Introduction
MicroRNAs (miRs) are a series of RNAs between 14 and
20 nt in length, which regulate the biological functions of cells
by acting on the translation of target proteins (1,2).
Several types of miR are significantly differentially expressed in
tumor tissues compared with normal tissues. miR-15 and miR-16 in
mononuclear cells in peripheral blood were shown to be absent or
deficient among >68% of patients with chronic lymphocytic
leukemia (3). The expression of
Let-7 was shown to be downregulated in lung cancer, while miR-143
and miR-145 were downregulated in colorectal cancer (4,5).
Therefore, it was suggested that these miRs may be closely
associated with the development and progression of cancer.
Lung cancer has rapidly increasing rates of
morbidity and mortality. According to a report by the World Health
Organization on global rates of cancer in 2008, lung cancer had the
highest rates of morbidity and mortality among different types of
tumor (6). On initial diagnosis,
certain patients present with tumor migration or other severe
complications, resulting in poor prognosis despite appropriate
treatment. Therefore, novel effective biomarkers for early
diagnosis and treatment are urgently required.
Since it was demonstrated that the expression of
let-7 is significantly reduced in lung cancer patients with
relatively poor prognosis, evidence on the association between miRs
and lung cancer has increased (4).
It has also been observed that the increased expression of miR-21
induces the proliferation of cancer cells in nude mice, indicating
enrichment of mRNAs encoding regulators of cell cycle checkpoints
(7).
However, few studies have been performed on the
expression and, in particular, the biofunctions of miR-125b in lung
cancer (8,9). In the present study, the expression
profile of human bronchial epithelial (16HBE) cells and lung cancer
(95D) cells were compared by miR microarray analysis. In addition,
bioinformatic analysis was performed to analyze the putative
targets of miR-125b and the possible regulatory mechanism. To
further examine the possible biological functions of miR-125b
involved in the development and progression of lung cancer, the
effects of miR-125b on the proliferation, apoptosis, cell cycle and
invasive ability of the 95D cell lines were investigated. The
results are the first, to the best of our knowledge, to demonstrate
the function of miR-125b in lung cancer cells.
Materials and methods
Cell culture and RNA preparation
The 95D and 16HBE cells were obtained from the
Chinese Academy of Sciences Type Culture Collection Cell Library
Committee (Shanghai, China). These cells were cultured in RPMI-1640
cell culture medium supplemented with 10% fetal bovine serum (FBS;
HyClone, GE Healthcare Life Sciences, Little Chalfont, UK), 100
units/ml penicillin and 100 units/ml streptomycin sulfate in a
humidified 5% CO2 incubator at 37°C. The total RNA was
isolated using TRIzol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA), resolved in nuclease-free water and preserved
at −80°C.
miR microarray analysis
The total RNA was purified to remove any mRNA prior
to sequencing. A GeneChip miRNA 2.0 array (Affymetrix, Santa Clara,
CA, USA) was used to analyze the miRNA expression profile and
one-way analysis of variance (ANOVA) was used to identify any
significant differences among the groups. P<0.05 was considered
to indicate a statistically significant difference. A fold change
>2 was used as the standard in identifying the differentially
expressed miRs.
miR bioinformatics analysis
The prediction of target candidates of miR-125b was
performed using miRGen targets on three authority target predicting
software programs, TargetScan 5.1 (http://www.targetscan.org/), Pictar (http:/pictar.mdc-berlin.de/) and miRanda (http://www.microrna.org/microrna/home.do). The
miR-125b targets were then entered into the DAVID Bioinformatics
Resources 6.7 software to perform Gene Ontology (GO) annotation. In
addition, DIANA-miRPath software (http://diana.cslab.ece.ntua.gr/pathways/) was used to
perform online gene enrichment analysis of the miR-125b putative
targets, which compared the set of miR-125b targets to all known
Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/) pathways. Subsequently, the
information containing the numbers and P-values, in the form ‘In
(P-value)’, of the enrichment genes in the specific pathways were
examined. The interaction and reaction networks between the genes
were also analyzed according to the pathway data provided by the
KEGG pathway database.
Transfection of anti-miR-125b
The transfections were performed using siPORT NeoFX
transfection reagent (Invitrogen Life Technologies) according to
the manufacturer’s instructions. The cells were trypsinized at the
logarithmic growth period and were resuspended in normal growth
medium at 1×105 cells/ml. The siPORT NeoFX transfection
reagent was diluted (1:30) in Opti-MEM I (Invitrogen Life
Technologies) and incubated for 10 min at room temperature.
Subsequently, anti-miR-125b (Invitrogen Life Technologies) and
anti-negative control (Anti-miR™ Negative Control #1, AM17010;
Invitrogen Life Technologies) were diluted in Opti-MEM I. The
transfection reagents were mixed and incubated at room temperature
for 10 min. The transfection complexes were dispensed into the
empty wells of a six-well culture plate. The cells were added to
the transfection complexes and were incubated at 37°C for 24 h.
Following transfection, reverse transcription quantitative
polymerase chain reaction (RT-qPCR) was performed to examine the
expression levels in the miR-125b-transfected cells and to confirm
the transfection efficiency. RT-qPCR analyses for miR-125b were
performed using TaqMan microRNA assays (Applied Biosystems Life
Technologies, Foster City, CA, USA).All reagents, primers and
probes were obtained from Applied Biosystems. The 15 μl RT reaction
consisted of 7 μl Master mix (100 mM dNTPs (with dTTP) 0.15 μl;
MultiScribe™ Reverse Transcriptase, 50 U/μl 1.00 μl; 10X Reverse
Transcription buffer 1.50 μl; RNase Inhibitor, 20 U/μl 0.19 μl;
Nuclease-free water 4.16 μl), 3 μl stem-loop RT primer (ID 000449),
and 5 μl RNA sample. The reactions were incubated on ice for 5 min
and followed by 1°C for 30 min, 42°C for 30 min, 85°C for 5 min and
held in 4°C. The 20 μl PCR reaction included product from RT
reaction (minimum 1:15 dilution) 1.33 μl; TaqMan MicroRNA assay
(20X) 1.00 μl (ID 000449); TaqMan 2X Universal PCR Master mix, No
AmpErase UNGa 10.00 μl; Nuclease-free water 7.67 μl. Amplification
was performed using the Step One Plus Detection System (Applied
Biosystems), and initiated at 95°C for 10 min, followed by 40
cycles of denaturation at 95°C for 15 sec, 60°C for 1 min and
annealing and extension at 60°C for 1 min.
Detection of cell proliferation
An MTT assay was used to determine the cell
proliferation. The cells (1×104) were seeded into each
well of a 96-well plate. The cells were transfected in triplicate
wells with 50 nM anti-miR-125b or anti-miR negative control and
were cultured in complete culture medium for 48 h. The culture
supernatants were then removed and 200 μl MTT solution (0.5 mg/ml;
Amresco LLC, Solon, OH, USA) was added to each well, followed by
incubation for 4 h at 37°C. The supernatants were then removed and
150 μl dimethyl sulfoxide (Sigma-Aldrich) was added into each well
and agitated to dissolve the formazan crystals. The optical density
(OD) was measured at 570 nm in an automatic enzyme standard
microtiter plate reader (MRX; Dynex Technologies, Chantilly, VA,
USA). The experiment was repeated three times.
Detection of cell apoptosis
The cells were analyzed for annexin V binding and
propidium iodide (PI) incorporation to distinguish between
apoptotic and necrotic cells. Cells (3×105) were seeded
into each well of a 96-well plate and transfected in triplicate
wells with 50 nM anti-miR-125b or anti-miR negative control,
followed by culture in complete culture medium for 48 h. The cells
were harvested, washed twice in ice-cold PBS, resuspended in 500 μl
binding buffer and stained for 30 min with Annexin V-fluorescein
isothiocyanate and PI. The stained cells were analyzed using a flow
cytometer (TY4124; BD Technologies, Durham, NC, USA) at an
excitation wavelength of 488 nm and emission wavelength of 525 nm
for fluorescein isothiocyanate fluorescence and 610 nm for PI
fluorescence. The experiment was repeated three times.
Cell cycle analysis
The cells (3×105) were seeded into each
well of a 96-well plate and transfected in triplicate wells with 50
nM anti-miR-125b or anti-miR negative control, followed by culture
in complete culture medium for 48 h. The cells were harvested,
washed in ice-cold PBS and fixed with 70% ice-cold ethanol for 24
h. Subsequently, the cells were resuspended in 100 μl RNase and
cultured for 30 min at 37°C. The suspension was mixed with 400 μl
PI for 30 min in the dark and the stained cells were analyzed using
a flow cytometer at an excitation wavelength of 488 nm and 610 nm
for PI fluorescence. The experiment was repeated three times.
Detection of cell invasion
Matrigel was thawed at 4°C overnight and then
diluted (5 mg/ml) in serum-free cold RPMI-1640 (Corning Inc.,
Corning, NY, USA) at a dilution of 1:8 and was transferred into the
upper chamber of a 24-well Transwell system (100 μl in each well).
The Transwell was incubated at 37°C for ~2–3 h for gelling.
Subsequently, 100 μl cell serum-free RMPI-1640 suspension
containing 1×104 anti-miR-125b-transfected cells were
added to the Matrigel. The lower chamber of the Transwell was
filled with 600 μl RPMI-1640 containing 10% FBS. The 24-well plate
was incubated at 37°C for 20–24 h. The cells which did not invade
and remained in the upper chamber of the Transwell were removed by
washing with PBS. Subsequently, 4% paraformaldehyde was used to
immobilize the cells that had invaded the lower chamber and
hematoxylin and eosin staining was performed. The cells in the
lower chamber of the Transwell were counted under a light
microscope (magnification, ×400; BX41; Olympus Corp., Tokyo,
Japan). The experiment was repeated three times.
Statistical analysis
Statistical analyses were performed using SPSS 13.0
software (SPSS, Inc., Chicago, IL, USA). Values are expressed as
the mean ± standard deviation. Analysis of variance and Dunnet’s
test were performed to compare the differences between the exposure
group and the control group. P<0.05 was considered to indicate a
statistically significant difference.
Results
Gene chip analysis of miRs in 95D cells
compared with those in 16HBE cells
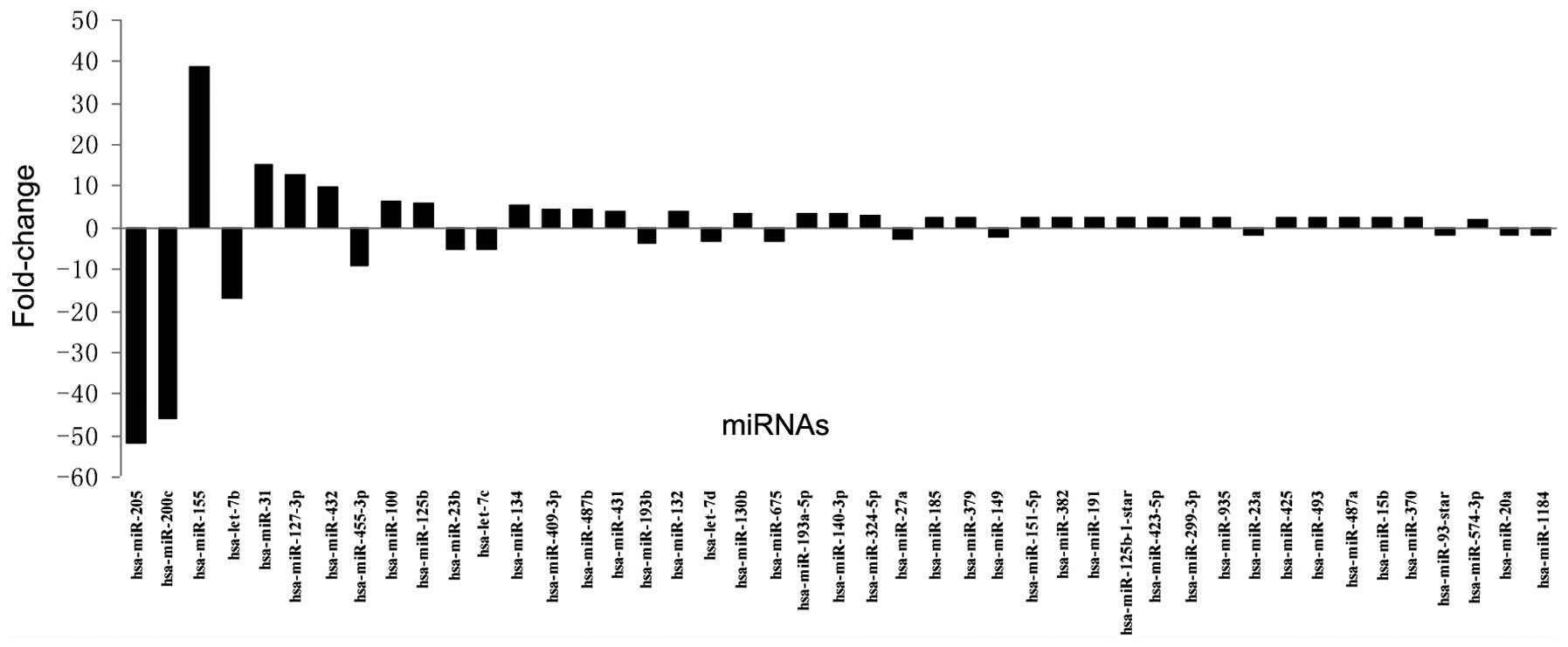
Gene chip analysis revealed that 45 miRs were
significantly differently expressed in the 95D cells compared with
the 16HBE cells, of which 30 were upregulated and 15 were
downregulated. The fold-changes in the dysregulated miRs are shown
in Fig. 1. Confirmation of the
expression of miR-125b by RT-qPCR supported the results of the gene
chip analysis (data not shown).
Bioinformatic analysis of miR-125b
Analysis was performed using miRGen Target software,
which involved the prediction of possible targets of miR-125b by
TargetScan, Pictar and miRanda. A total of 72 genes were identified
as possible putative targets of miR-125b (data not shown). The GO
terms of the biological process were further analyzed, revealing 21
associated functional annotations. The results demonstrated that
terms associated with the function of phosphorylation, including
the phosphate metabolic process and protein amino acid
phosphorylation, were annotated by several gene targets of
miR-125b. Stress-associated terms were also annotated by several
targets. Of note, among the 21 terms, the term ‘regulation of MAP
kinase kinase kinase (MAP3K) cascade’ was found (data not shown).
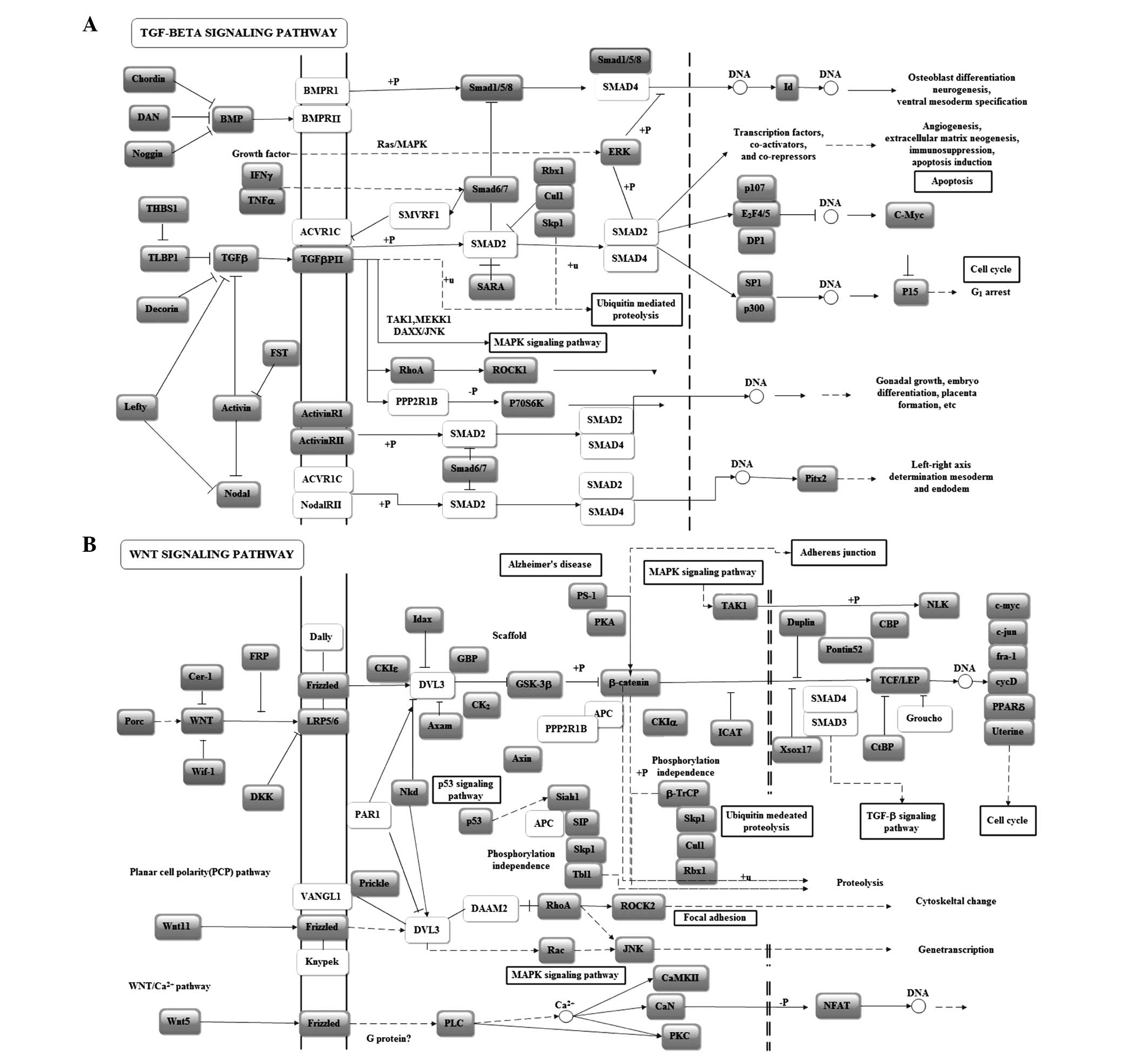
In the KEGG pathway analysis, the transforming growth factor
(TGF)-β signaling pathway scored highest (5.85). The Wnt signaling
pathway, closely associated with the development and progression of
cancer, also scored highly in the miR-125b enrichment pathway
analysis (Table I). To further
examine the impact of miR-125b on important pathways, pathway
graphs, indicating the regulatory sites and interactions, were
analyzed. In the TGF-β KEGG pathway graph, several genes at key
sites were found to be potentially regulated by miR-125b. ACVR1C is
involved in activating the TGF-β receptor and transmitting the
signal to SMAD2, which affects cell apoptosis and cell cycle by
activating a series of transcription factors (Fig. 2A). In the Wnt signaling KEGG
pathway graph, DVL3, a possible target gene of miR-125b, regulates
the mitogen-activated protein kinase (MAPK)-c-Jun N-terminal kinase
(JNK) apoptotic process. APC and PPP2R1B affect the adherence and
junctions between cells by regulating β-actin and are associated
with migration and invasion (Fig.
2B).
 | Table IKEGG pathway analysis of the putative
targets of microRNA-125b. |
Table I
KEGG pathway analysis of the putative
targets of microRNA-125b.
| KEGG pathway | KEGG pathway ID | −In (P-value) |
|---|
| Transforming growth
factor-β signaling pathway | hsa04350 | 5.85 |
| Renin-angiotensin
system | hsa04614 | 5.20 |
| Parkinson’s
disease | hsa05020 | 4.22 |
| Renin-angiotensin
system | hsa00602 | 3.95 |
| Polyunsaturated fatty
acid biosynthesis | hsa01040 | 3.95 |
| Pancreatic
cancer | hsa05212 | 3.72 |
| Notch signaling
pathway | hsa04330 | 3.28 |
| Wnt signaling
pathway | hsa04310 | 3.22 |
| Glycan
structures-biosynthesis 2 | hsa01031 | 3.16 |
| N-Glycan
biosynthesis | hsa00510 | 3.15 |
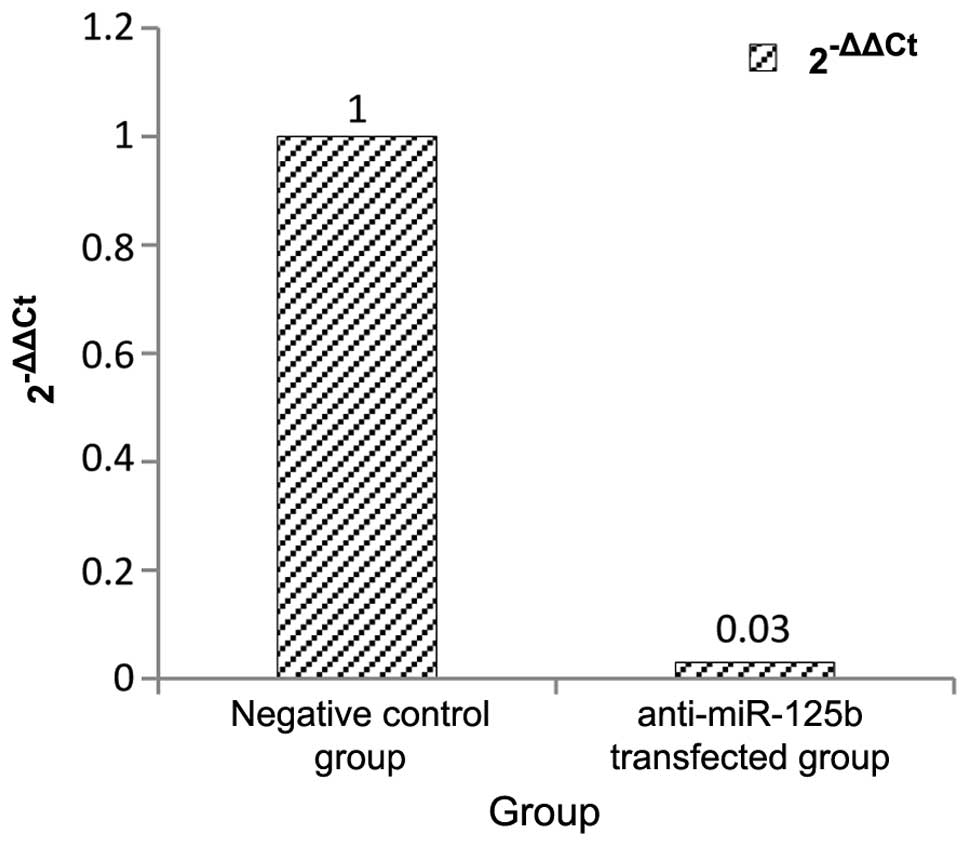
miR-125b transfection in 95D cells
Following anti-miR-125b transfection, miR-125b
levels were significantly inhibited to 3% of those in the negative
control (Fig. 3).
Effect of anti-miR-125b on the morphology
and proliferation of 95D cells
Under light microscopy, no significant differences
were observed in the morphology or density of the
anti-miR-125b-transfected 95D cells compared with those of the
cells in the negative control group. The MTT assay, used to examine
the effect of miR-125b downregulation on the viability of the 95D
cells, demonstrated no significant differences in the OD values
among the groups (P>0.05; Table
II).
 | Table IIEffect of downregulated miR-125b on
95D cell proliferation. |
Table II
Effect of downregulated miR-125b on
95D cell proliferation.
| Group | Optical density |
|---|
| Blank | 0.884±0.019 |
| Anti-negative
control-transfected | 0.869±0.010 |
|
Anti-miR-125b-transfected | 0.892±0.008 |
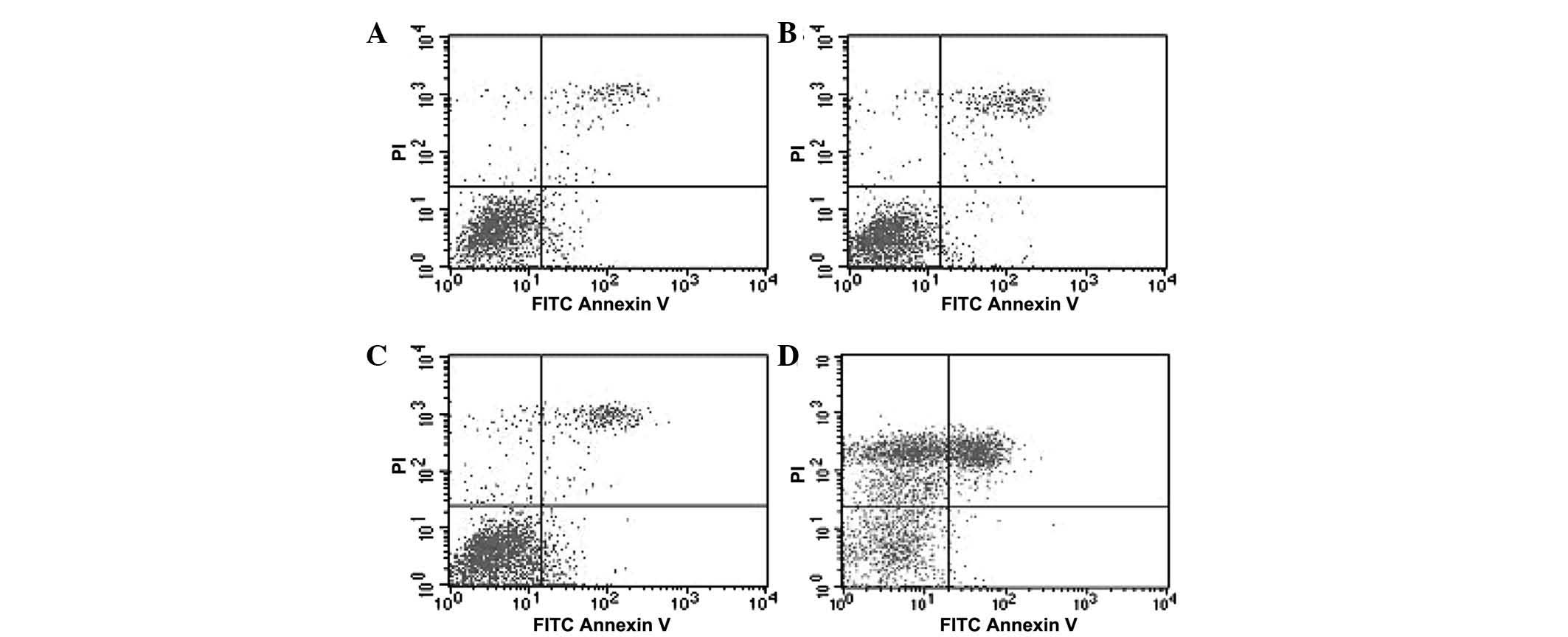
Apoptosis of anti-miR-125b-transfected
95D cells
PI and Annexin V were used to stain the 95D cells
and the apoptotic rate was determined using flow cytometry. No
significant difference was observed between the blank and the
negative control groups (P>0.05). The apoptotic rate of the
cells in the positive group was significantly higher than that of
cells in the other groups (P<0.05). Of note, the apoptotic rates
of the cells with downregulated miR-125b were significantly higher
compared with those in the negative control group (P<0.05;
Fig. 4 and Table III).
 | Table IIIEffect of downregulated miR-125b on
the apoptotic rate of 95D cells. |
Table III
Effect of downregulated miR-125b on
the apoptotic rate of 95D cells.
| Group | Apoptotic rate
(%) |
|---|
| Blank | 15.92±0.99 |
| Anti-negative
control-transfected | 16.39±1.11 |
|
Anti-miR-125b-transfected | 20.96±0.71a |
| Positive (0.1%
H2O2) | 30.42±2.50a |
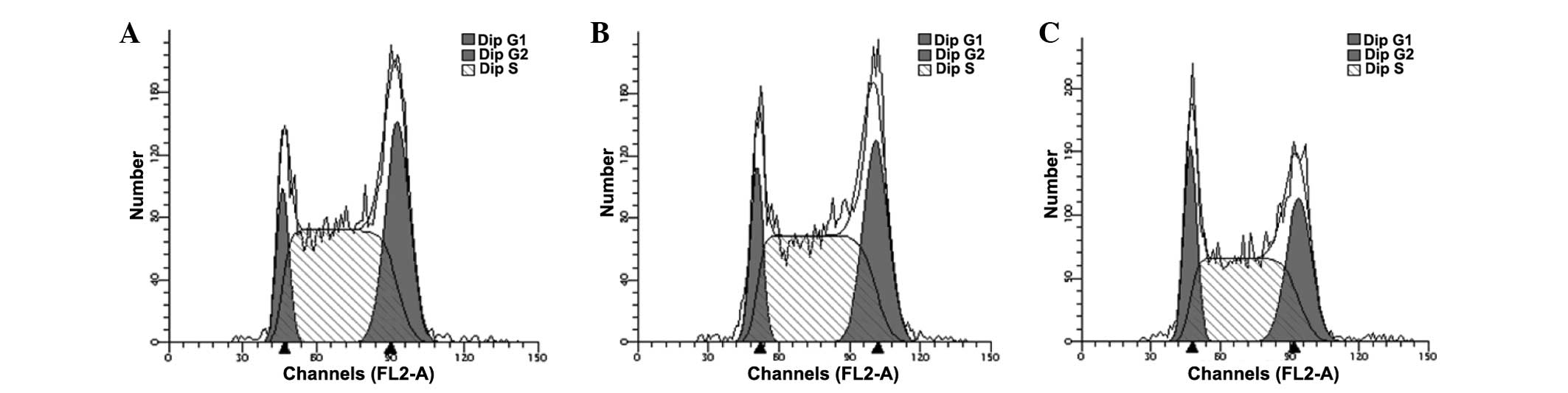
Cell cycle analysis of
anti-miR-125b-transfected 95D cells
The cell cycle of anti-miR-125b-transfected 95D
cells was assessed using flow cytometry. The results demonstrated
that downregulation of miR-125b in the 95D cells induced G1/S phase
arrest compared with the negative group (P<0.05; Fig. 5 and Table IV).
 | Table IVEffect of downregulated miR-125b on
the cell cycle of 95D cells. |
Table IV
Effect of downregulated miR-125b on
the cell cycle of 95D cells.
| Group | G1 (%) | S (%) | G2 (%) |
|---|
| Blank | 15.91±0.71 | 58.33±1.71 | 25.75±2.07 |
| Anti-negative
control-transfected | 15.24±2.33 | 57.54±1.43 | 27.22±1.51 |
| Anti-miR-125b
transfected | 18.27±0.17a | 53.86±0.82a | 27.87±0.95 |
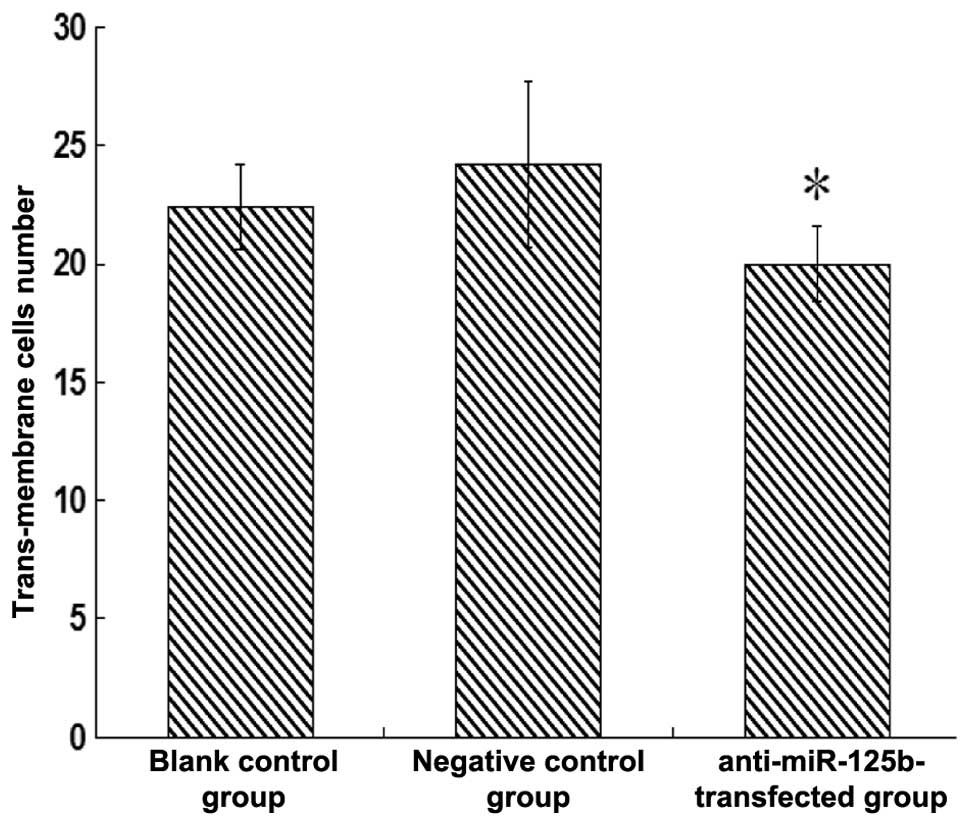
Invasive ability of the
anti-miR-125b-transfected 95D cells
A Transwell assay was used to evaluate the impact of
downregulated miR-125b on the invasive ability of 95D cells. No
significant change was observed in the number of invaded cells
between the negative control group and the blank group (P>0.05).
However, the number of invaded cells in the
anti-miR-125b-transfected group was significantly lower compared
with that in the negative control group (P<0.05; Fig. 6).
Discussion
In the present study, the expression of miR-125b was
observed to be significantly increased in lung cancer cells and
inhibition of miR-125b had marked effects on several biological
processes of lung cancer, including apoptosis, cell cycle and
invasion ability. Inhibition of the expression of miR-125b in the
95D cell line induced an increase in the apoptotic rate, caused
G1/S-arrest and reduced the invasive ability of the cells. These
three aspects are well known to be associated with tumor
development (10–12). During the development of cancer,
the apoptotic mechanism is dysregulated to a certain extent, which
reduces the number of apoptotic cells. The invasive ability is
increased, improving the migration of cancer cells and the cell
cycle is also affected, with an increased number of cells in the S
phase (DNA duplication phase) of the cell cycle (13). The results of the present study
also demonstrated that the deregulation of miR-125b resulted in
almost the opposite of an oncogenic effect, suggesting that
miR-125b may have an oncogenic function. These results are the
first, to the best of our knowledge, to elucidate the functions of
miR-125b in lung cancer.
The target prediction indicated 72 genes as
potential targets of miR-125b and, by clustering these 72 genes, GO
annotation and KEGG pathway analysis were performed. The results
demonstrated that miR-125b may be associated with TGF-β to
stimulate cancer growth. It has been previously suggested that
TGF-β is an effective factor in regulating cell growth, division
and migration (14), and that
TGF-β is overexpressed in lung cancer and can promote the malignant
transformation, invasion and migration of lung cancer, acting as an
oncogene (15). The potential
activation of TGF-β by miR-125b observed in the present study may
indicate that miR-125b has a possible cancer promoting role.
The present study also demonstrated that miR-125b is
involved in the MAPK signaling pathway, which is another important
cancer-associated pathway. KRAS, which is the key protein in the
RAS/RAF/MAPK process, regulates cell proliferation, cell division
and cell cycle, thus affecting the transformation and development
of cancer (16,17). The association between kRAS and
lung cancer has been confirmed by several studies (18). Several previous studies on MAP3K
have suggested that MAP3K not only regulates apoptosis, but also
affects the proliferation and invasion of cancer cells in the
development of cancer (19,20).
In addition, activation of JNK1 and JNK2 by the MAP3K family, which
can phosphorylate nuclear factor-κB inhibitors, may promote cancer
cell growth (21). The results of
the present study indicated the possible function of miR-125b in
cell proliferation, cell apoptosis and cell cycle and this
hypothesis was supported by the involvement of miR-125b in
regulating the Wnt signaling pathway, concerning β-catenin. The Wnt
signaling pathway, which regulates apoptosis, metabolism and other
biological processes, is also associated with the progression of
cancer (22). To briefly
summarize, miR-125b may act on several cancer-associated pathways
by regulating several potential targets, to promote the development
of lung cancer.
These results provide support for the
cancer-promoting role of miR-125b in the progression of lung
cancer. Although the function of miR-125b in lung cancer has not
been reported, several studies of other types of tumor have
reported miR-125b to function as an oncogenic or tumor-suppressive
miR.
In colorectal cancer, prostate carcinoma and
leukemia, miR-125b has been recognized as an oncogenic factor.
These studies found that upregulated miR-125b stimulated the growth
of HT29 colorectal cancer cells by inhibiting the expression of p53
(10). Shi et al (23) found that high expression levels of
miR-125b can inhibit cell apoptosis in prostate tumors by
downregulating B-cell lymphoma 2 (BCl-2)-antagonist/killer 1, a
member of the Bcl-2 family and in prostate carcinoma, miR-125b
improves cell growth (11). A
previous study on miR-125b in human U343 and U251 glioma cells
revealed that the overexpression of miR-125b can inhibit cell
apoptosis by reducing the levels of Bcl-2-modifying factor, a
pro-apoptotic protein (24). These
results indicated that miR-125b functions as an oncogene, which
mirror those of the present study. However, certain studies have
observed that the function of miR-125b was opposite of that of a
tumor suppressor gene. In a study investigating the impact of
miR-125b on SKBR3 breast carcinoma cells, miR-125b inhibited cell
proliferation (25). In another
study on hepatocellular carcinoma, the proliferative and clone
forming ability of hepatoma carcinoma cells were reduced and
S-phase arrest was observed (26)
in cells with low expression levels of miR-125b. Therefore, the
functions of miR-125b differed in various types of tumor,
suggesting different regulatory mechanisms of miR-125b. In lung
cancer, the regulatory mechanisms of the oncogenic effects of
miR-125b require further investigation.
In conclusion, the present study was the first to
investigate the biological functions of miR-125b in lung cancer
and, to the best of out knowledge, to demonstrate that suppression
of miR-125 expression induced apoptosis, G1/S phase arrest and, to
a certain extent, inhibited the invasive ability of lung cancer
cells. This may be involved in the regulation of several potential
targets at the key sites of TGF-β, MAPK, Wnt and other important
cancer-associated signaling pathways. By identifying the expression
and the functions of miR-125b in 95D cells, the present study
hypothesized that miR-125b may have an oncogenic role in the
development and progression of lung cancer. Further investigation
is required to identify the accurate targets of miR-125b and the
active sites in those significant pathways.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (nos. 81472939 and 81172618), the Qing
Lan Project (no. 2012), the 333 project of Jiangsu Province (no.
2012), the Liu Da Ren Cai Gao Feng Project of Jiangsu Province (no.
2013-WSW-053) and the Fundamental Research Funds for the Central
Universities (no. 2013).
References
|
1
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L,
Kipps T, Negrini M, Bullrich F and Croce CM: Frequent deletions and
down-regulation of microRNA genes miR15 and miR16 at 13q14 in
chronic lymphocytic leukemia. Proc Natl Acad Sci USA.
99:15524–15529. 2002. View Article : Google Scholar
|
|
4
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
Mitsudomi T and Takahashi T: Reduced expression of the let-7
microRNAs in human lung cancers in association with shortened
postoperative survival. Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Michael MZ, O’Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
6
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
7
|
Frezzetti D, De Menna M and Zoppoli P:
Upregulation of miR-21 by Ras in vivo and its role in tumor growth.
Oncogene. 30:275–286. 2011. View Article : Google Scholar
|
|
8
|
Yuxia M, Zhennan T and Wei Z: Circulating
miR-125b is a novel biomarker for screening non-small-cell lung
cancer and predicts poor prognosis. J Cancer Res Clin Oncol.
138:2045–2050. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Y, Chao Y, Fang Y, Wang J, Wang M,
Zhang H, Ying M, Zhu XX and Wang HF: MTA1 promotes the invasion and
migration of non-small cell lung cancer cells by downregulating
miR-125b. J Exp Clin Cancer Res. 32:332013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nishida N, Yokobori T, Mimori K, Sudo T,
Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H and Mori M: MicroRNA
miR-125b is a prognostic marker in human colorectal cancer. Int J
Oncol. 38:1437–1443. 2011.PubMed/NCBI
|
|
11
|
Pang Y, Young CY and Yuan H: MicroRNAs and
prostate cancer. Acta Biochim Biophys Sin (Shanghai). 42:363–369.
2010. View Article : Google Scholar
|
|
12
|
Bousquet M, Harris MH, Zhou B and Lodish
HF: MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA.
107:21558–21563. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dang XM, Ma AQ, Yang L, Hu H, Zhu B, Shang
D, Chen TJ and Luo Y: MicroRNA-26a regulates tumorigenic properties
of EZH2 in human lung carcinoma cells. Cancer Genet. 205:113–123.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-β family signaling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bacon AL, Farrington SM and Dunlop MG:
Mutation frequency in coding and non-coding repeat sequences in
mismatch repair deficient cells derived from normal human tissue.
Oncogene. 20:7464–7471. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Johnson GL and Lapadat R: Mitogen
activated protein kinases pathways mediated by ERK, JNK, and p38
protein kinases. Science. 298:1911–1912. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stacey DW: Cyclin D1 serves as a cell
cycle regulatory switch in actively proliferating cells. Curr Opin
Cell Biol. 15:158–163. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kranenburg O, Gebbink MF and Voest EE:
Stimulation of angiogenesis by Ras proteins. Biochim Biophys Acta.
1654:23–37. 2004.PubMed/NCBI
|
|
19
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weston CR and Davis RJ: The JNK signal
transduction pathway. Curr Opin Cell Biol. 12:14–21. 2002.
|
|
21
|
Cheung PC, Campbell DG, Nebreda AR and
Cohen P: Feedback control of the protein kinase TAK1 by
SAPK2a/p38α. EMBO J. 22:5793–5805. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Van SM, Randall J, Sorgew A, Williams LM,
Tennis M and Winn RA: Wnt signaling pathway and lung disease.
Transl Res. 151:175–180. 2008. View Article : Google Scholar
|
|
23
|
Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu
M, Tepper CG, Evans CP, Kung HJ and deVere White RW: An
androgen-regulated miRNA suppresses Bak1 expression and induces
androgen-independent growth of prostate cancer cells. Proc Natl
Acad Sci USA. 104:19983–19988. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin
XH and Ma X: MiR-125b expression affects the proliferation and
apoptosis of human glioma cells by targeting Bmf. Cell Physiol
Biochem. 23:347–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar
|
|
26
|
Liang L, Wong CM, Ying Q, Fan DN, Huang S,
Ding J, Yao J, Yan M, Li J, Yao M, Ng IO and He X: MicroRNA-125b
suppresses human liver cancer cell proliferation and metastasis by
directly targeting oncogene LIN28B2. Hepatology. 52:1731–1740.
2010. View Article : Google Scholar : PubMed/NCBI
|