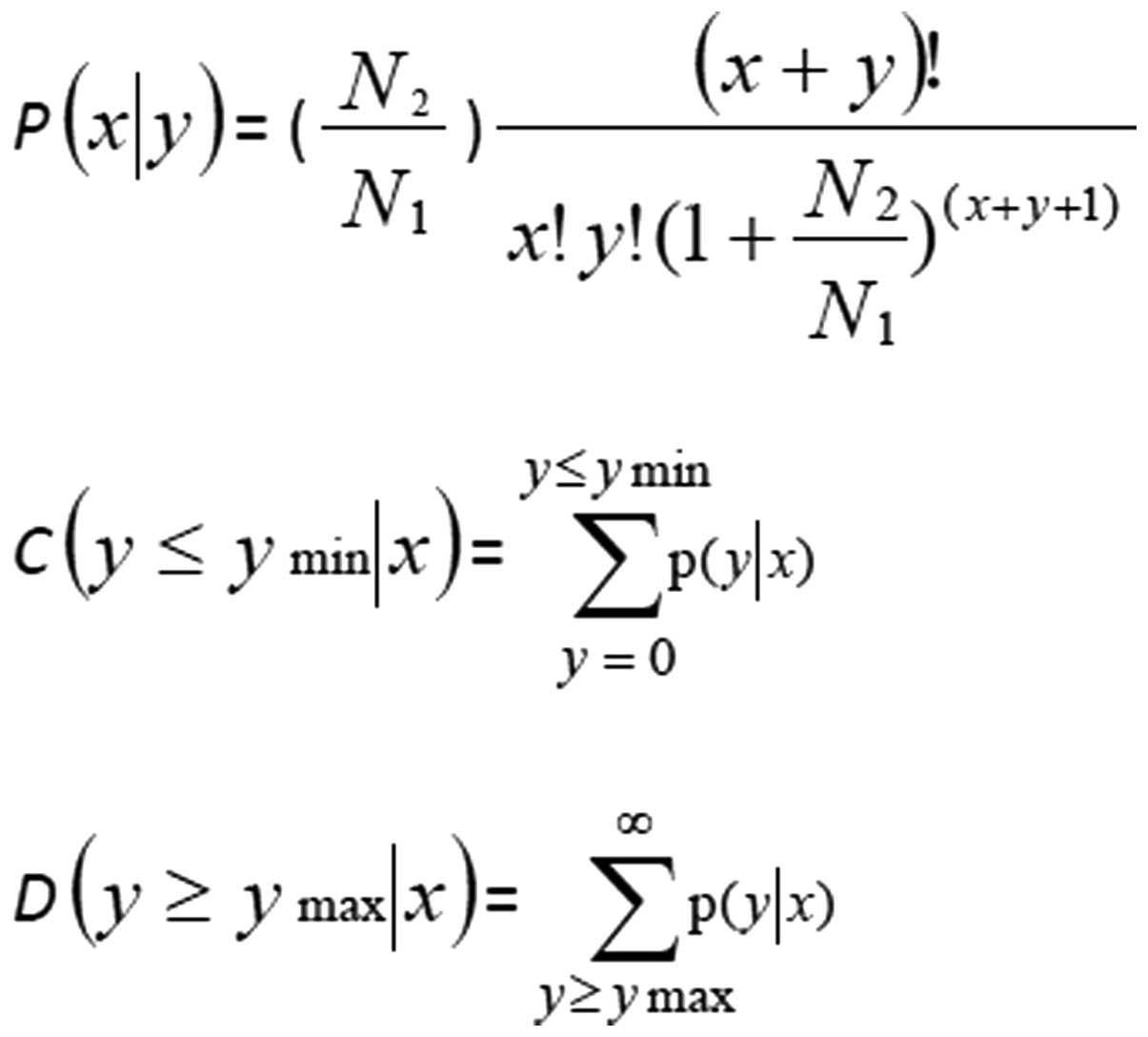Introduction
MicroRNAs (miRNAs) are small non-coding RNA
molecules, ~22 nucleotides long, which are widely distributed in
plants and animals (1,2). The primary miRNA transcripts are
cleaved by Drosha and Dicer enzymes to form mature miRNAs, which
serve as posttranscriptional negative gene regulators by either
cleaving mRNA or inhibiting translation (3,4).
Through such mechanisms, miRNAs are involved in various biological
processes, including organ development, tissue differentiation,
cell cycle regulation and cancer development (4–6).
Lung cancer is the predominant cause of
cancer-related mortality worldwide and its pathogenesis is closely
associated with tobacco smoking. Lung cancer is categorized into
two main histological groups: Non-small cell lung cancer (NSCLC)
and small cell lung cancer (SCLC). Regulation of miRNAs is closely
associated with tumor initiation, promotion and progression
(7). Recently, increasing evidence
has demonstrated that miRNAs may be associated with lung cancer. A
previous study revealed that an increase in hsa-miR-196a expression
levels was a characteristic molecular change in NSCLC (8,9), and
that hsa-miRNA-196a promoted NSCLC cell proliferation and invasion
via targeting HOXA5 (10).
Overexpression of miR-200 was shown to reduce the expression levels
of NSCLC prognostic biomarkers in H1299 and BEAS-2B cells (11). miR-449a, which inhibited migration
and invasion through targeting c-Met, was found to be
downregulated in NSCLC tissues and cell lines (12). Reduced expression of miR-101 was
associated with overexpression of EZH2 and exhibited
tumor-suppressive functions in NSCLC (13). This evidence suggests that miRNAs
are crucial in NSCLC; however, little is known regarding the
functions and regulation of miRNAs in SCLC.
In the present study, Solexa sequencing was used to
generate a large quantity of small RNA (sRNA) data for SCLC and
corresponding normal tissues. Sequence analysis was employed to
identify specific miRNAs associated with SCLC and these miRNAs were
validated by reverse transcription-quantitative polymerase chain
reaction (RT-qPCR).
Materials and methods
Samples and total RNA isolation
The experimental specimens were obtained with
informed consent between 2011 and 2012 from six patients at similar
stages of SCLC. The SCLC tissues and corresponding normal tissues
from healthy subjects were collected at the time of surgery and
prior to chemotherapy at the Second Affiliated Hospital of Harbin
Medical University (Harbin, China). This study was approved by the
Hospitals’ Ethical Review Committee. The isolated SCLC and
corresponding normal tissues were harvested in liquid nitrogen and
stored at −80°C. The total RNA was extracted from the SCLC tissues
and controls using Trizol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA).
sRNA library construction
Two sRNA libraries were constructed from the SCLC
tissues (LC sRNA library) and the corresponding normal tissues (NT
sRNA library) using methods described previously (14). Briefly, total RNA was purified by
electrophoretic separation on a 15% Tris/Borate/EDTA urea
denaturing PAGE gel (Sangon Biotech Co., Ltd., Shanghai, China) and
sRNA molecules in the range of 15–30 nucleotides (ligated to
adapters at the end of 5′ and 3′ sRNAs using T4 RNA ligase) were
enriched (Takara Biotechnology Co., Ltd., Dalian, China). The
adapter-ligated sRNAs were subsequently transcribed to
complementary DNA (cDNA) by Super-Script II Reverse Transcriptase
(Invitrogen Life Technologies) and then amplified by PCR. The PCR
products were purified and recovered, then subjected to deep
sequencing using an Illumina-Solexa 1G Genetic Analyzer (Illumina,
Inc., San Diego, CA, USA) at the Beijing Genomics Institute
(Shenzhen, China).
Differential expression analysis of
miRNAs involved in SCLC
The raw readings generated by deep sequencing were
analyzed through bioinformatics. These sequences were mapped to the
human genome using the short oligonucleotide alignment program
(15) with a perfect match.
Sequences matching ribosomal (r)RNAs, small cytoplasmic (sc)RNAs,
small nucleolar (sno)RNAs, snRNAs and transfer (t)RNAs in the NCBI
GenBank (http://www.ncbi.nih.gov/GenBank/) database and Rfam
(http://rfam.sanger.ac.uk/) database were
discarded. The conserved miRNAs were predicted by aligning to
miRBase 20.0 (http://www.mirbase.org/index.shtml); then the 187
conserved miRNAs (data not shown) were annotated by matching with
miRBase 20.0. TargetScan (http://www.targetscan.org) was used for annotation of
the targets of conserved miRNAs.
Differential expression levels of miRNAs were
analyzed as determined by the sequence readings of LC and NT sRNA
libraries. The abundance of miRNAs was normalized to one million
(normalized expression = actual miRNA count/total count of clean
readings × 1,000,000). The ratio between the two sRNA libraries was
calculated as follows: Ratio = miRNA normalized readings in LC
library/miRNA normalized readings in NT library. The P-values were
calculated according to the equation in Fig. 1. When the ratio was >1.5-fold or
<0.67-fold, and P<0.05, the miRNAs were considered to be up-
or downregulated respectively in SCLC.
Validation of differential expression
levels of miRNAs by RT-qPCR
sRNAs (<200 nt) were isolated from the SCLC
tissues (LC sRNA library) and the normal tissues using a mirVana
miRNA Isolation kit (Ambion, Carlsbad, CA, USA). The tissues used
for validation were the same as tissues used for the deep
sequencing. The sRNAs were polyadenylated by poly(A) polymerase
using the Poly(A) Tailing kit (Ambion) and then cDNA was
synthesized with a Quant reverse kit (Tiangen, Beijing, China)
(15). RT-qPCR was performed using
an ABI 7500 Fast Real-time PCR machine (Applied Biosystems, Foster
City, CA, USA) using an SYBR Premix Ex TaqTM kit (Takara Bio, Inc.,
Shiga, Japan) in a 20-μl reaction volume, containing 2 μl diluted
cDNA, 200 nM of each primer and 1× PCR Master mix. The
amplification conditions were provided by Takara Bio Inc.. The
expression levels of miRNAs were normalized to those of the U6
sRNA. Three replicates were analyzed for each sample. The primers
for the validated miRNAs are shown in Table I.
 | Table IPrimers used for reverse
transcription-polymerase chain reaction analysis. |
Table I
Primers used for reverse
transcription-polymerase chain reaction analysis.
| Member | microRNA sequence
(5′-3′) |
|---|
| hsa-miR-21-5p |
TAGCTTATCAGACTGATGTTGA |
| hsa-miR-22-3p |
AAGCTGCCAGTTGAAGAACTGT |
| hsa-miR-29a-3p |
TAGCACCATCTGAAATCGGTTA |
| hsa-miR-29a-5p |
ACTGATTTCTTTTGGTGTTCAG |
| hsa-miR-30a-3p |
CTTTCAGTCGGATGTTTGCAGC |
| hsa-miR-30a-5p |
TGTAAACATCCTCGACTGGAAG |
| hsa-miR-34a-3p |
CAATCAGCAAGTATACTGCCCT |
| hsa-miR-34a-5p |
TGGCAGTGTCTTAGCTGGTTGT |
| hsa-miR-99a-3p |
CAAGCTCGCTTCTATGGGTCTG |
| hsa-miR-99a-5p |
AACCCGTAGATCCGATCTTGTG |
| hsa-miR-100-3p |
CAAGCTTGTATCTATAGGTATG |
| hsa-miR-100-5p |
AACCCGTAGATCCGAACTTGTG |
| hsa-miR-101-5p |
CAGTTATCACAGTGCTGATGCT |
| hsa-miR-126-5p |
CATTATTACTTTTGGTACGCG |
| hsa-miR-134-3p |
CCTGTGGGCCACCTAGTCACCAA |
| hsa-miR-139-3p |
TGGAGACGCGGCCCTGTTGGAGT |
| hsa-miR-141-5p |
CATCTTCCAGTACAGTGTTGGA |
| hsa-miR-143-3p |
TGAGATGAAGCACTGTAGCTC |
| hsa-miR-145-5p |
GTCCAGTTTTCCCAGGAATCCCT |
| hsa-miR-148a-3p |
TCAGTGCACTACAGAACTTTGT |
| hsa-miR-152-5p |
AGGTTCTGTGATACACTCCGACT |
| hsa-miR-182-5p |
TTTGGCAATGGTAGAACTCACACT |
| hsa-miR-183-3p |
GTGAATTACCGAAGGGCCATAA |
| hsa-miR-185-5p |
TGGAGAGAAAGGCAGTTCCTGA |
| hsa-miR-192-3p |
CTGCCAATTCCATAGGTCACAG |
| hsa-miR-195-3p |
CCAATATTGGCTGTGCTGCTCC |
| hsa-miR-205-3p |
GATTTCAGTGGAGTGAAGTTC |
| hsa-miR-205-5p |
TCCTTCATTCCACCGGAGTCTG |
| hsa-miR-218-5p |
TTGTGCTTGATCTAACCATGT |
| hsa-miR-221-5p |
ACCTGGCATACAATGTAGATTT |
| hsa-miR-223-3p |
TGTCAGTTTGTCAAATACCCCA |
| hsa-miR-224-3p |
AAAATGGTGCCCTAGTGACTACA |
| hsa-miR-498 |
TTTCAAGCCAGGGGGCGTTTTTC |
| hsa-miR-557 |
GTTTGCACGGGTGGGCCTTGTCT |
| hsa-miR-623 |
ATCCCTTGCAGGGGCTGTTGGGT |
|
hsa-miR-1233-5p |
AGTGGGAGGCCAGGGCACGGCA |
| Poly(T)
adapter |
GCGAGCACAGAATTAATACGACTCACTATAGG(T)12VNa |
| Reverse primer |
GCGAGCACAGAATTAATACGAC |
| U6 small RNA |
GCAGGGGCCATGCTAATCTTCTCTGTATCG |
Statistical analysis
The correlation analysis between the miRNA
expression profile was conducted using bivariate correlation in
SPSS software (SPSS, Inc., Chicago, IL, USA). P-values were used to
reflect the significance of miRNA differential expression between
these two sRNA libraries. P<0.05 indicated a statistically
significant difference.
Results
Analysis of sRNAs
To identify the responsive miRNAs in the SCLC
patients, two sRNA libraries were constructed from the SCLC tissues
(LC sRNA library) and the normal tissues (NT sRNA library),
generating 25.1 million (LC) and 24.6 million (NT) raw readings.
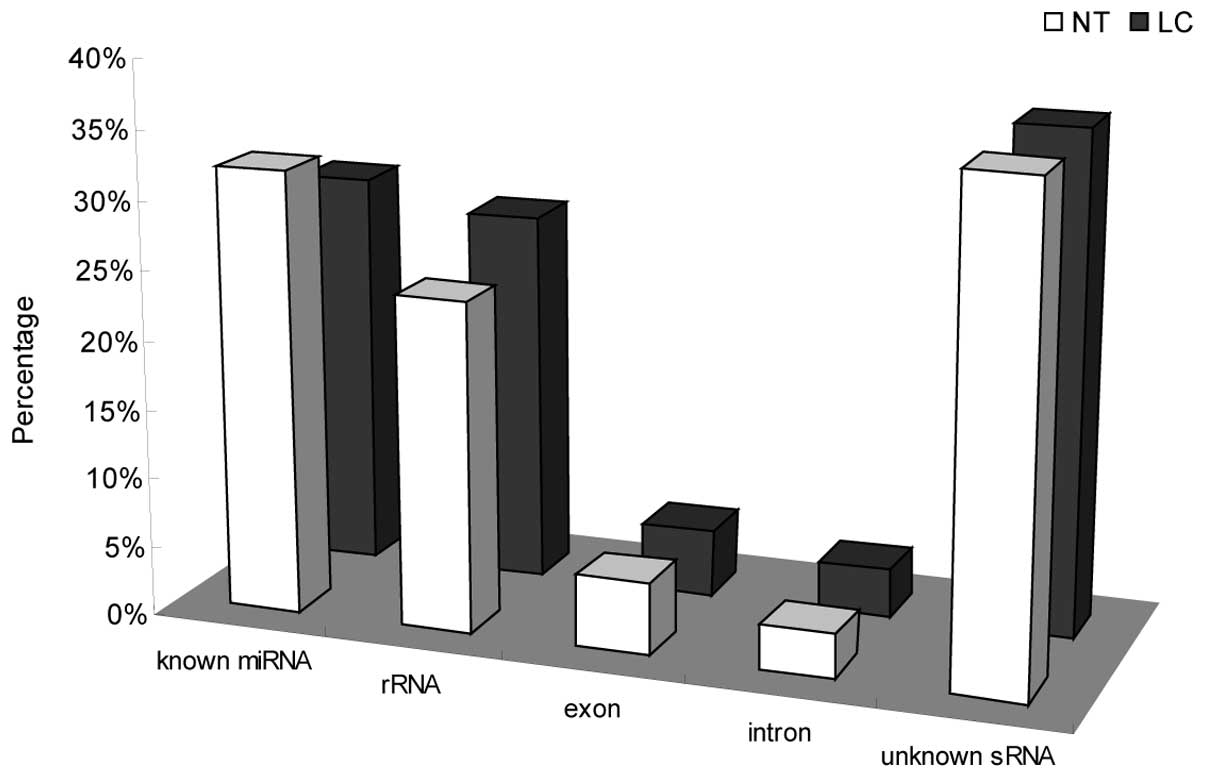
These sRNAs were classified into five categories: rRNAetc (rRNA,
scRNA, snoRNA, snRNA and tRNA), known miRNAs (miRNAs in miRBase
20.0), exon, intron and unknown sRNA (Fig. 2). The results revealed that the
proportion of total known miRNA readings decreased from 31.96% (NT)
to 28.63% (LC) in response to SCLC, suggesting that the known
miRNAs may be important in SCLC. However, the percentage of total
rRNAetc readings increased from 23.87% (NT) to 26.86% (LC),
implying that rRNAetc may exert certain functions in SCLC. Little
difference was identified in the percentages of introns and unknown
sRNAs between SCLC and normal tissue.
Identification of miRNAs in SCLC
Those miRNAs with a ratio of >1.5-fold or
<0.67-fold between the two libraries, and P<0.05 (Table II), were regarded as
differentially expressed miRNAs in SCLC. A total of 81
differentially expressed miRNAs were identified. The abundance of
these miRNAs varied between 4 and 4,343 readings. Of these, 18
corresponding miRNA*s were identified, including hsa-miR-29a-3p and
hsa-miR-29a-5p, and hsa-miR-30a-3p and hsa-miR-30a-5p (miRNA* is
another mRNA that derives from the same hairpin structure as that
of the original miRNA, and is complementary to the original miRNA).
Notably, the expression levels of these 18 miRNAs were found to be
associated with the corresponding miRNA*s, which exhibited similar
expression patterns. For example, hsa-miR-146a-3p was downregulated
and hsa-miR-146a-5p was also downregulated, implying that the
expression of miRNA*s followed the corresponding miRNAs. A total of
34 miRNAs were upregulated in SCLC and the remainder were
downregulated. The most notable changes occurred in
hsa-miR-520c-3p, which had the greatest reduction in expression
levels in SCLC with a ratio of 0.12 (LC/NT), and hsa-miR-148b-3p,
which exhibited the greatest increase with 7.39-fold more readings
sequenced in LC than in NT. These marked alterations in expression
levels, suggest that miRNAs are crucial in the response to
SCLC.
 | Table IIDifferentially expressed miRNAs in
small cell lung cancer. |
Table II
Differentially expressed miRNAs in
small cell lung cancer.
| Member | miRNA sequence
(5′-3′) | miRNA readings | Normalized
readings | Ratio LC/NT | Regulation | P-value |
|---|
|
|
|---|
| NT | LC | NT | LC |
|---|
| hsa-miR-7-5p |
UGGAAGACUAGUGAUUUUGUUGU | 323 | 653 | 13.527 | 29.373 | 2.17 | Upregulated | 0.00 |
| hsa-miR-21-5p |
UAGCUUAUCAGACUGAUGUUGA | 432 | 912 | 18.091 | 41.023 | 2.27 | Upregulated | 0.00 |
| hsa-miR-22-3p |
AAGCUGCCAGUUGAAGAACUGU | 1098 | 3312 | 45.982 | 148.979 | 3.24 | Upregulated | 0.00 |
|
hsa-miR-24-1-5p |
UGCCUACUGAGCUGAUAUCAGU | 334 | 766 | 13.987 | 34.456 | 2.46 | Upregulated | 0.00 |
|
hsa-miR-24-2-5p |
UGCCUACUGAGCUGAAACACAG | 121 | 312 | 5.067 | 14.034 | 2.77 | Upregulated | 0.00 |
| hsa-miR-24-3p |
UGGCUCAGUUCAGCAGGAACAG | 32 | 61 | 1.340 | 2.744 | 2.05 | Upregulated | 0.01 |
| has-miR-25-5p |
AGGCGGAGACUUGGGCAAUUG | 532 | 212 | 22.279 | 9.536 | 0.43 | Downregulated | 0.00 |
| has-miR-27a-3p |
UUCACAGUGGCUAAGUUCCGC | 763 | 232 | 31.953 | 10.436 | 0.33 | Downregulated | 0.00 |
| has-miR-27b-5p |
AGAGCUUAGCUGAUUGGUGAAC | 432 | 112 | 18.091 | 5.038 | 0.28 | Downregulated | 0.00 |
| hsa-miR-29a-3p |
UAGCACCAUCUGAAAUCGGUUA | 45 | 21 | 1.885 | 0.945 | 0.50 | Downregulated | 0.00 |
| hsa-miR-29a-5p |
ACUGAUUUCUUUUGGUGUUCAG | 344 | 121 | 14.406 | 5.443 | 0.38 | Downregulated | 0.00 |
|
hsa-miR-29b-1-5p |
GCUGGUUUCAUAUGGUGGUUUAGA | 321 | 182 | 13.443 | 8.187 | 0.61 | Downregulated | 0.00 |
|
hsa-miR-29b-2-5p |
CUGGUUUCACAUGGUGGCUUAG | 55 | 22 | 2.303 | 0.990 | 0.43 | Downregulated | 0.00 |
| hsa-miR-29c-3p |
UAGCACCAUUUGAAAUCGGUUA | 223 | 432 | 9.339 | 19.432 | 2.08 | Upregulated | 0.00 |
| hsa-miR-29c-5p |
UAGCACCAUUUGAAAUCAGUGUU | 231 | 421 | 9.674 | 18.937 | 1.96 | Upregulated | 0.00 |
| hsa-miR-30a-3p |
CUUUCAGUCGGAUGUUUGCAGC | 4343 | 788 | 181.877 | 35.446 | 0.19 | Downregulated | 0.00 |
| hsa-miR-30a-5p |
UGUAAACAUCCUCGACUGGAAG | 30 | 11 | 1.256 | 0.495 | 0.39 | Downregulated | 0.00 |
| hsa-miR-34a-3p |
CAAUCAGCAAGUAUACUGCCCU | 456 | 893 | 19.096 | 40.169 | 2.10 | Upregulated | 0.00 |
| hsa-miR-34a-5p |
UGGCAGUGUCUUAGCUGGUUGU | 21 | 32 | 0.879 | 1.439 | 1.64 | Upregulated | 0.00 |
| hsa-miR-34b-3p |
CAAUCACUAACUCCACUGCCAU | 321 | 983 | 13.443 | 44.217 | 3.29 | Upregulated | 0.00 |
| hsa-miR-34b-5p |
UAGGCAGUGUCAUUAGCUGAUUG | 11 | 32 | 0.461 | 1.439 | 3.12 | Upregulated | 0.00 |
| hsa-miR-34c-5p |
AGGCAGUGUAGUUAGCUGAUUGC | 1,110 | 432 | 46.485 | 19.432 | 0.42 | Downregulated | 0.00 |
| hsa-miR-96-3p |
AAUCAUGUGCAGUGCCAAUAUG | 121 | 433 | 5.067 | 19.477 | 3.84 | Upregulated | 0.00 |
| hsa-miR-96-5p |
UUUGGCACUAGCACAUUUUUGCU | 11 | 23 | 0.461 | 1.035 | 2.25 | Upregulated | 0.00 |
| hsa-miR-99a-3p |
CAAGCUCGCUUCUAUGGGUCUG | 22 | 9 | 0.921 | 0.405 | 0.44 | Downregulated | 0.00 |
| hsa-miR-99a-5p |
AACCCGUAGAUCCGAUCUUGUG | 454 | 233 | 19.013 | 10.481 | 0.55 | Downregulated | 0.00 |
| hsa-miR-100-3p |
CAAGCUUGUAUCUAUAGGUAUG | 89 | 45 | 3.727 | 2.024 | 0.54 | Downregulated | 0.00 |
| hsa-miR-100-5p |
AACCCGUAGAUCCGAACUUGUG | 21 | 10 | 0.879 | 0.450 | 0.51 | Downregulated | 0.02 |
| hsa-miR-101-5p |
CAGUUAUCACAGUGCUGAUGCU | 89 | 34 | 3.727 | 1.529 | 0.41 | Downregulated | 0.00 |
| hsa-miR-103-3p |
AGCAGCAUUGUACAGGGCUAUGA | 10 | 4 | 0.419 | 0.180 | 0.43 | Downregulated | 0.00 |
| hsa-miR-103-5p |
AGCUUCUUUACAGUGCUGCCUUG | 59 | 23 | 2.471 | 1.035 | 0.42 | Downregulated | 0.00 |
|
hsa-miR-106a-3p |
CUGCAAUGUAAGCACUUCUUAC | 123 | 321 | 5.151 | 14.439 | 2.80 | Upregulated | 0.00 |
|
hsa-miR-106a-5p |
AAAAGUGCUUACAGUGCAGGUAG | 41 | 12 | 1.717 | 0.540 | 0.31 | Downregulated | 0.00 |
|
hsa-miR-106b-3p |
CCGCACUGUGGGUACUUGCUGC | 212 | 32 | 8.878 | 1.439 | 0.16 | Downregulated | 0.00 |
|
hsa-miR-106b-5p |
UAAAGUGCUGACAGUGCAGAU | 213 | 123 | 8.920 | 5.533 | 0.62 | Downregulated | 0.00 |
|
hsa-miR-125a-3p |
ACAGGUGAGGUUCUUGGGAGCC | 432 | 232 | 18.091 | 10.436 | 0.58 | Downregulated | 0.00 |
| hsa-miR-125b |
UCACAAGUCAGGCUCUUGGGAC | 238 | 87 | 9.967 | 3.913 | 0.39 | Downregulated | 0.00 |
| hsa-miR-126-5p |
CAUUAUUACUUUUGGUACGCG | 344 | 119 | 14.406 | 5.353 | 0.37 | Downregulated | 0.00 |
| hsa-miR-134-3p |
CCUGUGGGCCACCUAGUCACCAA | 78 | 23 | 3.267 | 1.035 | 0.32 | Downregulated | 0.00 |
| hsa-miR-139-3p |
UGGAGACGCGGCCCUGUUGGAGU | 347 | 126 | 14.532 | 5.668 | 0.39 | Downregulated | 0.00 |
| hsa-miR-141-5p |
CAUCUUCCAGUACAGUGUUGGA | 812 | 421 | 34.005 | 18.937 | 0.56 | Downregulated | 0.00 |
| hsa-miR-143-3p |
UGAGAUGAAGCACUGUAGCUC | 238 | 434 | 9.967 | 19.522 | 1.96 | Upregulated | 0.00 |
| hsa-miR-145-5p |
GUCCAGUUUUCCCAGGAAUCCCU | 150 | 61 | 6.282 | 2.744 | 0.44 | Downregulated | 0.00 |
|
hsa-miR-146a-3p |
CCUCUGAAAUUCAGUUCUUCAG | 312 | 123 | 13.066 | 5.533 | 0.42 | Downregulated | 0.00 |
|
hsa-miR-146a-5p |
UGAGAACUGAAUUCCAUGGGUU | 58 | 11 | 2.429 | 0.495 | 0.20 | Downregulated | 0.00 |
|
hsa-miR-146b-3p |
UGCCCUGUGGACUCAGUUCUGG | 783 | 123 | 32.791 | 5.533 | 0.17 | Downregulated | 0.00 |
|
hsa-miR-146b-5p |
UGAGAACUGAAUUCCAUAGGCU | 328 | 138 | 13.736 | 6.207 | 0.45 | Downregulated | 0.00 |
|
hsa-miR-148a-3p |
UCAGUGCACUACAGAACUUUGU | 139 | 348 | 5.821 | 15.654 | 2.69 | Upregulated | 0.00 |
|
hsa-miR-148b-3p |
UCAGUGCAUCACAGAACUUUGU | 128 | 881 | 5.360 | 39.629 | 7.39 | Upregulated | 0.00 |
| hsa-miR-152-5p |
AGGUUCUGUGAUACACUCCGACU | 82 | 198 | 3.434 | 8.906 | 2.59 | Upregulated | 0.00 |
| hsa-miR-182-5p |
UUUGGCAAUGGUAGAACUCACACU | 873 | 323 | 36.560 | 14.529 | 0.40 | Downregulated | 0.00 |
| hsa-miR-183-3p |
GUGAAUUACCGAAGGGCCAUAA | 237 | 874 | 9.925 | 39.314 | 3.96 | Upregulated | 0.01 |
| hsa-miR-185-5p |
UGGAGAGAAAGGCAGUUCCUGA | 82 | 189 | 3.434 | 8.502 | 2.48 | Upregulated | 0.00 |
| hsa-miR-192-3p |
CUGCCAAUUCCAUAGGUCACAG | 762 | 218 | 31.911 | 9.806 | 0.31 | Downregulated | 0.00 |
| hsa-miR-195-3p |
CCAAUAUUGGCUGUGCUGCUCC | 832 | 283 | 34.843 | 12.730 | 0.37 | Downregulated | 0.00 |
| hsa-miR-205-3p |
GAUUUCAGUGGAGUGAAGUUC | 87 | 287 | 3.643 | 12.910 | 3.54 | Upregulated | 0.00 |
| hsa-miR-205-5p |
UCCUUCAUUCCACCGGAGUCUG | 18 | 38 | 0.754 | 1.709 | 2.27 | Upregulated | 0.00 |
|
hsa-miR-218-1-3p |
AUGGUUCCGUCAAGCACCAUGG | 238 | 48 | 9.967 | 2.159 | 0.22 | Downregulated | 0.00 |
|
hsa-miR-218-2-3p |
CAUGGUUCUGUCAAGCACCGCG | 384 | 871 | 16.081 | 39.179 | 2.44 | Upregulated | 0.00 |
| hsa-miR-218-5p |
UUGUGCUUGAUCUAACCAUGU | 248 | 438 | 10.386 | 19.702 | 1.90 | Upregulated | 0.00 |
| hsa-miR-221-5p |
ACCUGGCAUACAAUGUAGAUUU | 125 | 33 | 5.235 | 1.484 | 0.28 | Downregulated | 0.00 |
| hsa-miR-223-3p |
UGUCAGUUUGUCAAAUACCCCA | 137 | 433 | 5.737 | 19.477 | 3.39 | Upregulated | 0.00 |
| hsa-miR-224-3p |
AAAAUGGUGCCCUAGUGACUACA | 221 | 873 | 9.255 | 39.269 | 4.24 | Upregulated | 0.00 |
| hsa-miR-375 |
UUUGUUCGUUCGGCUCGCGUGA | 82 | 238 | 3.434 | 10.706 | 3.12 | Upregulated | 0.00 |
| hsa-miR-383-3p |
ACAGCACUGCCUGGUCAGA | 98 | 21 | 4.104 | 0.945 | 0.23 | Downregulated | 0.00 |
| hsa-miR-425-5p |
AAUGACACGAUCACUCCCGUUGA | 23 | 82 | 0.963 | 3.688 | 3.83 | Upregulated | 0.00 |
| hsa-miR-451a |
AAACCGUUACCAUUACUGAGUU | 322 | 187 | 13.485 | 8.412 | 0.62 | Downregulated | 0.00 |
| hsa-miR-497-5p |
CAGCAGCACACUGUGGUUUGU | 87 | 32 | 3.643 | 1.439 | 0.40 | Downregulated | 0.01 |
| hsa-miR-498 |
UUUCAAGCCAGGGGGCGUUUUUC | 93 | 328 | 3.895 | 14.754 | 3.79 | Upregulated | 0.00 |
|
hsa-miR-520a-3p |
AAAGUGCUUCCCUUUGGACUGU | 76 | 187 | 3.183 | 8.412 | 2.64 | Upregulated | 0.00 |
|
hsa-miR-520a-5p |
CUCCAGAGGGAAGUACUUUCU | 233 | 431 | 9.758 | 19.387 | 1.99 | Upregulated | 0.00 |
| hsa-miR-520b |
AAAGUGCUUCCUUUUAGAGGG | 34 | 123 | 1.424 | 5.533 | 3.89 | Upregulated | 0.00 |
|
hsa-miR-520c-3p |
AAAGUGCUUCCUUUUAGAGGGU | 213 | 23 | 8.920 | 1.035 | 0.12 | Downregulated | 0.02 |
|
hsa-miR-520d-5p |
CUACAAAGGGAAGCCCUUUC | 54 | 31 | 2.261 | 1.394 | 0.62 | Downregulated | 0.00 |
| hsa-miR-557 |
GUUUGCACGGGUGGGCCUUGUCU | 354 | 123 | 14.825 | 5.533 | 0.37 | Downregulated | 0.00 |
| hsa-miR-623 |
AUCCCUUGCAGGGGCUGUUGGGU | 232 | 73 | 9.716 | 3.284 | 0.34 | Downregulated | 0.00 |
| hsa-miR-654-3p |
UAUGUCUGCUGACCAUCACCUU | 32 | 51 | 1.340 | 2.294 | 1.71 | Upregulated | 0.00 |
| hsa-miR-654-5p |
UGGUGGGCCGCAGAACAUGUGC | 82 | 18 | 3.434 | 0.810 | 0.24 | Uownregulated | 0.00 |
|
hsa-miR-1233-5p |
AGUGGGAGGCCAGGGCACGGCA | 63 | 26 | 2.638 | 1.170 | 0.44 | Downregulated | 0.02 |
RT-qPCR validation
To validate the expression levels of the miRNAs
involved in SCLC, RT-qPCR was performed on 36 miRNA sequences from
the SCLC tissues and the corresponding normal tissues (Fig. 3). Of these 36 miRNAs, 6 miRNAs and
the corresponding miRNA*s were identified, including hsa-miR-29a-3p
and hsa-miR-29a-5p, and hsa-miR-30a-3p and hsa-miR-30a-5p (Fig. 2A). This indicates that the mature
miRNA and corresponding miRNA* exhibit similar expression patterns
and are important in SCLC. The results revealed similar abundance
profiles with Solexa sequencing and RT-qPCR analysis; they
indicated that hsa-miR-21-5p was upregulated in the SCLC tissues.
However, a few differences regarding the ratio (LC/NT) between
RT-qPCR and Solexa sequencing were identified; for example, the
ratio of hsa-miR-224-3p was 4.24 with sequencing data, but the
RT-qPCR ratio was only 3.64, possibly since different data
normalization protocols of the two methods were provided. The
sequencing was normalized to the whole abundance of all miRNAs
sequenced by Solexa, while the RT-qPCR was normalized to the
expression levels of U6 sRNA.
Annotation of the targets of miRNAs
involved in SCLC
The targets of the miRNAs involved in SCLC were
annotated by the TargetScan database. The functions of the targets
of these miRNAs were involved in organ development, tissue
differentiation, cell apoptosis and defense, signal transduction
and the electron transfer chain. For example, the target of
hsa-miR-29 encodes vascular endothelial growth factor A, which has
various effects, including mediating increased vascular
permeability, vasculogenesis and endothelial cell growth, promoting
cell migration and inhibiting apoptosis (17,18).
The target of hsa-miR-375 encodes phosphoinositide-dependent
kinase-1, which is important in the signaling pathways activated by
several growth factors and hormones, including insulin (19). F-box and WD repeat
domain-containing 7 encoded by hsa-miR-223 may regulate intestinal
cell lineage commitment (20). The
various functions of these targets implied that these responsive
miRNAs exert crucial regulatory functions in SCLC. However, no
functional annotations were identified for certain miRNAs,
including hsa-miR-185 and hsa-miR-224 (Table III).
 | Table IIITargets of differentially expressed
microRNAs in small cell lung cancer. |
Table III
Targets of differentially expressed
microRNAs in small cell lung cancer.
| Member | Target
proteins |
|---|
| hsa-miR-7 | UBX domain protein
2B, spermatogenesis associated 2, phosphoinositide-3-kinase,
catalytic, Δ polypeptide, ubiquilin 4, SH2 domain containing 5 |
| hsa-miR-21 | Zinc finger protein
367, G protein-coupled receptor 64, PHD finger protein 14,
polybromo 1, vinculin, retinitis pigmentosa 2 (X-linked
recessive) |
| hsa-miR-22 | Glutamate receptor,
metabotropic 5, coiled-coil domain containing 67,
fucosyltransferase 9 (α (1,3) fucosyltransferase), H3 histone,
family 3B |
| hsa-miR-24 | Calcitonin
receptor, mannose-P-dolichol utilization defect 1, kinase
suppressor of ras 2, CKLF-like MARVEL transmembrane domain
containing 4, spermatogenesis associated, serine-rich 2-like |
| has-miR-25 | CD69 molecule,
folliculin interacting protein 1, actin, α, cardiac muscle 1; F-box
and WD repeat domain containing 7; aspartate β-hydroxylase |
| hsa-miR-29 | PIK3R1, Vascular
endothelial growth factor A, PIK3RZ |
| hsa-miR-30 | MAPKS, NRG3, KRAS,
PIK3CD, RARB, KRAS, CCNEZ, ITGA6, peroxisome proliferator-activated
receptor γ, coactivator 1β, makorin ring finger protein 3 |
| hsa-miR-34 | Cyclin E, p53;
hyperpolarization activated cyclic nucleotide-gated potassium
channel 3, family with sequence similarity 76, member A; neuron
navigator 3 |
| hsa-miR-96 | Leucine-rich
repeats and calponin homology domain containing 2; adenylate kinase
3; spindlin 1; dihydrolipoamide S-acetyltransferase |
| hsa-miR-99 | THAP domain
containing, apoptosis associated protein 2; kelch repeat and BTB
(POZ) domain containing 8; ependymin related protein 1
(zebrafish) |
| hsa-miR-101 | transportin 1;
family with sequence similarity 108, member C1; family with
sequence similarity 108, member C1; FLJ20160 protein |
| hsa-miR-103 | Dicer 1,
ribonuclease type III; forkhead box P1; HIV-1 Rev binding protein;
eukaryotic translation initiation factor 5; armadillo repeat
containing 1 |
| hsa-miR-106 | Carnitine
O-octanoyltransferase; zinc finger with KRAB and SCAN domains 1;
RAB22A, member RAS oncogene family; cytochrome b reductase 1 |
| hsa-miR-125 | StAR-related lipid
transfer domain containing 13; zinc finger protein 792; SH3 domain
and tetratricopeptide repeats 2; zinc finger and SCAN domain
containing 29 |
| hsa-miR-126 | ITGA6, CRK, SPRED1;
epidermal growth factor-like domain 7; protein tyrosine
phosphatase, non-receptor type 9; low density lipoprotein
receptor-related protein 6; F-box protein 33 |
| hsa-miR-139 | TATA element
modulatory factor 1; USP6 N-terminal like; T-box 1; early B-cell
factor 1; protein prenyltransferase α subunit repeat containing 1;
S phase cyclin A-associated protein in the ER |
| hsa-miR-141 | Transmembrane
protein 170B; zinc finger E-box binding homeobox 2; RAN binding
protein 6; zinc finger RNA binding protein; protein kinase,
cAMP-dependent, catalytic, β |
| hsa-miR-143 | Solute carrier
family 30 (zinc transporter), member 8; vasohibin 1; homeodomain
interacting protein kinase 2; development and differentiation
enhancing factor-like 1 |
| hsa-miR-145 | Epidermal growth
factor receptor, IGF-1R; family with sequence similarity 108,
member C1; SLIT-ROBO Rho GTPase activating protein 2; ATP-binding
cassette, sub-family E (OABP), member 1; tripartite
motif-containing 2 |
| hsa-miR-146 | Zinc finger protein
826; TNF receptor-associated factor 6; zinc finger and BTB domain
containing 2; neuro-oncological ventral antigen 1; chemokine
binding protein 2 |
|
hsa-miR-148/152 | Cholecystokinin B
receptor; ATPase, H+ transporting, lysosomal accessory
protein 2; oxysterol binding protein-like 11; chloride channel
6 |
| hsa-miR-182 | Regulator of
G-protein signaling 17; microphthalmia-associated transcription
factor; ARP2 actin-related protein 2 homolog (yeast);
microfibrillar-associated protein 3; neurocalcin delta |
| hsa-miR-183 | VIL2-coding-protein
Ezrin; A kinase anchor protein (gravin) 12; phosphatidylinositol
glycan anchor biosynthesis, class X; neurotrophic tyrosine kinase,
receptor, type 2; profilin 2; SLAIN motif family, member 1 |
| hsa-miR-192 | IKAROS family zinc
finger 2 (Helios); IKAROS family zinc finger 2 (Helios); poly(A)
binding protein, cytoplasmic 4 (inducible form); dihydrolipoamide
branched chain transacylase E2 |
| hsa-miR-205 | RAB11 family
interacting protein 1 (class I); RAB11 family interacting protein 1
(class I); cell division cycle 2-like 6 (CDK8-like); AP2 associated
kinase 1; lysophosphatidylcholine acyltransferase 1 |
| hsa-miR-218 | Chromosome 3 open
reading frame 70; solute carrier family 1 (glial high affinity
glutamate transporter), member 2; glucuronic acid epimerase; one
cut homeobox 2; stress-associated endoplasmic reticulum protein
1 |
| hsa-miR-221 | Sorting nexin 4;
regulator of G-protein signaling 6; osteopetrosis associated
transmembrane protein 1; v-kit Hardy-Zuckerman 4 feline sarcoma
viral oncogene homolog |
| hsa-miR-223 | F-box and WD repeat
domain containing 7; myosin VB; adenomatous polyposis coli; ras
homolog gene family, member B; solute carrier family 4, sodium
bicarbonate cotransporter, member 4 |
| hsa-miR-375 | TSC1,
Phosphoinositide-dependent kinase-1, SOCS4, SPRY3, PRKX; solute
carrier family 16, member 2 (monocarboxylic acid transporter 8);
short stature homeobox 2; RAS, dexamethasone-induced 1 |
| hsa-miR-383 | NCK-associated
protein 1; striatin, calmodulin binding protein 3; solute carrier
family 35 (UDP-N-acetylglucosamine transporter), member A3; mal,
T-cell differentiation protein 2 |
| hsa-miR-425 | Nuclear fragile X
mental retardation protein interacting protein 2; family with
sequence similarity 133, member B; forkhead box J3; sterile α motif
and leucine zipper containing kinase AZK |
| hsa-miR-451a | Tuberous sclerosis
1; chromosome 11 open reading frame 30; chromosome 11 open reading
frame 30; GATA zinc finger domain containing 2B |
| hsa-miR-498 | Chromosome X open
reading frame 1; chromosome 9 open reading frame 5; CD2-associated
protein; glutamate receptor, ionotrophic, AMPA 3 |
| hsa-miR-520 | Family with
sequence similarity 102, member B; DIRAS family, GTP-binding
RAS-like 2; sodium channel, voltage-gated, type III, β; homeodomain
interacting protein kinase 2; YTH domain family, member 3 |
| hsa-miR-623 | Myotubularin
related protein 7; acyl-CoA synthetase medium-chain family member
2A; adaptor-related protein complex 3, μ subunit; septin 11; ring
finger protein 169 |
| hsa-miR-654 | Sorbin and SH3
domain containing 1; calcium channel, voltage-dependent, T type,
α1I subunit; transducin (β)-like 1X-linked; pregnancy-associated
plasma protein A, pappalysin 1; pregnancy-associated plasma protein
A, pappalysin 1; WD repeat domain 26 |
Discussion
SCLC is a leading cause of cancer-related morality
worldwide. There are numerous reports regarding the roles of miRNAs
in NSCLC (10,12,21,22),
however, little is known regarding the regulatory functions of
miRNAs in SCLC. In the present study two sRNA libraries were
constructed from SCLC and normal tissues. Sequence analysis and
RT-qPCR validation identified 81 miRNAs differentially expressed in
SCLC. This may provide useful information for improving the
diagnosis, prevention and treatment of this disease. Notably, the
expression levels of miRNA*s were associated with the corresponding
miRNAs and revealed similar expression patterns. This may be due to
miRNAs and the corresponding miRNA*s being derived from the same
precursor (23). In addition, more
than half the miRNAs associated with SCLC were downregulated and
the total readings of known miRNAs were reduced (Fig. 2), implying that the corresponding
targets become activated and trigger defense mechanisms against
illness. However, further studies are required to characterize the
miRNAs involved in SCLC.
Only a few studies have been published regarding the
miRNAs involved in SCLC. Miko et al (24) employed microarray and RT-qPCR
analyses to determine the miRNA expression profile in primary SCLC,
and the result revealed that at least 24 miRNAs were differentially
expressed between the normal lung and primary SCLC tumor tissues. A
previous study performed gene expression profiling of
drug-resistant SCLC cells by combining miRNA and cDNA expression
analyses, and identified 61 significantly differentially expressed
miRNAs (25). However, in the
present study, a relatively complete analysis of miRNAs involved in
SCLC at a genome-wide level was performed and certain miRNAs were
reported to be involved in SCLC for the first time, including
hsa-miR451. In addition, there were a few differences in the
results of these studies. For example, hsa-miR-223 was
downregulated in the previous study (25) while upregulated in the present
study in SCLC tissues; this may be due to different methods for
profiling miRNAs.
The functions of the targets of miRNAs involved in
SCLC were diverse and revealed inhibitory roles in the regulation
of the corresponding targets. In the present study, the expression
levels of hsa-miR-34-c, hsa-miR-126 and hsa-miR-145 were
downregulated, while the expression levels of hsa-miR-183 was
upregulated (Table II). The
targets of hsa-miR-34, cyclin E and p53, are associated with the
cell cycle and tumor formation (26,27);
the low expression of hsa-miR-34 may result in upregulation of
cyclin E and p53, which is crucial in the development of SCLC. The
epidermal growth factor receptor (EGFR) encoded by the target of
hsa-miR-145 may inhibit cancer growth (28); the downregulation of hsa-miR-145 in
SCLC may generate an increase in the expression levels of EGFR and
thus promote cancer growth. The targets of hsa-miR-126 encode
epidermal growth factor-like domain 7 (EGFL7), which is involved in
cellular responses, such as cell migration and blood vessel
formation (29). The
downregulation of hsa-miR-126 may activate EGFL7 and advance tumor
growth in vivo. The VIL2-coding-protein Ezrin, a known
target of hsa-miR-183 is involved in migration and invasion
(30); overexpression of
hsa-miR-183 may repress VIL2-coding-protein Ezrin and inhibit
migration and invasion of lung cancer cells, perhaps since defense
mechanisms against illness have been initiated. Therefore, the
evidence suggests that these identified miRNAs are important in
SCLC.
In conclusion, 81 miRNAs involved in SCLC have been
identified via deep sequencing and the profiles for a subset of
miRNAs were validated by RT-qPCR. The functions for the target
genes of these miRNAs were analyzed. These findings contribute to
the understanding of the function of posttranscriptional regulation
of miRNA in SCLC development and progression, which is essential
for improving the diagnosis, prevention and treatment of this
disease.
Abbreviations:
|
miRNA
|
microRNA
|
|
sRNA
|
small RNA
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
SCLC
|
small cell lung cancer
|
|
NSCLC
|
non-small cell lung cancer
|
|
EGFR
|
epidermal growth factor receptor
|
References
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carrington JC and Ambros V: Role of
microRNAs in plant and animal development. Science. 301:336–338.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kosik KS: MicroRNAs and cellular
phenotypy. Cell. 143:21–26. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hui AB, Shi W, Boutros PC, et al: Robust
global micro-RNA profiling with formalin-fixed paraffin-embedded
breast cancer tissues. Lab Invest. 89:597–606. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun M, Liu XH, Li JH, Yang JS, et al:
MiR-196a is upregulated in gastric cancer and promotes cell
proliferation by downregulating p27(kip1). Mol Cancer Ther.
11:842–852. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu XH, Lu KH, Wang KM, et al:
MicroRNA-196a promotes non-small cell lung cancer cell
proliferation and invasion through targeting HOXA5. BMC Cancer.
12:3482012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pacurari M, Addison JB, Bondalapati N, et
al: The microRNA-200 family targets multiple non-small cell lung
cancer prognostic markers in H1299 cells and BEAS-2B cells. Int J
Oncol. 43:548–560. 2013.PubMed/NCBI
|
|
12
|
Luo W, Huang B, Li Z, et al: MicroRNA-449a
Is downregulated in non-small cell lung cancer and inhibits
migration and invasion by targeting c-Met. PLoS One. 8:e647592013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang JG, Guo JF, Liu DL, Liu Q and Wang
JJ: MicroRNA-101 exerts tumor-suppressive functions in non-small
cell lung cancer through directly targeting enhancer of zeste
homolog 2. J Thorac Oncol. 6:671–678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hafner M, Landgraf P, Ludwig J, et al:
Identification of microRNAs and other small regulatory RNAs using
cDNA library sequencing. Methods. 44:3–12. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi R and Chiang VL: Facile means for
quantifying microRNA expression by real-time PCR. Biotechniques.
39:519–525. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li R, Li Y, Kristiansen K and Wang J:
SOAP: short oligonucleotide alignment program. Bioinformatics.
24:713–714. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mackenzie F and Ruhrberg C: Diverse roles
for VEGF-A in the nervous system. Development. 139:1371–1380. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stockmann C, Doedens A, Weidemann A, et
al: Deletion of vascular endothelial growth factor in myeloid cells
accelerates tumorigenesis. Nature. 456:814–818. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamada T, Katagiri H, Asano T, et al: Role
of PDK1 in insulin-signaling pathway for glucose metabolism in
3T3-L1 adipocytes. Am J Physiol Endocrinol Metab. 282:E1385–E1394.
2002.PubMed/NCBI
|
|
20
|
Sancho R, Jandke A, Davis H, et al: F-box
and WD repeat domain-containing 7 regulates intestinal cell lineage
commitment and is a haploinsufficient tumor suppressor.
Gastroenterology. 139:929–941. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang XC, Wang W, Zhang ZB, et al:
Overexpression of miRNA-21 promotes radiation-resistance of
non-small cell lung cancer. Radiat Oncol. 8:1462013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Allen E, Xie Z, Gustafson AM, et al:
Evolution of microRNA genes by inverted duplication of target gene
sequences in Arabidopsis thaliana. Nat Genet. 36:1282–1290.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miko E, Czimmerer Z, Csánky E, et al:
Differentially expressed microRNAs in small cell lung cancer. Exp
Lung Res. 35:646–664. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo L, Liu Y, Bai Y, Sun Y, Xiao F and Guo
Y: Gene expression profiling of drug-resistant small cell lung
cancer cells by combining microRNA and cDNA expression analysis.
Eur J Cancer. 46:1692–1702. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu X, Sempere LF, Galimberti F,
Freemantle SJ, et al: Uncovering growth-suppressive MicroRNAs in
lung cancer. Clin Cancer Res. 15:1177–1183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhong M, Ma X, Sun C and Chen L: MicroRNAs
reduce tumor growth and contribute to enhance cytotoxicity induced
by gefitinib in non-small cell lung cancer. Chem Biol Interact.
184:431–438. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun Y, Bai Y, Zhang F, Wang Y, et al:
miR-126 inhibits non-small cell lung cancer cells proliferation by
targeting EGFL7. Biochem Biophys Res Commun. 391:1483–1489. 2010.
View Article : Google Scholar
|
|
30
|
Wang G, Mao W and Zheng S: MicroRNA-183
regulates Ezrin expression in lung cancer cells. FEBS Lett.
582:3663–3668. 2008. View Article : Google Scholar : PubMed/NCBI
|