Introduction
Hyperhomocysteinemia (HHcy) is a pathology caused by
abnormal methionine metabolism, due to a combination of genetic and
environmental factors. The dysfunction of three enzymes,
methylenetetrahydrofolate reductase (1), cystathionine synthetase (CBS)
(2) and methionine synthase,
together with a deficiency of metabolic cofactors, such as folic
acid, vitamin B6 and B12, contributes to the occurrence of HHcy
(3).
Long-term HHcy can cause toxicity to endothelial
cells, stimulate the proliferation of vascular smooth muscle cells,
induce thrombosis and disrupt fat, sugar and protein metabolism.
Epidemiological data have shown that HHcy increases Alzheimer’s
disease (AD)-related morbidity, and may act as an independent AD
risk factor (4). A study by Wang
et al (5) has shown that
HHcy can induce apoptosis of mouse hippocampal neurons. However,
the exact mechanism of how HHcy causes neuronal damage remains to
be explored (5).
Quantitative proteomics is an important molecular
technique used to quantify and identify all the proteins expressed
by the genome. Quantitative proteomics may therefore facilitate the
understanding of changes that occur in multiple proteins upon
hippocampal neuronal injury. Isobaric tags for relative and
absolute quantification (iTRAQ™) were developed by Applied
Biosystems Incorporation (Foster City, CA, USA) in 2004. Differing
from two-dimensional gel electrophoresis, iTRAQ labels global
peptides and uses a non-gel-based method for absolute quantitation
of proteins in up to four samples at one time. In this study,
samples were labeled with four independent isobaric tags (from 114
to 117), the fragmentation by tandem mass spectrometry (MS/MS) was
analyzed and the four different samples with peak areas were
quantified (1).
iTRAQ is a quantitative method for up to four
different samples. An iTRAQ label is incorporated in the N-termini
and lysine residues of the peptide. By tandem mass spectrometry,
mass-to-charge ratios (114–117), according to the wave height and
area, can be identified from the protein and analysis of
quantitative information with a different protein processing iTRAQ
gives quantitative information on the protein and its
post-translational processing (6,7).
In the present study, the proteome changes caused by
HHcy in mouse hippocampal neurons using the iTRAQ technology were
investigated. Functional analysis was further used to examine the
role of components from tau proteins in the neuronal damage induced
by homocysteine.
Materials and methods
Animal model
All animals were housed at the Experimental Animal
Center, Tongji University (Shanghai, China). This study was
performed according to the recommendations in the Guide for the
National Science Council of the Republic of China. The protocol was
approved by the Animal Care and Use Committee of The Tenth People’s
Hospital of Tongji University (permit number: 2011–0111; Shanghai,
China). C57BL/6 mice, weighing 18–20 g, were randomly divided into
two groups (10 mice/group); the control and 1% methionine groups,
following adaptive feeding for 2 days. Serum Hcy levels were
detected by high performance liquid chromatography (HPLC) (8) Briefly, samples were reduced with
tri-n-butylphosphine, proteins were precipitated with
trichloroacetic acid (10%) and derivatized with ammonium
7-fluorobenzo-2-oxa-1,3-diazole-4-sulfonate. The derivatives were
separated by reversed-phase high-performance liquid chromatography
followed by fluorescence detection.
Sample preparation for MS analysis
The hippocampi of mice from the model and control
groups were removed and dissected. The two groups were then
subsequently further divided randomly into two groups, five in each
group, with the same body weight. Hippocampal neurons were
harvested in Tris-buffered saline (TBS) and centrifuged at 500 × g.
Total hippocampal neuronal protein was extracted by centrifugation
at 13,000 × g for 5 min following lysis in extraction buffer (50 mM
Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium
deoxycholate, 30 mM NaF and 1 mM protease inhibitor) (9). The homogenate was designated to the
four groups to be labeled as 114 (control group 1), 115 (control
group 2), 116 (model group 1) and 117 (model group 2) by iTRAQ
reagents.
Liquid chromatography-MS/MS analysis on
LTQ-Orbitrap (Applied Biosystems)
Liquid chromatography was performed using the
Shimadzu Corporation (Kyoto, Japan) 2D-nano-HPLC and the ABI
MALDI-TOF-TOF 4700 mass spectrometer (Applied Biosystems). The
iTRAQ labeling kit (Applied Biosystems) was utilized for labeling
proteins.
The four chemically identical iTRAQ reagents, 114,
115, 116 and 117, have the same overall mass. Each label is
composed of a peptide reactive group (N-hydroxysuccinimide-ester)
and an isobaric tag of 145 Da that consists of a balancer
(carbonyl) and a reporter group (based on N-methylpiperazine). The
four tags vary in their stable isotope compositions such that the
reporter group has a mass of 114–117 Da and the balancer of 28–31
Da. The fragmentation site between the balancer and the reporter
group is responsible for the generation of reporter ions in the
region of 114–117 m/z (10).
Protein identification was performed using Mascot
(Matrix Science, London, UK) using the following search parameters:
Taxonomy: Mus musculus; enzyme, trypsin; fixed
modifications, carboxyamidomethylation of cysteine and oxidation of
methionine variable modifications, 95%; missed cleavages allowed,
one; peptide tolerance, 100 ppm; MS/MS tolerance, 100 ppm; peptide
charge, +0.1 Da (11).
Analysis of protein expression by western
blotting
Western blotting was performed for examining changes
of proteins. Equal quantities of protein were separated by SDS-PAGE
and transferred onto nitrocellulose membranes. The membranes were
pre-incubated in blocking TBS buffer [containing 5% (V/V) non-fat
dried milk] for 1 h at room temperature and then probed with a
primary antibody overnight. Following washing with buffer, the
blots were incubated with the secondary antibodies
(peroxidase-conjugated anti-rabbit immunoglobulin G) at room
temperature for 1 h. Blots were developed using the enhanced
chemiluminescence system (GE Healthcare Bioscience, Pittsburgh, PA,
USA), following the manufacturer’s instructions (12). Densitometric analysis was performed
using ImageJ software (National Institutes of Health, Bethesda, MD,
USA). Anti-mouse primary antibodies used were as follows:
Actinin-α-1 (Actn1), fibronectin 1 (FN1), integrin-β-1 (Itgb1),
myosin heavy chain 9 non-muscle (Myh9), microtubule-associated
protein tau (Tau), Rab11a member RAS oncogene family,
calcium/calmodulin-dependent protein kinase II δ (Camk2d),
calcium/calmodulin-dependent protein kinase II β (Camk2b), ATPase
Ca++ transporting plasma membrane 1 (Atp2b1) and β-actin
(Actb) (Abcam, Cambridge, MA, USA).
Gene-ontology (GO) analysis
GO analysis was used to analyze the main function of
the differentially expressed genes based on the GO database, which
provides the key functional classification for the National Center
for Biotechnology Information. Fisher’s exact and χ2
test were implemented to classify the GO category, and the false
discovery rate (FDR) was calculated for P-value correction. The
smaller the FDR, the smaller the error in judging the P-value
(13,14).
Ingenuity® Pathway Analysis
(IPA)
IPA (Qiagen, Redwood City, CA, USA) was performed to
identify the significant pathways of the differentially expressed
genes according to the Kyoto Encyclopedia of Genes and Genomes
(KEGG, Kanehisa Laboratories, Kyoto, Japan), Biocarta (San Diego,
CA, USA) and Reactome (http://www.reactome.org) annotation. The Fisher’s
exact test and χ2 test were used to select the most
statistically significant pathways, and the threshold of
significance was defined by P-value and FDR. The enrichment was
calculated as the equation mentioned above (15,16).
Statistical analysis
Differences in GO terms and pathway analysis between
the genes up-regulation or down-regulation by HHcy and normal mice
were assessed using the χ2 test or Fisher’s exact test.
A two-sided P<0.05 was considered a statistically significant
difference.
Results
Detection of differentially expressed
proteins using the iTRAQ technique
By using the iTRAQ technique, 52 down-regulated and
44 upregulated proteins were detected in the hippocampi of mice
with HHcy. Western blot analysis was used to further examine
protein expression levels of Actn1, Tau, FN1, Itgb1, Myh9, Rab11a,
Thy-1 cell surface antigen, filamin A-α and Atp2b1, and the levels
showed consistency with the iTRAQ data. The western blot analysis
results showed that protein levels of Actn1, FN1, Tau and Atp2b1
were higher in the HHcy group than those in the control group,
whereas Itgb1, Myh9, Rab11a, Camk2b and Camk2d were downregulated
by HHcy (Fig. 1).
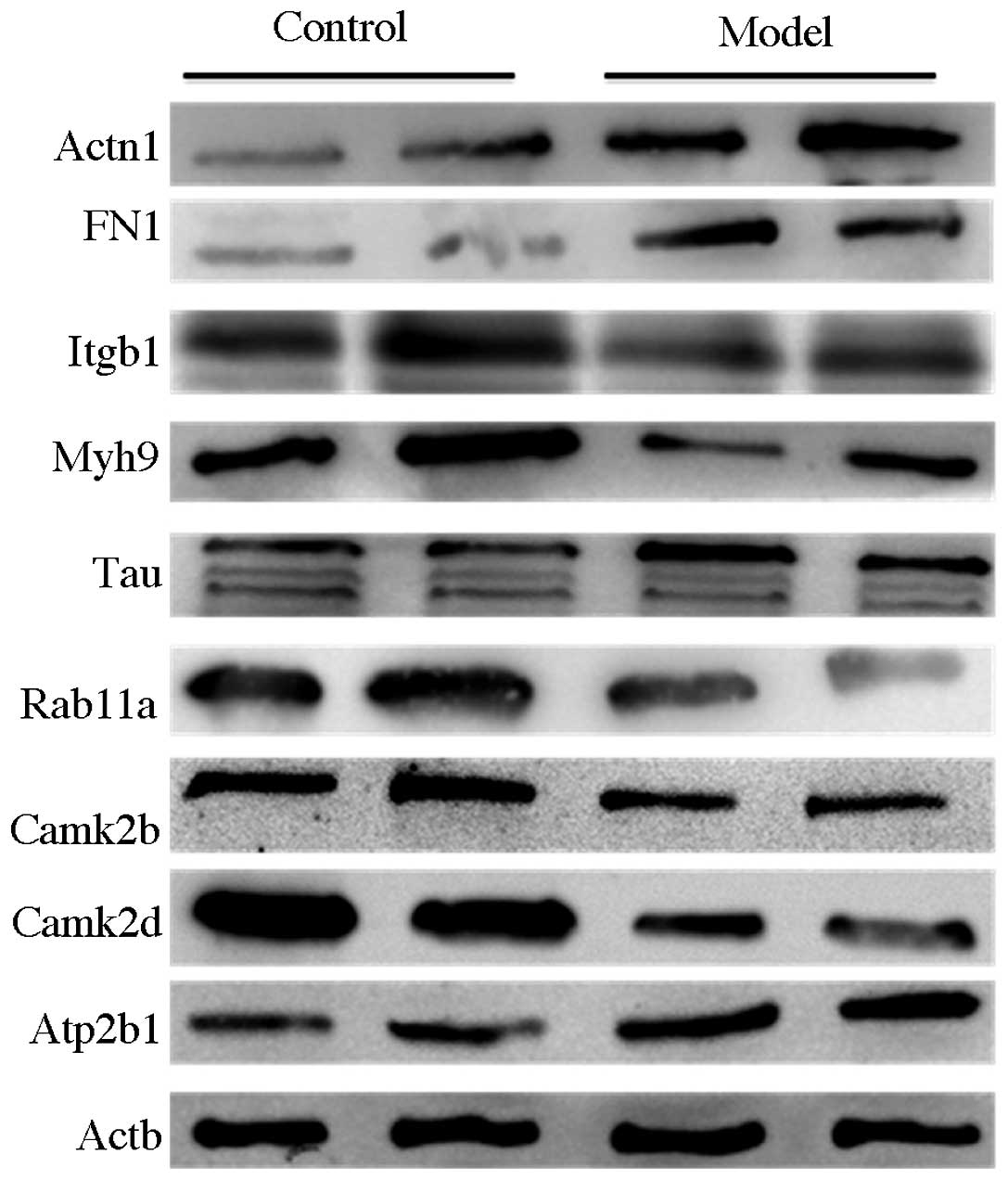 | Figure 1Western blot analysis results of
differentially expressed proteins in normal mice (control) and
hyperhomocysteinemia mice (model). Itgb1, Myh9, Rab11a, Camk2b and
Camk2d were downregulated, and Actn1, FN1, Tau and Atp2b1 were
upregulated in the model mice compared with the control mice.
Actn1, actinin-α-1; FN1, fibronectin 1; Itgb1, integrin-β-1; Myh9,
myosin heavy chain 9 non-muscle; Tau, microtubule-associated
protein tau; Rab11a, Rab11a member RAS oncogene family; Camk2d,
calcium/calmodulin-dependent protein kinase II δ; Camk2b,
calcium/calmodulin-dependent protein kinase II β; Atp2b1, ATPase
Ca++ transporting plasma membrane 1; Actb, β-actin. |
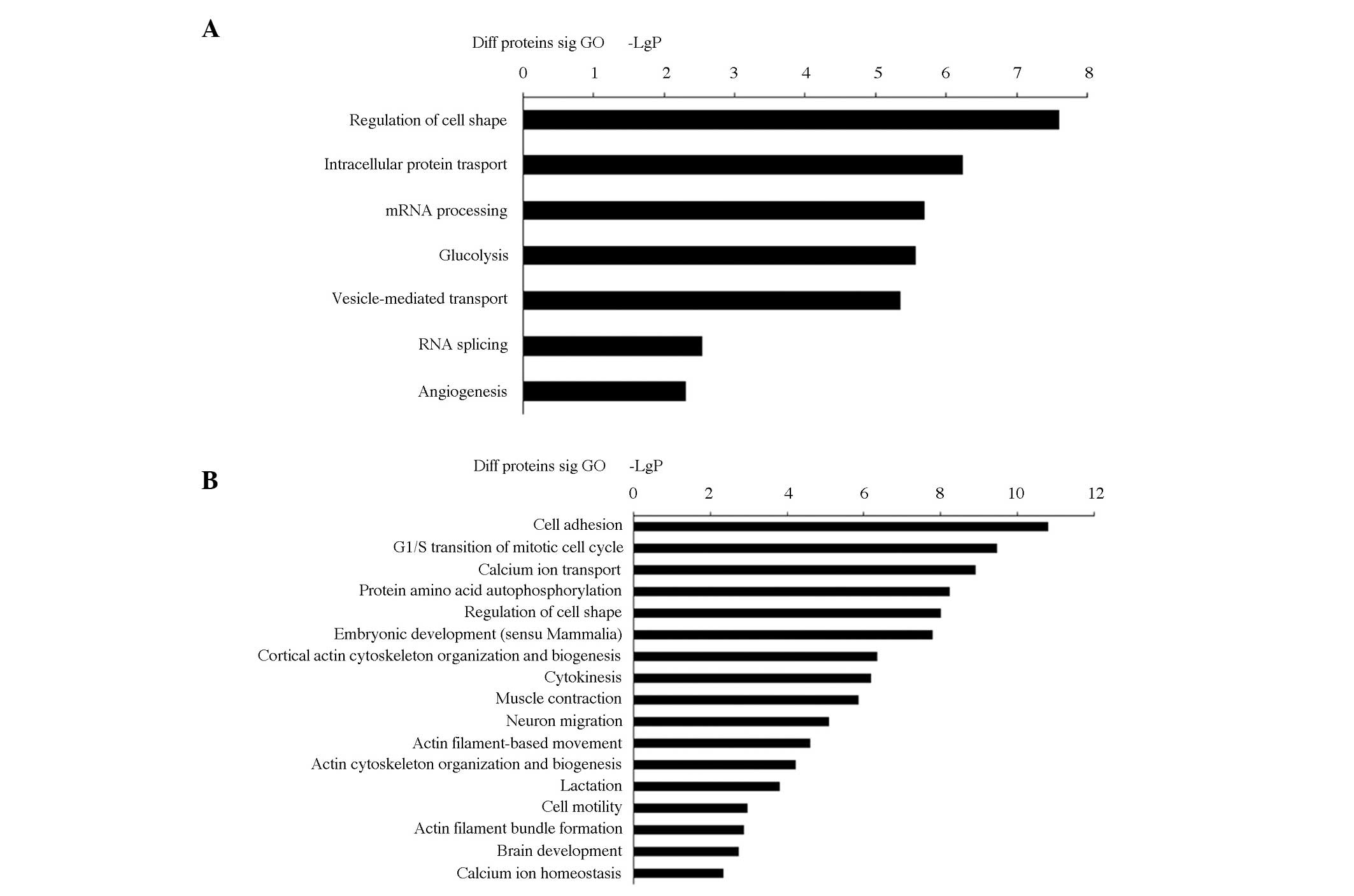
GO analysis
GO analysis was performed to test whether molecular
functions and biological processes were significantly associated
with those genes bearing the greatest difference between HHcy and
normal mice. The most enriched terms for each group can be seen in
Fig. 2. Genes that were
up-regulated at the protein level showed relevance to energy
metabolism, cell differentiation and signal transducer activity.
The following GO categories were included: Regulation of cell shape
(GO:0008360); intracellular protein transport (GO:0006886); mRNA
processing (GO:0006397); glycolysis (GO:0006096); vesicle-mediated
transport (GO:0016192); and RNA splicing (GO:0008380); angiogenesis
(GO:0001525); (Fig. 2A). Enriched
GO terms associated with genes that were downregulated at the
protein level were involved in: Cell adhesion (GO:0007155); G1/S
transition of mitotic cell cycle (GO:0000082); calcium ion
transport (GO:0006816); protein amino acid autophosphorylation
(GO:0046777); regulation of cell shape (GO:0008360); embryonic
development (sensu Mammalia) (GO:0001701); cortical actin
cytoskeleton organization and biogenesis (GO:0030866); cytokinesis
(GO:0000910); muscle contraction (GO:0006936); neuron migration
(GO:0001764); actin filament-based movement (GO:0030048); actin
cytoskeleton organization and biogenesis (GO:0030036); lactation
(GO:0007595); cell motility (GO:0006928); actin filament bundle
formation (GO:0051017); brain development (GO:0007420); and calcium
ion homeostasis (GO:0006874) (Fig.
2B).
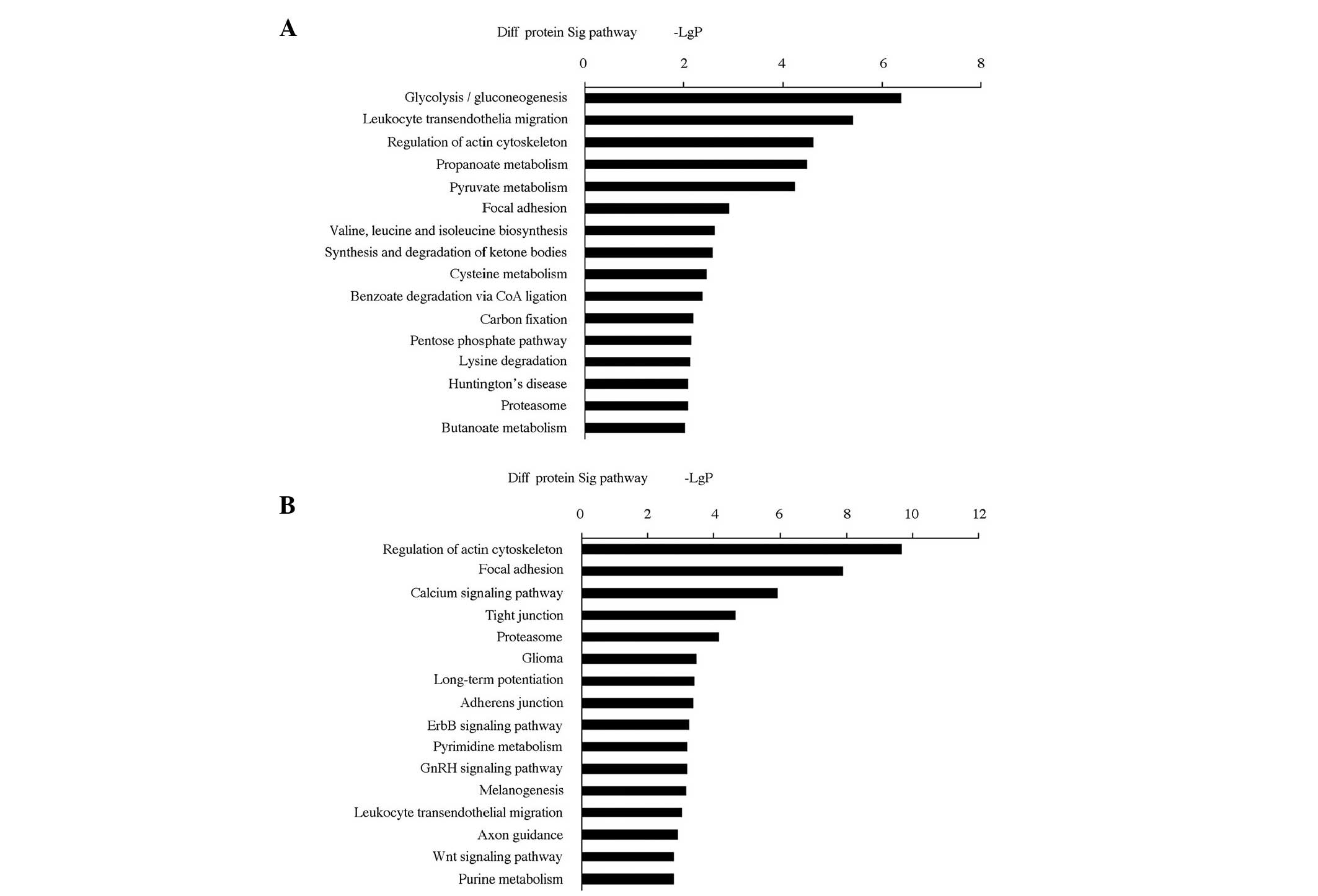
Pathway analysis
For further characterization of the functional
significance of the differentially expressed proteins, a systematic
analysis to discover gene classifiers and pathways that were
significantly enriched between HHcy and normal mice, was performed.
More than 15 signaling pathways were detected and considered
statistically significant (P<0.05), indicating that Hcy affects
numerous cytokines and signaling molecules involved in numerous
signaling procedures or pathways. The biological pathways are shown
in Fig. 3A (pathways derived from
upregulated proteins) and Fig. 3B
(pathways derived from downregulated proteins). Pathway analysis
showed that hypermethylated genes were implicated in the following
pathways: i) Cytokine-cytokine receptor interaction; ii) regulation
of the actin cytoskeleton; iii) MAPK signaling pathway; iv) calcium
signaling pathway; v) adipocytokine signaling pathway; vi) retinol
metabolism; vii) Janus kinase/signal transducers and activators of
transcription signaling pathway; viii) peroxisome
proliferator-activated receptor signaling pathway; ix)
phosphatidylinositol signaling system. Functional networks derived
from hypomethylated genes included: i) Cytokine-cytokine receptor
interaction; ii) cell cycle; iii) MAPK signaling pathway; iv) fatty
acid metabolism; v) transforming growth factor β (TGFβ) signaling
pathway; vi) adipocytokine signaling pathway; and vii) Wnt
signaling pathway. Furthermore, the KEGG database was used to
facilitate construction of gene networks according to associations
among these genes, proteins and compounds in the database. Based on
this computed signaling network, PPAR, MAPK and ras homolog family
member A were identified as being central to the establishment of
this pathway network (Fig. 4).
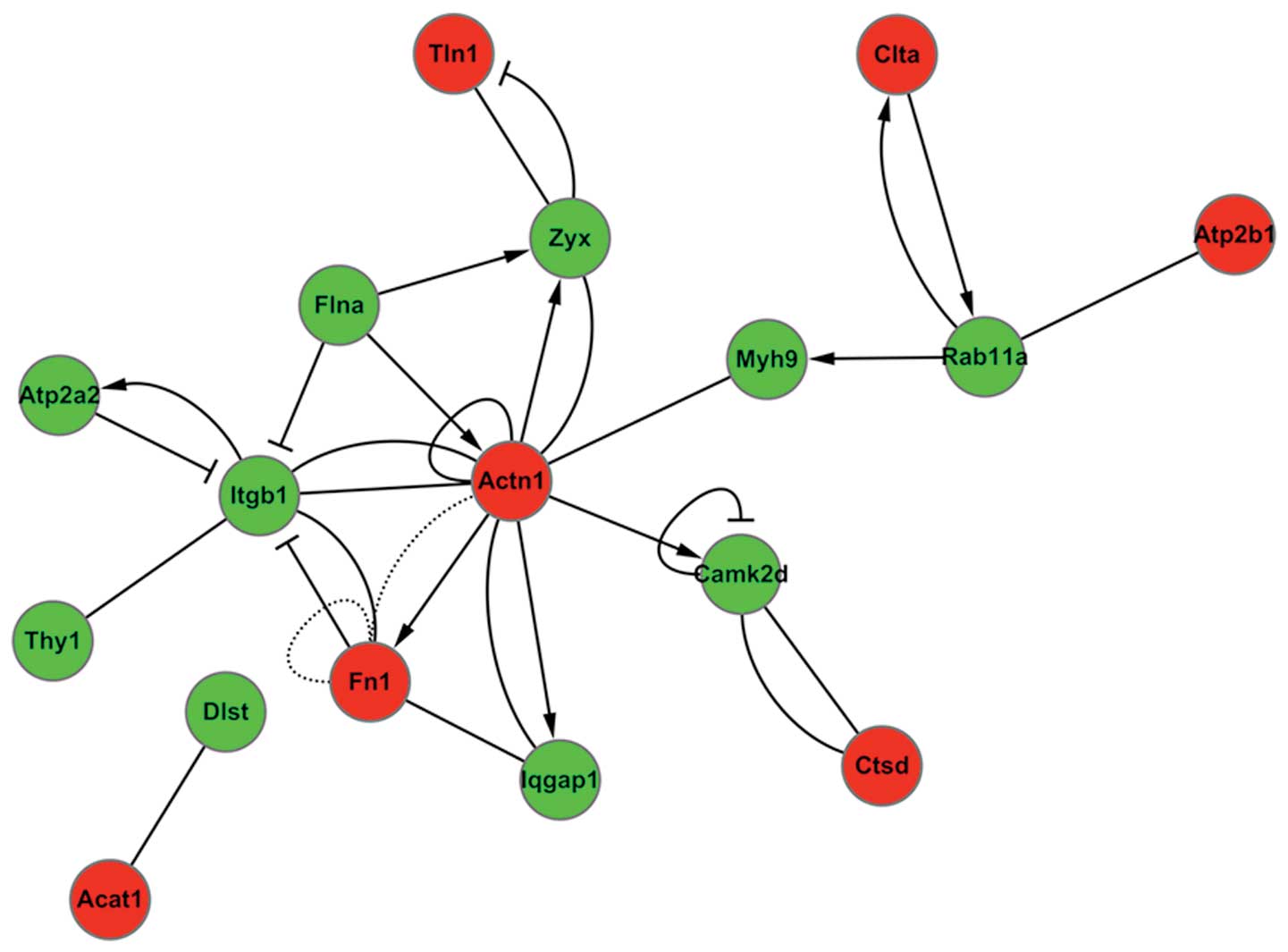 | Figure 4Interactions within the differentially
regulated proteins were identified using biological network
analysis utilities in Ingenuity Pathway Analysis. Green circles
represent downregulated proteins and red circles represent
upregulated proteins. Actn1, actinin-α-1; FN1, fibronectin 1;
Itgb1, integrin-β-1; Myh9, myosin heavy chain 9 non-muscle; Tau,
microtubule-associated protein tau; Rab11a, Rab11a member RAS
oncogene family (Rab11a); Camk2d, calcium/calmodulin-dependent
protein kinase II δ; Atp2b1, ATPase Ca++ transporting
plasma membrane 1; Atp2a2, ATPase, Ca++ transporting,
cardiac muscle, slow twitch 2; Thy-1, Thy-1 cell surface antigen;
Dlst, dihydrolipoamide S-succinyltransferase; Acat1, acetyl-CoA
acetyltransferase 1; Iqgap1, IQ motif containing GTPase activating
protein 1; Flna, filamin A-α; Ctsd, cathepsin D; Zyx, zyxin; Tln1,
talin 1; Clta, clathrin, light chain A. |
Discussion
Numerous epidemiological studies have shown that
elevated total homocysteine in the plasma is correlated with an
increased risk of vascular diseases, including cardiovascular,
peripheral vascular and cerebral vascular diseases (17). HHcy is characterized by a
perturbation of methionine metabolism due to an enzymatic or
vitamin deficiency (18). HHcy
disrupts the coagulation system and is therefore observed to have
prothrombotic effects (19). High
levels of homocysteine can result in neurological abnormalities,
including, cerebral atrophy and seizures (20). Additionally, ectopic administration
of homocysteine in the brain has been shown to enhance neuronal
degeneration (21), likely due to
the effect of elevated levels of homocysteine causing an
accumulation of neuronal DNA damage (22). On a molecular level, increased
homocysteine can cause a deficiency of methyl donors, which may
result in misincorporation of uracil and oxidative damage to DNA
bases (23).
The results of this study demonstrated that several
of the differentially expressed proteins mediated by HHcy are
involved in nerve injury, including: Non-POU domain containing
octamer-binding; structure specific recognition protein 1 (response
to DNA damage stimulus, DNA repair), FN1; annexin A7 (Anxa7); ezrin
(Ezr); Myh9; Myh10 (regulation of cell shape); thioredoxin
reductase 1 (cell proliferation), Camk2b; Camk2d; Itgb1 (G1/S
transition of mitotic cell cycle), Myh10; and capping protein
(actin filament) muscle Z-line α-2 (neuron migration). Other
biological processes identified were the regulation of RNA
splicing; angiogenesis; cell redox homeostasis; calcium ion
transport and actin filament depolymerization.
The IPA results suggested that the differentially
expressed proteins induced by HHcy had involvement in multiple
biological signaling pathways, such as the calcium,
gonadotropin-releasing hormone and Wnt signaling pathways. Camk2d
and Camk2b belong to the serine/threonine protein kinase family and
to the Ca2+/calmodulin-dependent protein kinase
subfamily. Calcium signaling is crucial for maintaining synaptic
plasticity of glutamatergic nerve endings. It was observed that
Camk2 was downregulated in hippocampal neurons by HHcy. It has been
previously shown that Camk2 functions in the following biological
processes: Hypoxia response (24);
cell cycle (25); regulation of
extracellular signal-regulated kinase 1 (ERK1) and ERK2 cascade
(26); and regulation of Rac
protein signal transduction (27).
Under hypoxic conditions, Camk2 can activate E1A binding protein
p300, which is necessary in the transcription of erythropoietin
(EPO) and endothelial nitric oxide synthase (eNOS) (24). Thus, the downregulation of Camk2
may lead to the deficiency of EPO and eNOS, both of which are
important neurotrophic factors. Another important role of Camk2 is
the activation of B-cell CLL/lymphoma 2 expression, which results
in the inhibition of apoptosis under various pathological
conditions (24,27).
Notably, proteins involved in the genesis of
neurodegenerative diseases were observed to be upregulated by high
Hcy expression. One of these proteins was Tau, with various
transcripts differentially expressed in the nervous system,
depending on the stage of neuronal maturation and neuronal types
(28). It has been shown that the
genetic mutations of Tau are associated with several
neurodegenerative disorders, including Alzheimer’s (29) and Pick’s disease (30), frontotemporal dementia (31,32),
cortico-basal degeneration (33)
and progressive supranuclear palsy (34). Tau has multiple phosphorylation
sites, and the numerous phosphorylated forms have different
functions. Tau phosphorylation may affect microtubule stability and
axonal transport, dendritic positioning and synaptic health, cell
signaling at plasma membranes, protection of DNA from cell
stressors, tau release and pathologic propagation (35). In this study, total Tau was
identified to be upregulated, yet the specific phosphorylation
sites remain to be further explored.
In conclusion, this study provides valuable insight
into proteomic changes following induction of HHcy, and offers
indicators as to the molecular effects of HHcy injury on
hippocampal neurons. Further functional studies are likely to be
informative on the roles of the differentially expressed proteins
mediated by HHcy in hippocampal neurons.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (81171163) and Key Projects of
Science and Technology Commission of Shanghai (11411952100).
References
|
1
|
Ross PL, Huang YN, Marchese JN, Williamson
B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S,
Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson
A and Pappin DJ: Multiplexed protein quantitation in
Saccharomyces cerevisiae using amine-reactive isobaric
tagging reagents. Mol Cell Proteomics. 3:1154–1169. 2004.
|
|
2
|
Mangoni AA, Zinellu A, Carru C, Attia JR
and McEvoy M: Serum thiols and cardiovascular risk scores: a
combined assessment of transsulfuration pathway components and
substrate/product ratios. J Transl Med. 11:992013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ito T and Jensen RT: Association of
long-term proton pump inhibitor therapy with bone fractures and
effects on absorption of calcium, vitamin B12, iron, and magnesium.
Curr Gastroenterol Rep. 12:448–457. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhuo JM, Wang H and Praticò D: Is
hyperhomocysteinemia an Alzheimer’s disease (AD) risk factor, an AD
marker, or neither? Trends Pharmacol Sci. 32:562–571. 2011.
|
|
5
|
Wang X, Cui L, Joseph J, Jiang B, Pimental
D, Handy DE, Liao R and Loscalzo J: Homocysteine induces
cardiomyocyte dysfunction and apoptosis through p38 MAPK-mediated
increase in oxidant stress. J Mol Cell Cardiol. 52:753–760. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yin HR, Zhang L, Xie LQ, Huang LY, Xu Y,
Cai SJ, Yang PY and Lu HJ: Hyperplex-MRM: a hybrid multiple
reaction monitoring method using mTRAQ/iTRAQ labeling for multiplex
absolute quantification of human colorectal cancer biomarker. J
Proteome Res. 12:3912–3919. 2013. View Article : Google Scholar
|
|
7
|
Yu Y, Pan X, Ding Y, Liu X, Tang H, Shen
C, Shen H and Yang P: An iTRAQ based quantitative proteomic
strategy to explore novel secreted proteins in metastatic
hepatocellular carcinoma cell lines. Analyst. 138:4505–4511. 2013.
View Article : Google Scholar
|
|
8
|
Akoglu B, Kindl P, Weber N, Trojan J,
Caspary WF and Faust D: Polymorphisms in the
methylenetetrahydrofolate reductase gene are determinant for
vascular complications after liver transplantation. Eur J Clin
Nutr. 62:430–435. 2008. View Article : Google Scholar
|
|
9
|
Zhang L, Zhao Q, Chen CH, Qin QZ, Zhou Z
and Yu ZP: Synaptophysin and the dopaminergic system in hippocampus
are involved in the protective effect of rutin against
trimethyltin-induced learning and memory impairment. Nutr Neurosci.
26:2013.(Epub ahead of print).
|
|
10
|
Boehm AM, Pütz S, Altenhöfer D, Sickmann A
and Falk M: Precise protein quantification based on peptide
quantification using iTRAQ. BMC Bioinformatics. 8:2142007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Galea CA, High AA, Obenauer JC, Mishra A,
Park CG, Punta M, Schlessinger A, Ma J, Rost B, Slaughter CA and
Kriwacki RW: Large-scale analysis of thermostable, mammalian
proteins provides insights into the intrinsically disordered
proteome. J Proteome Res. 8:211–226. 2009. View Article : Google Scholar
|
|
12
|
Zhang M, Huang K, Zhang Z, Ji B, Zhu H,
Zhou K, Li Y, Yang J, Sun L, Wei Z, He G, Gao L, He L and Wan C:
Proteome alterations of cortex and hippocampus tissues in mice
subjected to vitamin A depletion. J Nutr Biochem. 22:1003–1008.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese
JC, Richardson JE, Ringwald M, Rubin GM and Sherlock G: Gene
ontology: tool for the unification of biology. The Gene Ontology
Consortium. Nat Genet. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dupuy D, Bertin N, Hidalgo CA, Venkatesan
K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A,
Carvunis AR, Pulak R, Shingles J, Reece-Hoyes J, Hunt-Newbury R,
Viveiros R, Mohler WA, Tasan M, Roth FP, Le Peuch C, Hope IA,
Johnsen R, Moerman DG, Barabási AL, Baillie D and Vidal M:
Genome-scale analysis of in vivo spatiotemporal promoter activity
in Caenorhabditis elegans. Nat Biotechnol. 25:663–668. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schlitt T, Palin K, Rung J, Dietmann S,
Lappe M, Ukkonen E and Brazma A: From gene networks to gene
function. Genome Res. 13:2568–2576. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yi M, Horton JD, Cohen JC, Hobbs HH and
Stephens RM: WholePathwayScope: a comprehensive pathway-based
analysis tool for high-throughput data. BMC Bioinformatics.
7:302006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He B, Gai L, Gai J, Qiao H, Zhang S and
Guan Z: Coronary plaques identified by coronary computed tomography
angiography and the risk factors for major adverse cardiac events:
a correlation analysis. Nan Fang Yi Ke Da Xue Xue Bao.
32:1400–1406. 2012.(In Chinese).
|
|
18
|
Selicharová I, Kořínek M, Demianová Z,
Chrudinová M, Mládková J and Jiráček J: Effects of
hyperhomocysteinemia and betaine-homocysteine S-methyltransferase
inhibition on hepatocyte metabolites and the proteome. Biochim
Biophys Acta. 1834:1596–1606. 2013.PubMed/NCBI
|
|
19
|
Kolodziejczyk-Czepas J, Talar B, Nowak P,
Olas B and Wachowicz B: Homocysteine and its thiolactone impair
plasmin activity induced by urokinase or streptokinase in vitro.
Int J Biol Macromol. 50:754–758. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Willette AA, Gallagher C, Bendlin BB,
McLaren DG, Kastman EK, Canu E, Kosmatka KJ, Field AS, Alexander
AL, Colman RJ, Voytko ML, Weindruch RH, Coe CL and Johnson SC:
Homocysteine, neural atrophy, and the effect of caloric restriction
in rhesus monkeys. Neurobiol Aging. 33:670–680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tagliari B, Zamin LL, Salbego CG, Netto CA
and Wyse AT: Homocysteine increases neuronal damage in hippocampal
slices receiving oxygen and glucose deprivation. Metab Brain Dis.
21:273–278. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuwahara K, Nanri A, Pham NM, Kurotani K,
Kume A, Sato M, Kawai K, Kasai H and Mizoue T: Serum vitamin B6,
folate, and homocysteine concentrations and oxidative DNA damage in
Japanese men and women. Nutrition. 29:1219–1223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matté C, Mackedanz V, Stefanello FM,
Scherer EB, Andreazza AC, Zanotto C, Moro AM, Garcia SC, Gonçalves
CA, Erdtmann B, Salvador M and Wyse AT: Chronic
hyperhomocysteinemia alters antioxidant defenses and increases DNA
damage in brain and blood of rats: protective effect of folic acid.
Neurochem Int. 54:7–13. 2009.PubMed/NCBI
|
|
24
|
Melvin A, Mudie S and Rocha S: Further
insights into the mechanism of hypoxia-induced NFκB. (corrected).
Cell Cycle. 10:879–882. 2011.
|
|
25
|
Brassac T, Castro A, Lorca T, Le Peuch C,
Dorée M, Labbé JC and Galas S: The polo-like kinase Plx1 prevents
premature inactivation of the APC(Fizzy)-dependent pathway in the
early Xenopus cell cycle. Oncogene. 19:3782–3790. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Burger LL, Haisenleder DJ, Aylor KW and
Marshall JC: Regulation of intracellular signaling cascades by GNRH
pulse frequency in the rat pituitary: roles for CaMK II, ERK, and
JNK activation. Biol Reprod. 79:947–953. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Arsenault RJ, Kogut MH and He H: Combined
CpG and poly I:C stimulation of monocytes results in unique
signaling activation not observed with the individual ligands. Cell
Signal. 25:2246–2254. 2013. View Article : Google Scholar
|
|
28
|
Zaheer S, Thangavel R, Wu Y, Khan MM,
Kempuraj D and Zaheer A: Enhanced expression of glia maturation
factor correlates with glial activation in the brain of triple
transgenic Alzheimer’s disease mice. Neurochem Res. 38:218–225.
2013.PubMed/NCBI
|
|
29
|
Zhao Y and Zhao B: Oxidative stress and
the pathogenesis of Alzheimer’s disease. Oxid Med Cell Longev.
2013:3165232013.
|
|
30
|
Mattsson N, Zetterberg H, Bianconi S,
Yanjanin NM, Fu R, Mansson JE, Porter FD and Blennow K: Miglustat
treatment may reduce cerebrospinal fluid levels of the axonal
degeneration marker tau in niemann-pick type C. JIMD Rep. 3:45–52.
2012. View Article : Google Scholar
|
|
31
|
Chaunu MP, Deramecourt V, Buée-Scherrer V,
Le Ber I, Brice A, Ehrle N, El Hachimi K, Pluot M, Maurage CA,
Bakchine S and Buée L: Juvenile frontotemporal dementia with
parkinsonism associated with tau mutation G389R. J Alzheimers Dis.
37:769–776. 2013.PubMed/NCBI
|
|
32
|
Dang TN, Dobson-Stone C, Glaros EN, Kim
WS, Hallupp M, Bartley L, Piguet O, Hodges JR, Halliday GM, Double
KL, Schofield PR, Crouch PJ and Kwok JB: Endogenous progesterone
levels and frontotemporal dementia: modulation of TDP-43 and Tau
levels in vitro and treatment of the A315T TARDBP mouse model. Dis
Model Mech. 6:1198–1204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Casseron W, Azulay JP, Guedj E, Gastaut JL
and Pouget J: Familial autosomal dominant cortico-basal
degeneration with the P301S mutation in the tau gene: an example of
phenotype variability. J Neurol. 252:1546–1548. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iwasaki Y, Mori K, Ito M, Tatsumi S,
Mimuro M and Yoshida M: An autopsied case of progressive
supranuclear palsy presenting with cerebellar ataxia and severe
cerebellar involvement. Neuropathology. 33:561–567. 2013.
|
|
35
|
Hashiguchi M and Hashiguchi T:
Kinase-kinase interaction and modulation of tau phosphorylation.
Int Rev Cell Mol Biol. 300:121–160. 2013. View Article : Google Scholar : PubMed/NCBI
|