Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies worldwide. HCC incidence continues to increase,
ranking as the second leading cause of cancer-related mortality
among males in China (1). Similar
to other cancer types, HCC is characterized by an evident
multistage process of tumor progression (2). In spite of the developments in
surgical treatment strategies and options made available in recent
years, the overall prognosis of HCC patients remains extremely
poor, and this is largely due to the high frequency of recurrence
or metastasis following surgery (3). Thus, the capacity to subjectively
predict the risk of recurrence and subsequent prognosis is
essential to guide surgery and chemotherapy.
The oncogene DEK was originally identified as
one of the parts of the DEK-CAN fusion gene, arising from
the translocation (6;9) in a subtype of acute myeloid leukemia
patients (4). The overexpression
of DEK has been observed in several malignancies including
melanoma, bladder cancer, glioblastoma, retinoblastoma and HCC
(5–9). The increasing list of tumor types
demonstrating high and easily detectable DEK protein expression
indicates the potential of using DEK as a tumor marker (9). A previous study has characterized
DEK as a potential urinary and tissue biomarker for
transitional cell carcinoma of the bladder of various grades,
stages and progression (10).
Furthermore, since the amount of DEK expressed in immature
cells is greater than that in differentiated cells, it could assist
in assessing the differentiation potential of tumor cells (11). Previous studies have identified
that DEK overexpression may be linked with the progression
of breast cancer, and that DEK may possibly be used not only
as a therapeutic target, but also as a breast cancer biomarker for
the early diagnosis and prognostic evaluation of breast cancer
(12–14).
While DEK overexpression has been observed in
HCC, little is known about its clinical significance in this
context (6,15–16).
In the present study, we examined the expression level of
DEK mRNA and protein in HCC surgical specimens and matched
normal hepatic tissues, and then analyzed the correlation between
DEK expression and clinicopathological characteristics and
patient prognosis.
Materials and methods
Patients and specimens
Fifty-five pairs of HCC tissues and their adjacent
non-cancerous tissues were obtained from patients who had undergone
surgical resection at Xiamen Zhongshan Hospital affiliated to
Xiamen University, China, between July 2007 and November 2010. The
histological diagnoses and tumor grades of all samples were
confirmed by an experienced pathologist. The tumor grading was
determined based on the Edmondson-Steiner classification (17). Patient characteristics collected
for analysis included age, gender, tumor size, Edmondson-Steiner
grade, hepatitis history, number of tumor nodules and presence of
portal venous invasion. None of the patients engaged in this study
received chemotherapy or radiotherapy prior to surgery. Follow-up
data was obtained following hepatic resection for all 55 patients.
The follow-up period was defined as the interval between the date
of the surgery and that of the patient’s mortality or the last
follow-up. Mortality from other causes was treated as censored
cases. This study was authorized by the Research Ethics Committee
of Xiamen Zhongshan Hospital affiliated to Xiamen University.
Informed consent was obtained from all of the patients. The tissues
were collected according to local ethical guidelines and approved
beforehand by all participants and the Human Investigation
Committee of The Medical College of Xiamen University (Xiamen,
China).
All samples were managed anonymously on
the basis of ethical and legal standards
For RNA preparation, the resected samples were
placed in liquid nitrogen as soon as possible following resection,
and stored at −80°C. The remaining tissues were used for routine
histopathological examination.
Isolation of total RNA and first strand
cDNA synthesis
Total RNA was isolated from frozen tumor and matched
normal tissues using TRIzol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA) according to the manufacturer’s instructions.
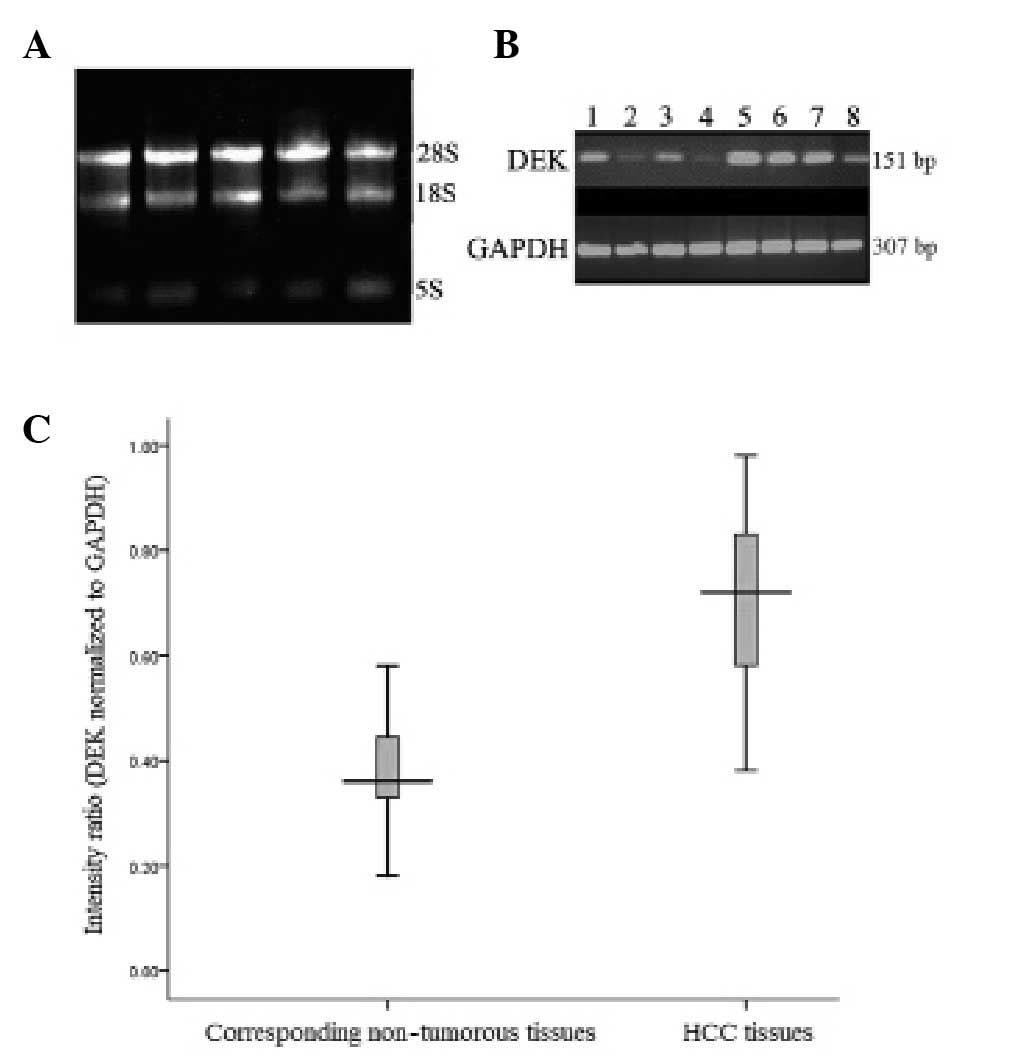
RNA quality was confirmed by agarose gel electrophoresis. Only RNA
without DNA contamination and biodegradable 26S rRNA was prepared
for succeeding cDNA synthesis (Fig.
1A). Following photometric quantification, 2 μg total RNA was
used for a 20 μl reverse transcription (RT) reaction containing 4
μl 5× first strand buffer (Promega Corporation, Madison, WI, USA),
2 μl 0.1 M DTT, 1 μl oligo(dT)15 primer (10 mM), 1 μl
dNTPs (10 mM) and diethylpyrocarbonate-treated water. The reaction
mixture was incubated for 1 h at 42°C, heated for 5 min at 95°C and
placed for 5 min in an ice bath; the first strand of cDNA was
either stored at 4°C or used immediately for polymerase chain
reaction (PCR).
PCR
Primer sequences were as follows:
5′-TCTGTGAGGTTCTTGATTTGGA-3′ (forward) and
5′-CTGTTCCGTTCCTTTTTACTGC-3′ (reverse) for DEK;
5′-ACCTGACCTGCCGTCTAGAA-3′ (forward) and 5′-TCCACCACCCTGTTGCTGTA-3′
(reverse) for GAPDH. PCR analysis was performed under the following
conditions: 4 min at 94°C, 30 cycles of denaturation for 20 sec at
94°C, annealing for 30 sec at 60°C, extension for 40 sec at 72°C,
and finally 5 min at 72°C. The amplification products were analyzed
on a 1.5% agarose gel. The intensity ratio was the relative
expression of DEK normalized to that of GAPDH.
Immunohistochemical staining
A total of 55 HCC tissues and matched normal liver
tissues were evaluated by immunohistochemistry. Multiple 5-μm
sections were prepared from formalin-fixed, paraffin-embedded
tissue blocks. Sections were placed overnight at 4°C with mouse
anti-human DEK monoclonal antibody (1:100, (Proteintech Group,
Inc., Chicago, IL, USA). The streptavidin-biotin peroxidase complex
tertiary system (Boster Bio, Pleasanton, CA, USA) was applied in
accordance with the manufacturer’s specifications. Sections were
counter-stained using hematoxylin, dehydrated via gradient alcohol
and fixed for observation. All sections were examined independently
by two pathologists who were unaware of the patients’ clinical
status. With regard to the percentage of DEK-positive hepatocytes,
immunohistochemical staining was scored as follows: 0, <5%
positive; 1+, 5–25% positive; 2+, 26–50% positive; and 3+, >50%
positive. Only nuclear expression was considered to be positive
staining. Tissues with moderate to strong nuclear staining were
designated as the DEK high expression group (scores 2+ and 3+).
Tissues designated as the DEK low expression group were either
devoid of any nucleus staining or scored as 0 or 1+.
Statistical analysis
SPSS version 16.0 for Windows (SPSS Inc., Chicago,
IL, USA) was used for all data analyses. The paired-samples t-test
was used to compare the intensity ratio between HCC tissues and
matching non-cancerous livers. Pearson’s χ2 test was
used to assess differences in the rate of positive immunostaining
between hepatic cancer tissues and adjacent tissues. An independent
Student’s t-test and Pearson’s χ2 test were used to
evaluate the correlations between DEK mRNA and protein
expression and clinicopathological variables of HCCs, respectively.
The Kaplan-Meier method was employed to analyze patient survival,
and the differences in survival were evaluated using the log-rank
test. The Cox proportional-hazards model was used to confirm
factors independently associated with survival. All P-values were
based on two-sided statistical analyses, and P<0.05 was
considered to indicate a statistically significant difference.
Results
Significantly increased DEK mRNA and
protein expression in HCC tissues
The expression of DEK mRNA was analyzed in 55
HCC samples and matched normal hepatic tissues. Higher expression
levels of DEK mRNA were observed in 50 of the HCC tissue
samples compared with the matched non-tumor hepatic tissues.
DEK mRNA levels in HCC tissues were significantly higher
than those in corresponding non-tumorous livers (0.707±0.157 versus
0.391±0.116; t=18.3, P<0.001; Fig.
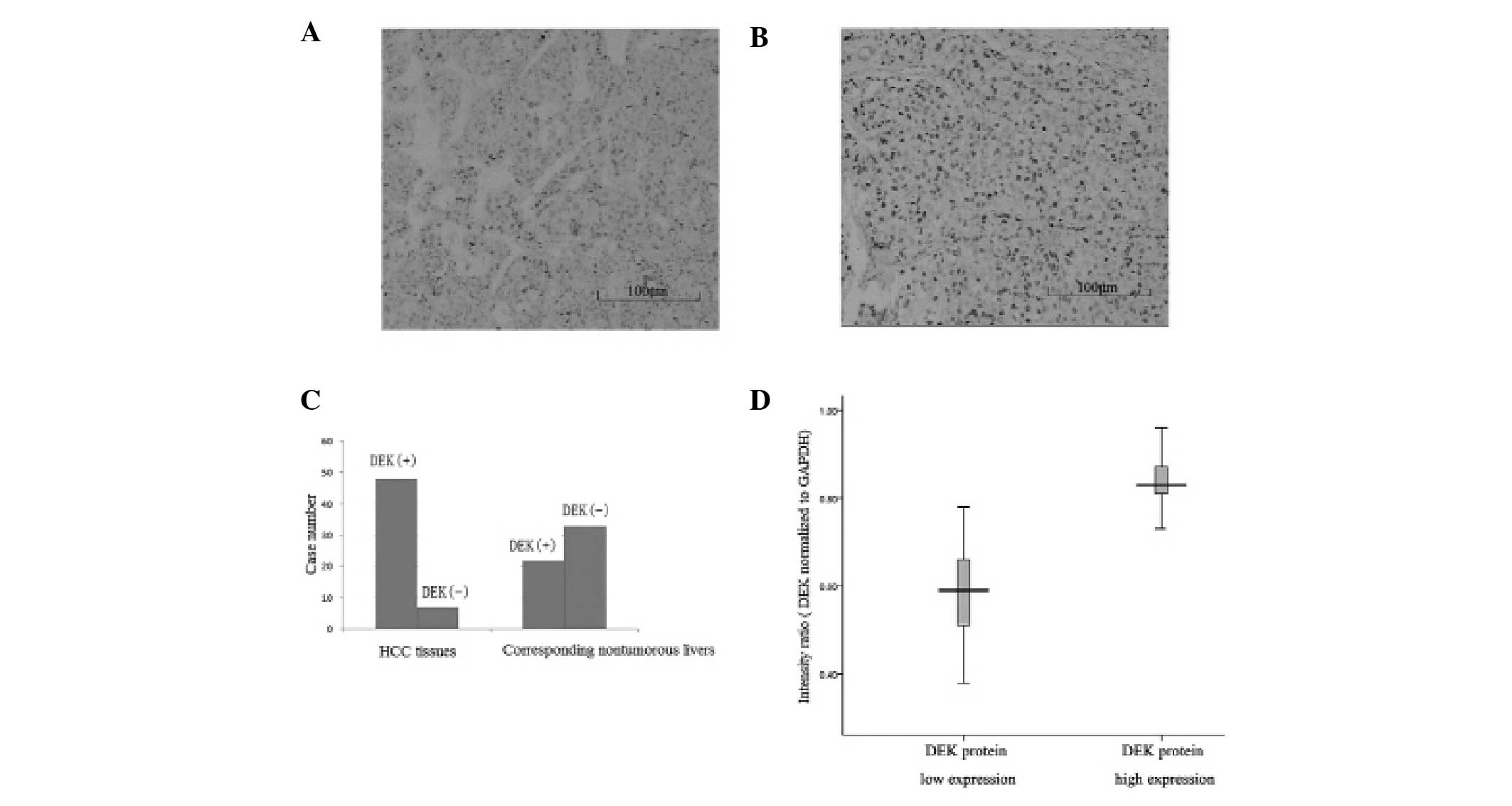
1B and C). By immunohistochemical analysis, the percentage of
positive DEK expression in HCC tissues was significantly higher
than that in corresponding normal livers [87.3% (48 of 55) versus
40.0% (22 of 55); P=0.002; Fig.
2C]. Moreover, the DEK protein levels in HCC tissues were
significantly higher than those in the corresponding non-tumorous
livers (P<0.01, Fig. 2A and B).
Furthermore, an increase in nuclear expression of DEK was observed
in HCC tumor tissues, both in intensity and in the positive
proportion.
On the basis of the immunohistochemistry data, the
55 HCC cases were divided into the low expression group (score 0 or
1+; n=29) and the high expression group (score 2+ or 3+; n=26). The
DEK mRNA levels in the DEK high expression group were
significantly higher than those in the low expression group
(0.841±0.073 versus 0.587±0.106; P<0.001; Fig. 2D).
Taken together, these data confirm the
overexpression of DEK at the transcription and translation
level in human HCC.
Correlation between DEK expression and
clinicopathological characteristics
The immunohistochemistry data were analyzed for the
correlation of DEK protein expression with clinicopathological
features. The expression of DEK protein showed a positive
correlation with tumor size (P=0.001), Edmondson-Steiner grade
(P=0.025) and portal venous invasion (P=0.002). However, no
significant correlation was observed between DEK protein expression
and other clinical characteristics, including age (P=0.143), gender
(P=0.385), hepatitis history (P=0.589), number of tumor nodules
(P=0.795) and liver cirrhosis (P=0.418) (Table I). The mRNA levels were compared
with the clinical data and found to be associated with the same
clinical variables as observed in the protein level analysis.
 | Table ICorrelations between DEK protein
expression and clinicopathological variables in 55 cases of
hepatocellular carcinoma. |
Table I
Correlations between DEK protein
expression and clinicopathological variables in 55 cases of
hepatocellular carcinoma.
| | DEK expression | | |
|---|
| |
| | |
|---|
| Clinicopathological
features | n | Low | High | χ2 | P-value |
|---|
| Gender |
| Male | 43 | 24 | 19 | 0.753 | 0.385 |
| Female | 12 | 5 | 7 | | |
| Age (years) |
| <50 | 26 | 11 | 15 | 2.148 | 0.143 |
| ≥50 | 29 | 18 | 11 | | |
| Tumor size
(cm) |
| ≤5 | 23 | 18 | 5 | 10.34 | 0.001a |
| >5 | 32 | 11 | 21 | | |
| Hepatitis
history |
| Yes | 44 | 24 | 20 | 0.292 | 0.589 |
| No | 11 | 5 | 6 | | |
| Edmondson-Steiner
grade |
| 1 and 2 | 14 | 11 | 3 | 5.033 | 0.025a |
| 3 and 4 | 41 | 18 | 23 | | |
| Portal venous
invasion |
| Absent | 15 | 13 | 2 | 9.531 | 0.002a |
| Present | 40 | 16 | 24 | | |
| Tumor nodule
no. |
| Solitary | 39 | 21 | 18 | 0.067 | 0.795 |
| Multiple (≥2) | 16 | 8 | 8 | | |
| Liver
cirrhosis |
| Absent | 11 | 7 | 4 | 0.656 | 0.418 |
| Present | 44 | 22 | 22 | | |
Follow-up and prognostic value of
DEK
The Kaplan-Meier method was used to analyze the
correlation of DEK expression level with the prognosis of
HCC patients. The results indicated that the DEK high expression
group had a shorter median survival time than the low expression
group (23 versus 39 months), and the overall survival rates for the
low and high expression groups were significantly different
(P=0.003, log-rank test; Fig.
3).
To elucidate factors that may predict survival
following hepatic resection, univariate and multivariate Cox
regression analyses were applied. In the univariate analysis, DEK
expression level [hazard risk (HR), 2.273; P=0.022),
Edmondson-Steiner grade (HR, 1.542; P=0.039) and portal venous
invasion (HR, 3.145; P=0.015) were all significantly associated
with survival. However, age, gender, tumor size, hepatitis history,
number of tumor nodules and liver cirrhosis were not significantly
correlated with survival (Table
II). In the multivariate analysis, DEK expression level (HR,
2.974; P=0.017), Edmondson-Steiner grade (HR, 2.065; P=0.026) and
portal venous invasion (HR, 1.967; P=0.028) were identified to be
independent prognostic factors of survival (Table II).
 | Table IIMultivariate analysis using a Cox
proportional-hazards regression model for hepatocellular carcinoma
patients. |
Table II
Multivariate analysis using a Cox
proportional-hazards regression model for hepatocellular carcinoma
patients.
| | Univariate
analysis | Multivariate
analysis |
|---|
| |
|
|
|---|
| Clinicopathological
variables | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Edmondson-Steiner
grade | | | 0.039 | | 0.026 |
| 1 and 2 | 14 | 1 | | 1 | |
| 3 and 4 | 41 | 1.542
(1.026–2.746) | | 2.065
(1.185–2.934) | |
| Portal venous
invasion | | | 0.015 | | 0.028 |
| Absent | 15 | 1 | | 1 | |
| Present | 40 | 3.145
(1.943–5.627) | | 1.967
(1.173–2.836) | |
| DEK | | | 0.022 | | 0.017 |
| Low
expression | 29 | 1 | | 1 | |
| High
expression | 26 | 2.273
(1.474–3.643) | | 2.974
(1.725–4.139) | |
Discussion
DEK was originally identified in the fusion
protein DEK-CAN, resulting from the recurrent t(6;9)
translocation in a subset of acute myeloid leukemia patients
(4). In addition, previous studies
have suggested that DEK is ubiquitously expressed in the
majority of mammalian cells (18–19).
Subsequently, DEK was reported to be frequently upregulated
in aggressive human tumors including melanoma, glioblastoma,
retinoblastoma, bladder cancer and HCC (5–8,15–16).
DEK has been proven to boost tumorigenesis in numerous
cancer cell types through its function of intervening with cell
division; inhibiting cell differentiation, senescence and apoptosis
(20–22).
In the present study, we measured DEK mRNA
and protein expression levels in 55 HCC tissue samples paired with
adjacent non-cancerous tissues. The study revealed that levels of
DEK mRNA were significantly higher in HCC tissues than in
the matching non-cancerous tissues (P<0.001). Although the
present study had a higher number of clinical cases, the result was
consistent with that from a previous HCC study, which demonstrated
that DEK mRNA levels were higher in 4 of 5 primary HCCs
compared with matched non-tumorous liver tissues (13). Lü et al also demonstrated
that DEK mRNA levels in HCC tissues were higher than those
in paraneoplastic tissues (6).
Furthermore, DEK protein expression was analyzed by
immunohistochemistry in the same 55 paired specimens. The
percentage of cells with positive DEK expression in HCC tissues was
significantly higher than that in corresponding non-tumorous livers
(87.3 versus 40.0%; P=0.002), comparable with the results from a
previous study of DEK protein expression in HCC samples (14). The immunohistochemistry results
were in agreement with the mRNA analyses in this study. An increase
in nuclear expression of DEK was observed in HCC tumor tissues,
both in intensity and in the positive percentage of cells.
In the current study, we divided the HCC cases into
low (n=29) and high (n=26) expression groups based on
immunohistochemistry scores. DEK mRNA levels in the high
expression group were significantly higher than those in the low
expression group (P<0.001). Taken together, these results
suggest that overexpression of DEK may be a common event in
HCC tumorigenesis.
In this study, the DEK expression data
obtained from RT-PCR and immunohistochemistry were analyzed for
correlation with clinicopathological features. The results revealed
that the expression levels of DEK mRNA and protein were
correlated with pathological grade, tumor size and portal venous
invasion. Significant differences in the expression levels of
DEK mRNA and protein were observed when comparing grade 1
and 2 HCC with grade 3 and 4 HCC. These results are in agreement
with a previous HCC study which reported that the level of
DEK mRNA was correlated with the histological grade
(15). However, Lü et al
reported that the difference in expression of DEK mRNA
between well- and poorly differentiated HCC was not statistically
significant (P>0.05) (6).
In the present study, the results demonstrated that
increased expression of DEK protein was significantly correlated
with poor patient outcomes. However, age, gender, tumor size,
hepatitis history, number of tumor nodules and liver cirrhosis had
no effect on overall survival (P>0.05), whereas DEK expression,
Edmondson-Steiner grade and portal venous invasion were significant
predictors of overall survival (P<0.05). The association of DEK
expression with overall survival was consistent with a previous
study which demonstrated that increased DEK expression was
significantly correlated with poor survival in breast cancer
patients (12).
A number of tumorigenic functions of DEK may
contribute to the inverse correlation between DEK expression
and survival of HCC patients. First, DEK is involved in the
inhibition of differentiation and facilitation of cellular
transformation (20). Second,
DEK inhibits cell apoptosis through its interference with
p53 functions (24). Lastly,
DEK overexpression has been reported to extend cellular life
span, supporting the role of DEK as a senescence inhibitor
(25).
In summary, overexpression of DEK in human
HCC is significantly correlated with the prognosis and
differentiation potential of HCC, suggesting that DEK may
serve as a useful prognostic marker. Further studies should be
carried out to investigate the precise function and molecular
mechanism of DEK in the progression of HCC.
Acknowledgements
The authors sincerely thank the patients and their
families for their cooperation and participation in the study, as
well as the staff involved with the study for data collection and
management.
References
|
1
|
Murray CJ and Lopez AD: Mortality by cause
for eight regions of the world: Global Burden of Disease Study.
Lancet. 349:1269–1276. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sakamoto M: Pathology of early
hepatocellular carcinoma. Hepatol Res. 37(Suppl 2): S135–S138.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tung-Ping Poon R, Fan ST and Wong J: Risk
factors, prevention, and management of postoperative recurrence
after resection of hepatocellular carcinoma. Ann Surg. 232:10–24.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
von Lindern M, Fornerod M, van Baal S, et
al: The translocation (6;9), associated with a specific subtype of
acute myeloid leukemia, results in the fusion of two genes, dek and
can, and the expression of a chimeric, leukemia-specific dek-can
mRNA. Mol Cell Biol. 12:1687–1697. 1992.PubMed/NCBI
|
|
5
|
Carro MS, Spiga FM, Quarto M, et al: DEK
expression is controlled by E2F and deregulated in diverse tumor
types. Cell Cycle. 5:1202–1207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lü ZL, Luo DZ and Wen JM: Expression and
significance of tumor-related genes in HCC. World J Gastroenterol.
11:3850–3854. 2005.PubMed/NCBI
|
|
7
|
Kappes F, Khodadoust MS, Yu L, et al: DEK
expression in melanocytic lesions. Hum Pathol. 42:932–938. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khodadoust MS, Verhaegen M, Kappes F, et
al: Melanoma proliferation and chemoresistance controlled by the
DEK oncogene. Cancer Res. 69:6405–6413. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng J, Kohler ME, Chen Q, et al: Serum
from mice immunized in the context of Treg inhibition identifies
DEK as a neuroblastoma tumor antigen. BMC Immunol. 8:42007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Datta A, Adelson ME, Mogilevkin Y, et al:
Oncoprotein DEK as a tissue and urinary biomarker for bladder
cancer. BMC Cancer. 11:2342011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wise-Draper TM, Morreale RJ, Morris TA, et
al: DEK proto-oncogene expression interferes with the normal
epithelial differentiation program. Am J Pathol. 174:71–81. 2009.
View Article : Google Scholar :
|
|
12
|
Liu S, Wang X, Sun F, et al: DEK
overexpression is correlated with the clinical features of breast
cancer. Pathol Int. 62:176–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Abba MC, Sun H, Hawkins KA, et al: Breast
cancer molecular signatures as determined by SAGE: correlation with
lymph node status. Mol Cancer Res. 5:881–890. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Privette Vinnedge LM, McClaine R, Wagh PK,
et al: The human DEK oncogene stimulates β-catenin signaling,
invasion and mammosphere formation in breast cancer. Oncogene.
30:2741–2752. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kondoh N, Wakatsuki T, Ryo A, et al:
Identification and characterization of genes associated with human
hepatocellular carcinogenesis. Cancer Res. 59:4990–4996.
1999.PubMed/NCBI
|
|
16
|
Lin LJ and Chen LT: The role of DEK
protein in hepatocellular carcinoma for progression and prognosis.
Pak J Med Sci. 29:778–782. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edmondson H and Steiner P: Primary
carcinoma of the liver: a study of 100 cases among 48,900
necropsies. Cancer. 7:462–503. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sitwala KV, Adams K and Markovitz DM: YY1
and NF-Y binding sites regulate the transcriptional activity of the
dek and dek-can promoter. Oncogene. 21:8862–8870. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu HG, Scholten I, Gruss C and Knippers R:
The distribution of the DEK protein in mammalian chromatin. Biochem
Biophys Res Commun. 358:1008–1014. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Grasemann C, Gratias S, Stephan H, et al:
Gains and overexpression identify DEK and E2F3 as targets of
chromosome 6p gains in retinoblastoma. Oncogene. 24:6441–6449.
2005.PubMed/NCBI
|
|
21
|
Evans AJ, Gallie BL, Jewett MA, et al:
Defining a 0.5-mb region of genomic gain on chromosome 6p22 in
bladder cancer by quantitative-multiplex polymerase chain reaction.
Am J Pathol. 164:285–293. 2004. View Article : Google Scholar
|
|
22
|
Castellano G, Torrisi E, Ligresti G, et
al: The involvement of the transcription factor Yin Yang 1 in
cancer development and progression. Cell Cycle. 8:1367–1372. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wise-Draper TM, Mintz-Cole RA, Morris TA,
et al: Overexpression of the cellular DEK protein promotes
epithelial transformation in vitro and in vivo. Cancer Res.
69:1792–1799. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wise-Draper TM, Allen HV, Jones EE, et al:
Apoptosis inhibition by the human DEK oncoprotein involves
interference with p53 functions. Mol Cell Biol. 26:7506–7519. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wise-Draper TM, Allen HV, Thobe MN, et al:
The human DEK proto-oncogene is a senescence inhibitor and an
upregulated target of high-risk human papillomavirus E7. J Virol.
79:14309–14317. 2005. View Article : Google Scholar : PubMed/NCBI
|