Introduction
Hirudin is a human thrombin-specific inhibitor, the
natural form of which was extracted from the salivary gland of
medicinal leech Hirudo medicinalis. It is composed of 65–66
amino acid residues with a molecular weight of ~6,800 Da. Hirudin
has three intramolecular disulfide bonds and a tyrosine sulfation
at the 63 site, to acquire a native conformation and antithrombin
activity (1). As a human
thrombin-specific inhibitor, hirudin has an increasing number of
applications in the clinic (2,3). As
the production of hirudin from Hirudo is relatively
inefficacious, at present, almost all of the therapeutic hirudin is
generated by recombinant bacteria.
Though recombinant hirudin (rHirudin) has
antithrombin activity and has been expressed in several expression
systems, including Escherichia (E.) coli,
Saccharomyces and Hansenula polymorpha (4), the antithrombin activity of rHirudin
appears to be lower than that of the native form, which is
extracted from Hirudo. This may be because rHirudin does not
bear any sulfate on the 63 tyrosine amino acid residue, as the
expression systems used recently appear to lack the
sulfotransferase accessory, particularly in the E. coli
expression system (2).
In order to improve the bioactivity of rHirudin,
several modification methods have been applied and
site-specifically substituted hirudins have been produced (5). In these studies,
L-phenylalanine-modified dodecapeptide was synthesized at position
63 using an Fmoc solid-phase synthesis strategy. The dodecapeptide
prolonged the thrombin time to two times more than the recombinant
ones in vitro (6). Liu
et al (5) incorporated the
unnatural amino acid of sulfo-tyrosine into hirudin on 63 sites
using the MjSTyrRS/tRNATyr cua
orthogonal pair in an E coli expression system. The
site-specifically modified hirudin was purified and functional
analysis demonstrated that the sulfation at site 63 of hirudin was
a crucial factor in the formation of the hirudin/thrombin complex.
It interacted directly with Lys 81 and Tyr 76 of human thrombin,
formed a strong salt bridge and a new hydrogen bond network. This
was a marked difference from that of the rHirudin/thrombin complex.
The sulfate group was able to augment the affinity of sulfohirudin
with thrombin and gave a lower inhibition constant Ki of
25 fM than the rHirudin/thrombin complex with a Ki of
307 fM (5). The prolonged thrombin
time of the carboxymethyl-phenylalanine-modified peptide and the
enhanced affinity of sulfated hirudin for human thrombin suggested
that the different group residues that existed at the 63 site
appeared to be important factors affecting hirudin biofunction.
Boronic acids are known to form complexes with amino
acids or hydroxamic acids and have been employed as moieties in
ligands for the selective recognition of sugars and have also been
used as potent serine protease inhibitors (7). In addition, boronates have clinical
utility as boron neutron capture agents for selectively targeting
tumor cells (8). The novel
chemical properties of boronic acids as components of proteins
allows for selective chemistry on the protein surface, including
oxidation, reduction, Suzuki coupling reactions, as well as the
formation of covalent boronic esters with polyhydroxylated
compounds. Proteins modified by addition of boronic acid moieties
may be used in combination with a polyhydroxylated solid support to
purify native protein sequences in a one-step affinity purification
procedure. Furthermore, the ability of boronic acids to bind diols
and reactive serine residues allows for the development of
boronate-containing antibodies that specifically recognize and
covalently bind to various glycoproteins or proteases. It may also
be possible to form intramolecular serine-boronate crosslinks in
proteins to enhance stability (9,10).
The unique chemical characteristics of boronates may allow for the
in vivo labeling of boronate-containing proteins with
polyhydroxylated reporter molecules.
In the present study, using an codon-expanding
method, the boronic-amino acid of boronophenylalanine was
site-specifically incorporated into target proteins. Previously,
Liu and Schultz (11) have
successfully used and evolved
MjSTyrRS/tRNATyr cua orthogonal pairs
to substitute the tyrosine at site 63 with sulfotyrosine. The
affinity of sulfohirudin was evidently enhanced as compared with
that of rHirudin. The orthogonal pair
MjBTyrRS/tRNATyr cua is able to
recognize and incorporate boronophenylalanine efficiently into
proteins of interest in the E. coli expression system
(12) and is not recognized by
endogenous tRNAs and aminoacyl-tRNA synthetases. Using this
orthogonal pair, the 63 tyrosine of rHirudin was substituted with
unnatural amino acids (NAAs) in order to give the protein specific
new biofunctions.
Furthermore, it has been reported that hirudin may
inhibit the proliferation of fibroblasts and have an important role
in wound healing (13). The
fibroblast cell line L929 was considered as a primary source of
extracellular matrix components. L020 cells have an important
function in wound healing, as they were demonstrated to have a
critical role in regulating the turnover of the extracellular
matrix and have been widely used in cell functional studies
(14). In the present study, the
effects of the boronophenylalanine-modified hirudin on the
proliferation of fibroblasts cells was examined and compared with
that of rHirudin. The enhanced bioactivity and convenient synthetic
procedure suggested a feasible method for the site-specific
modification of hirudin.
Materials and methods
Plasmid construction and strains
The plasmid
pEVOL-MjBTyrRS/tRNATyr cua was used as
the boronophenylalanine incorporation vector. The plasmid
pBAD-gIII-hirudin was used as a template to construct
pBAD-gIII-hirudin-TAG-6xhis tag. All of these plasmids were
generously donated by Professor W Liu group of the A&M
University (College Station, TX, USA). The gene sequence of hirudin
was in accordance with the National Center for Biotechnology
Information GenBank entry (accession no., GI: 208479; https://www.ncbi.nlm.nih.gov/genbank/).
Using a modified Quickchange strategy (15) to construct the new expression
plasmid, a TAG sequence at the 5′-end was designed to substitute
the original tyrosine codon TAC. The sequence of the forward primer
was 5′-TAGCTGCAATGACTCGAG ATCTG-3′ and the reverse primer sequence
was 5′-TTCTTC CGGAATTTCTTCAAAATCGCC-3′. To fuse a 6xhis sequence at
the 3′-end, the forward primer was 5′-ATGGTG
ATGTTGCAGCTAATTCTTCCGGAATTTC TTC-3′ and the reverse primer was
5′-CACCATCACTGACTCGAGATC TGCAGCTGGTAC-3′, there were three
histidine codons (underlined) in each primer. The constructed
plasmids were confirmed by DNA sequencing.
Top10 Electrocomp E. coli cells Novagen
(Madison, WI, USA) were used to express hirudins with
site-specifically incorporated NNAs. The cells transformed with
plasmids pBAD-gIII-hirudin-6xhis and pEVOL were used as controls to
express rHirudin.
Expression and purification of the
proteins
A single colony of the recombinant E. coli
Top 10 cells harboring the pBAD-gIII-hirudin-TAG-6xhis tag and
pEVOL was collected and cultured at 37°C overnight in 5 ml
Luria-Bertani (LB) medium containing ampicillin (100 μg
ml−1) and chloramphenicol (34 μg ml−1)
(Takara Biotechnology Co., Ltd., Dalian, China). The cultures were
diluted 1:100 in a 250-ml flask with 50 ml LB and grown at 37°C
with agitation at 4.3 × g until the optical density at 600 nm
(OD600) reached 0.6. Then, l-arabinose was added to a
final concentration of 0.2%, followed by a further 16 h incubation
at 37°C to induce rHirudin expression.
To express the site-specifically modified hirudins,
a single colony of the recombinant E. coli Top 10 cells
harboring the pEVOL-MjBTyrRS/tRNATyr
cua mutant and pBAD-gIII-hirudin-TAG-6xhis tag plasmids were
cultured at 37°C overnight in 5 ml LB containing ampicillin (100 μg
ml−1) and chloramphenicol (34 μg ml−1). The
culture was diluted 1:100 in a 250-ml flask with 50 ml LB.
Boronophenylalanine (1 mM; Sigma-Aldrich, St. Louis, MO, USA) was
added to the medium and grown at 37°C, 4.3 × g until
OD600 reached 0.6. l-arabinose was added to the medium
to a final concentration of 0.2%, following another 16-h induction
at 37°C.
Next, the medium was collected and concentrated
through the ultra-filter apparatus with 3 kD cut-off membranes.
After filtering through a 0.45 μm filter, the samples were loaded
on an AKTA-purifier system (Amersham Biosciences, Uppsala, Sweden).
It was equipped with a nickel nitrilotriacetic acid (Ni-NTA)
affinity column which was pre-equilibrated with buffer A (50 mM
NaH2PO4, 300 mM NaCl, pH 8.0). Proteins of
interest were eluted using elution buffer (50 mM
NaH2PO4, 300 mM NaCl, 300 mM imidazole, pH
8.0) at an elution rate of 0.4 ml/min. The fractions with
anticoagulant activity were collected, concentrated and dialyzed
against 25 mM Tris-HCl, 10 mM NaCl and 5% glycerol (pH 7.6) for 3 h
at 4°C. The dialysis buffer was changed three times prior to sample
concentration. Each of the concentrated samples were loaded on a
gel filtration column (Superdex 200 10/300 GL; GE Healthcare,
Pittsburgh, PA, USA) for further purification. The proteins of
interest were eluted with 25 mM Tris-HCl, 125 mM NaCl and 2 mM KCl
(pH 7.6) at an elution rate of 0.2 ml/min. The collected samples
were stored at 4°C for further detection.
Tricine/SDS-PAGE and matrix-assisted
laser desorption time-of-flight (MALDI-TOF) analysis
Tricine/SDS-PAGE analysis of the purified protein
samples was performed according to Schägger (16). The low-molecular weight marker was
obtained from Fermentas (Vilnius, Lithuania). The gels were stained
with Coomassie Brilliant R-250. Mass analysis of the purified
proteins was performed using MALDI-TOF mass spectrometry on an ABI
4800 MALDI/TOF analyzer (Applied Biosystems, Foster City, CA,
USA).
Antithrombin activity
Antithrombin activity was assessed by titration of a
thrombin solution and the thrombin neutralizing activity of
hirudins was determined in antithrombin units (ATU) (17), Briefly, 200 μl standard solution of
0.05% bovine fibrinogen (Ameresco, Inc., Columbus, OH, USA) was
mixed with 10 μl hirudin samples and 5 μl thrombin [47 National
Institute of Health (NIH) units; Ameresco, Inc.], mixed gently and
left to stand for 1 min at 37°C. If the fibrin did not clot, the
hirudin samples had 47 ATU. The hirudin samples were diluted to 1
mg/ml in the working buffer (50 mM Tris, pH 7.5, 100 mM NaCl;
Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) in a series
of gradient as 1:100, 1:200, 1:500, 1:1,000, 1:3,000 and 1:6,000. A
total of 10 μl diluted solution each were incubated with 200 μl
0.05% bovine fibrinogen in Tris-HCl buffer (pH 7.4) at 37°C, 5 μl
(47 NIH units) of standard thrombin solution was added
progressively at intervals of 1 min and mixed gently. The end point
of the titration was considered reached when a fibrin clot formed
within 1 min.
Protein analysis
Protein concentration was determined using a
bicinchoninic acid protein assay kit (Thermo Fisher Scientific,
Columbia, SC, USA) according to the manufacturer’s instruction.
Fibroblast proliferation test
The fibroblast cell line L929 (#CCL-1; American Type
Culture Collection, Manassas, VA, USA) was cultured in RPMI-1640
supplemented with 100 mg l−1 benzylpenicillin, 100 mg
l−1 streptomycin and 10% fetal calf serum (Gibco Life
Technologies, Grand Island, NY, USA). Cells were seeded in 96-well
dishes (5×103 cells/well) and incubated under sterile
conditions at 37°C in a humidified atmosphere containing 5%
CO2. A total of 16 h later, 1, 0.5, 0.25, 0.125 and 0
ATU μl−1 hirudin solutions, respectively, were added to
the medium. The solution concentration was 1.158 μg ml−1
for the boronopheylalanine-modified hirudin and 1.475 μg
ml−1 for rHirudin. Cell proliferation was assessed using
an MTT assay following 24 h (18).
Statistical analysis
SPSS software, version 20 (IBM SPSS, Armonk, NY,
USA) was used for all statistical analyses. Data are expressed as
the mean ± standard deviation. P<0.05 was considered to indicate
a statistically significant difference compared with the
hirudin-untreated sample.
Results
Plasmid construction
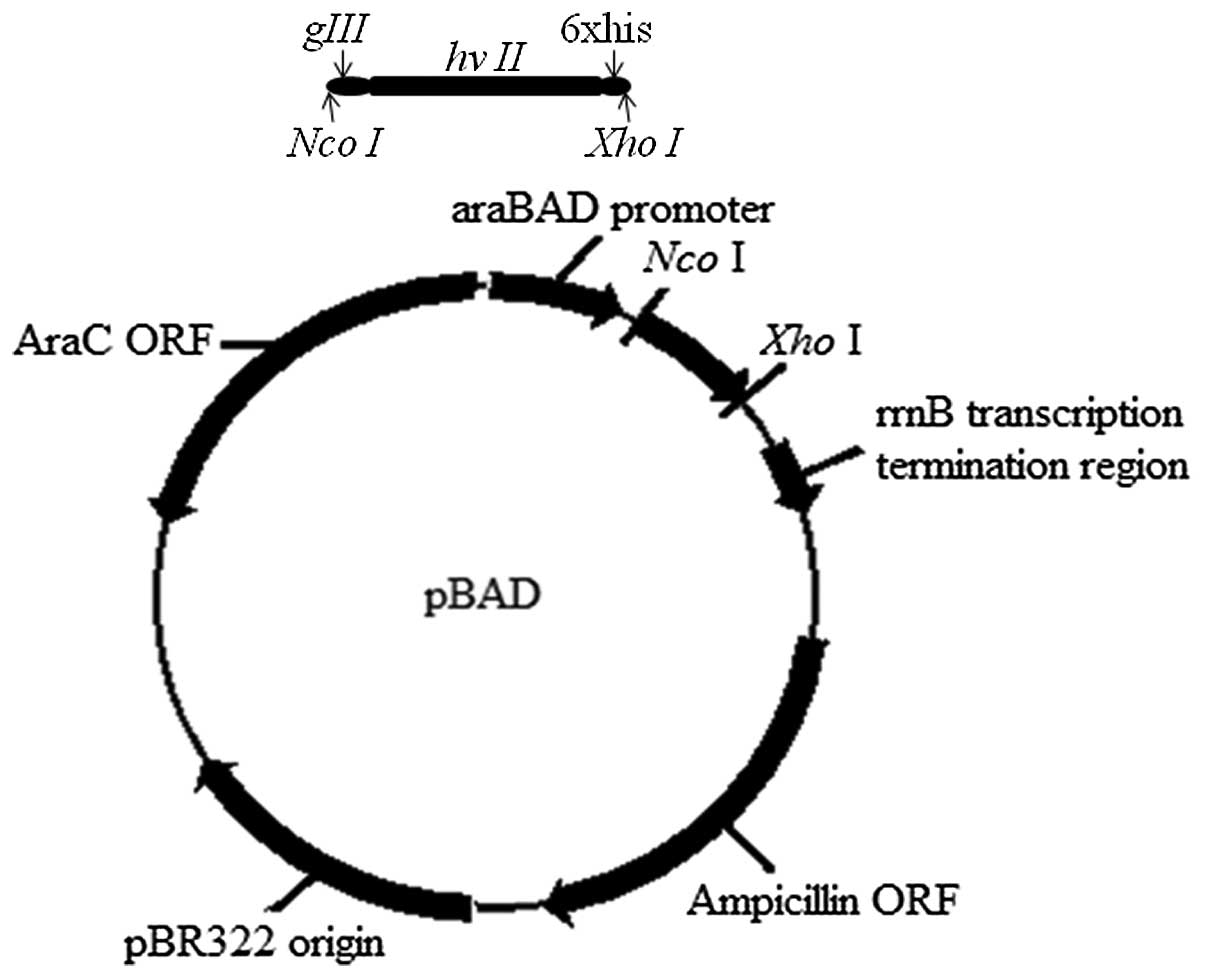
The gIII signal peptide nucleotide sequence and
hirudin gene hv II were fused with 6xhis codons at the 3′-end of
the hirudin gene on the expression vector of pBAD (Fig. 1). Next, a TAG mutation was
introduced into the fusion gene at position 63 of the hirudin gene
to substitute the original TAC codon (Fig. 1B). The constructed plasmids were
confirmed by DNA sequencing.
Expression of
boronophenylalanine-modified hirudin and rHirudin
The plasmids
pEVOL-MjBTyrRS/tRNATyr cua and
pBAD-gIII-hv II-TAG-6xhis were co-transformed into E. coli
Top 10 and named as Top 10(pBAD-hv II-TAG-pEVOL-B). The translation
of the fusion protein was started at the start codon of the gIII
signal peptide at the 3′-terminal and followed by the hirudin gene
and a 6xhis-hirudin, which was controlled by the araB promoter.
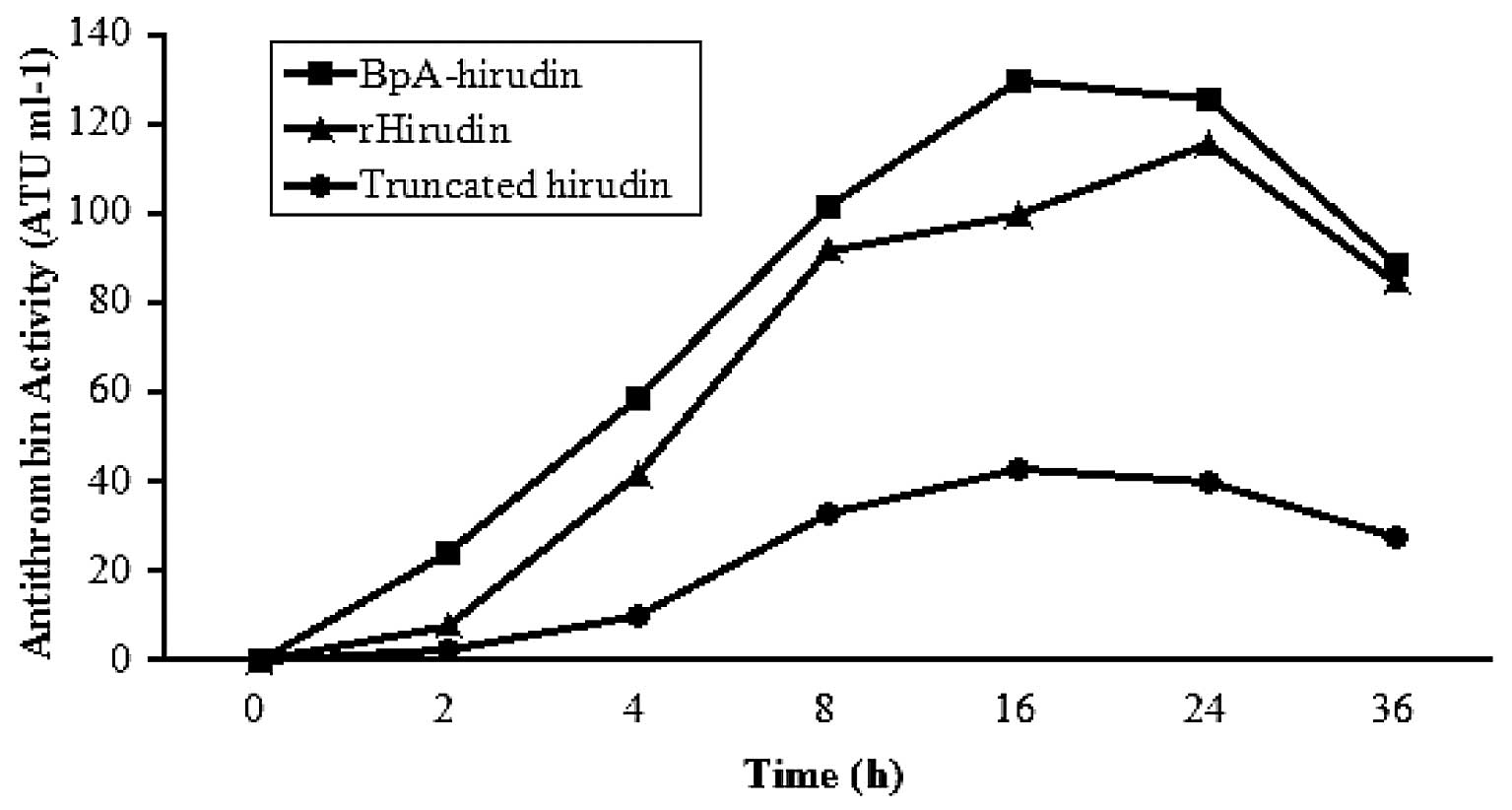
The expression of the recombinant vectors was
induced by 0.2% arabinose and 1 mM boronophenylalanine. The time
course of hirudin expression was examined using a thrombin
inhibitory assay. The medium exhibited antithrombin activity
following 2 h. The boronophenylalanine-modified hirudin had an
antithrombin activity of ~130 ATU ml−1 culture medium
following 16 h, and that of rHirudin was ~100 ATU ml−1
after 16 h. The truncated derivative was about 40 ATU
ml−1 at 16 h. After this, the antithrombin activity of
the three types of hirudins decreased with time, particularly after
24 h (Fig. 2).
Purification of hirudin products
The fractions which exhibited antithrombin inhibitor
activity were collected and eluted with 300 mM NaCl-buffer B from
the Ni-NTA affinity column. Following concentration, a further
purification step was performed with an Superdex-G250 filtration
column (Superdex 200 10/300 GL; GE Healthcare). The proteins of
interest were eluted out with 25 mM Tris-HCl, 125 mM NaCl and 2 mM
KCl (pH 7.6), with an elution rate of 0.2 ml/min. The collected
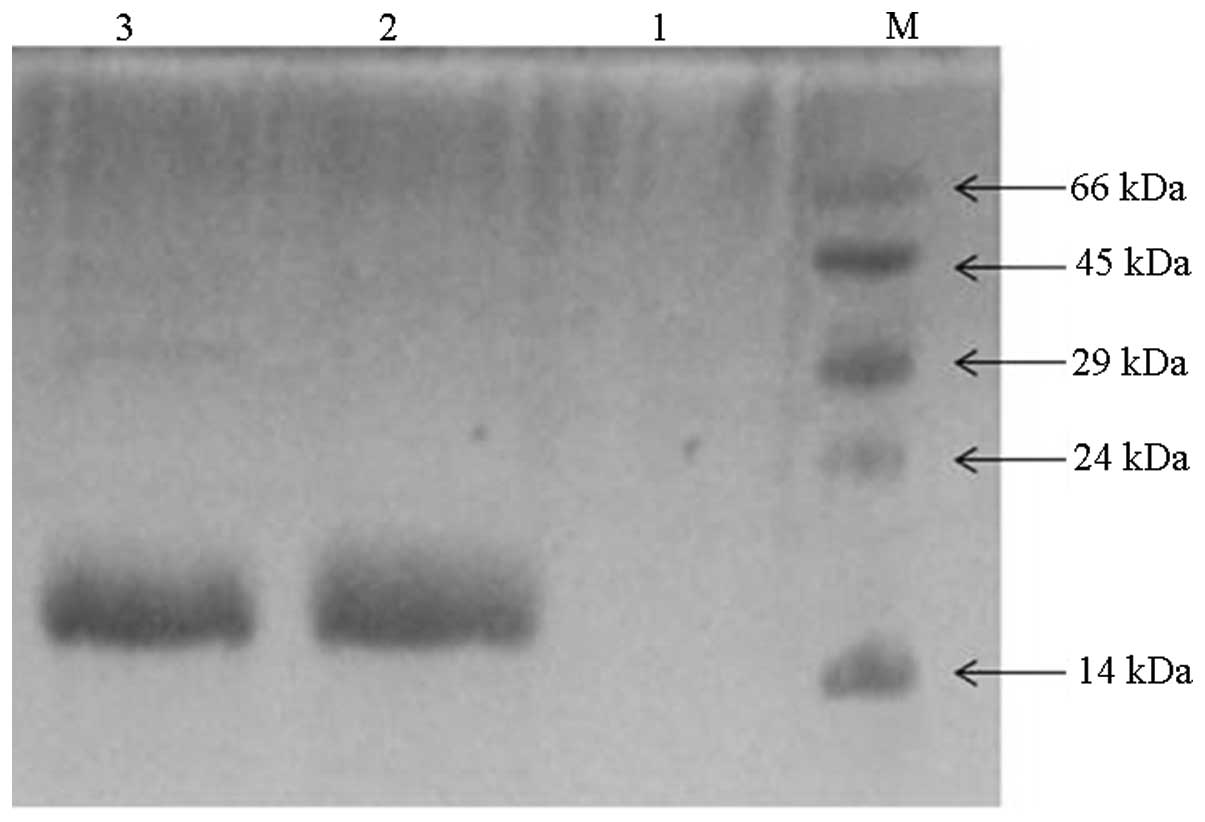
samples demonstrated single bands of ~14 kDa on the coomassie
brilliant blue stained tricine-PAGE (Fig. 3 lane 2 and 3), implying the
formation of homodimers as reported by previous studies (19). Purification was performed on
Ni-NTA, which only has affinity for positively charged compounds.
The hirudins fused with a 6xhis tag at the C-terminus bound to the
solid phase of the column and were then eluted, as were the protein
samples of rHirudin (lane 2) and the full length
boronophenylalanine-incorporated hirudin (lane 3). The negative
control confirmed that there appeared to be no protein from the
PAGE-gel, as revealed in lane 1. No boronopheylalanine was added to
the negative control culture medium, so the introduced TAG codon
may not be translated into the NAA of boronopheylalanine and a
truncated polypeptide was expressed in this case. The expressed
truncated hirudin peptide was composed of 62 amino acid residues
counted from the N-terminus, but lacked the hexapeptides at the
C-terminus, and was not able to bind to the Ni-NTA column, so there
was no protein sample eluted (lane 1).
The purity was ~93% for the purified hirudins
detected by trincine-PAGE. The yield of the boronophenylalanine
site-specifically modified hirudin was 10 mg l−1 and
that of the rHirudin was 19 mg l−1.
Authenticity analysis using
MALDI-TOF
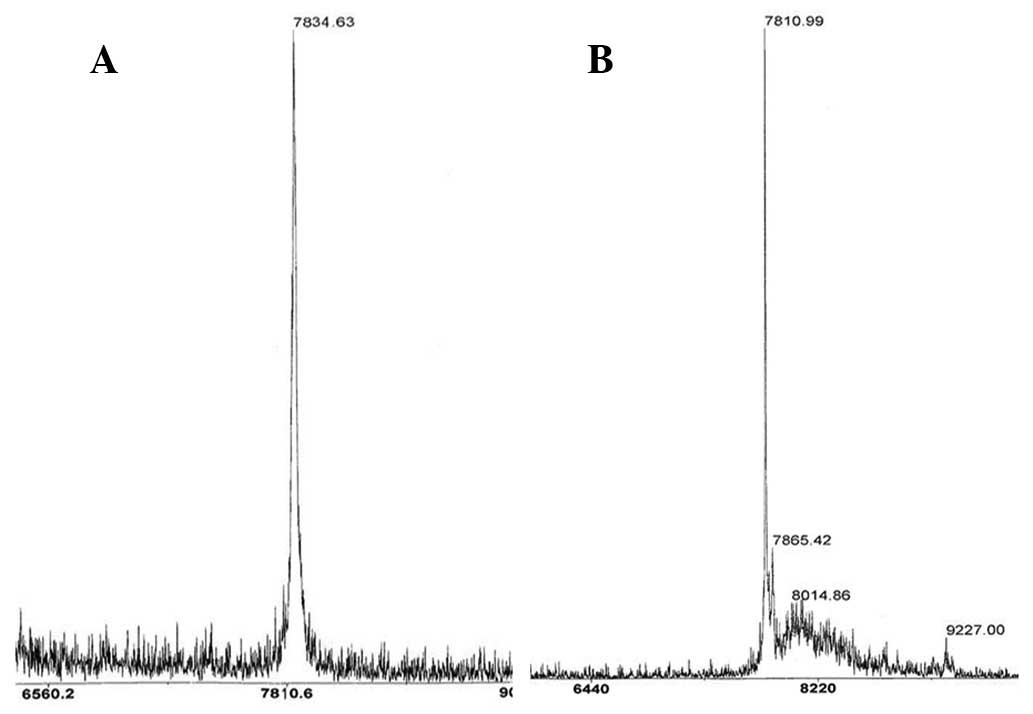
The detected molecular weight of
boronophenylalanine-incorporated hirudin was 7,834.6 kDa as
determined by MALDI-TOF mass spectrometry. The calculated
theoretical molecular weight was 7,834.5 Da, which matched with the
experimental result. As for the rHirudin, the MALDI-TOF peak
indicated a molecular weight of ~7,810.9 Da (Fig. 4A and B) and the theoretical
molecular weight was 7810.5 Da, which confirmed the identity of the
proteins.
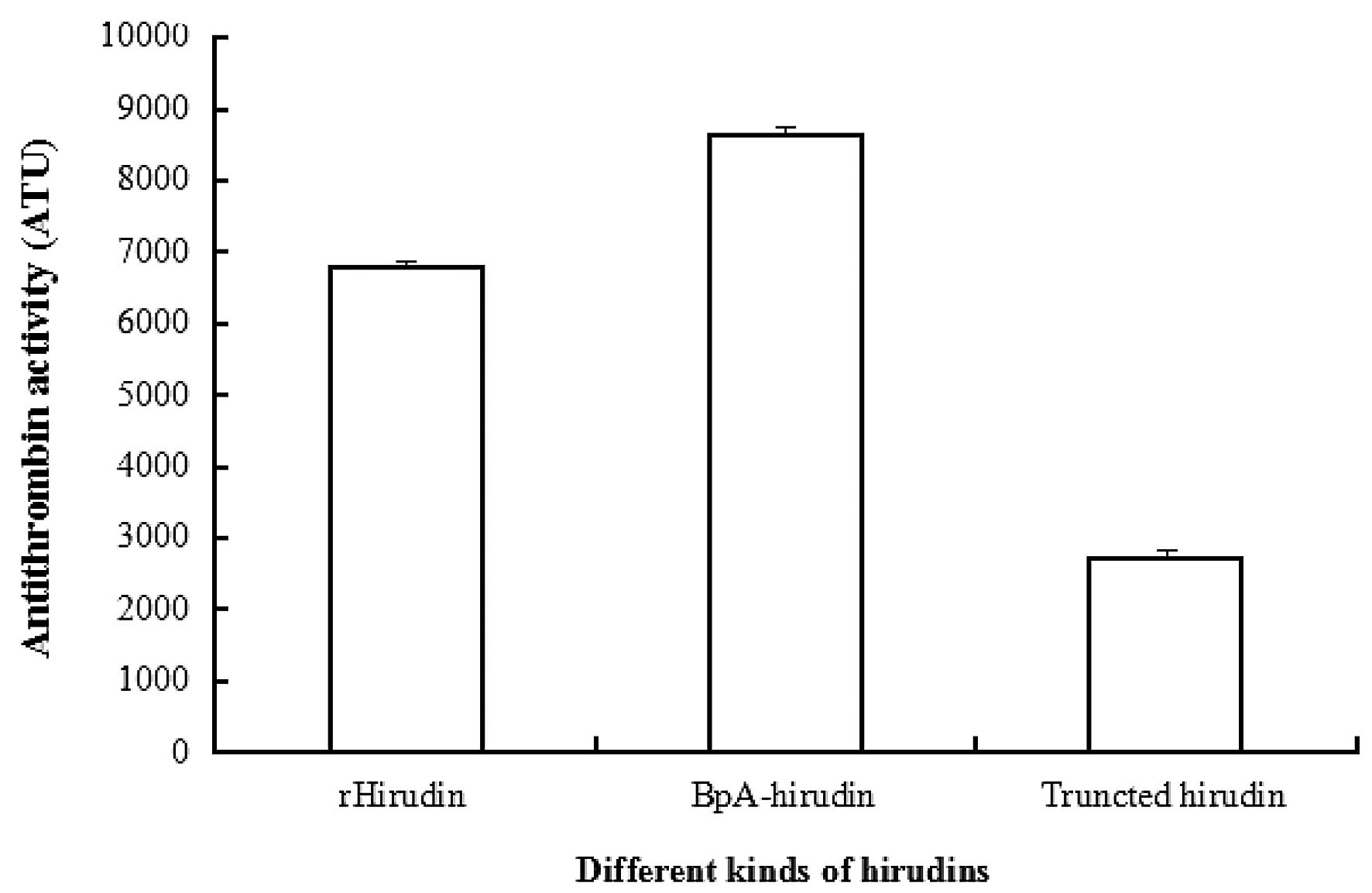
Antithrombin activity assay
The antithrombin activity of rHirudin following
purification in the present study was 6,778 ATU mg−1
protein, the antithrombin activity of the
boronophenylalanine-modified hirudin was 8,639 ATU mg−1
protein following purification. The antithrombin activity of the
site 63 boronophenylalanine-modified hirudin was enhanced compared
with that of rHirudin (Fig.
5).
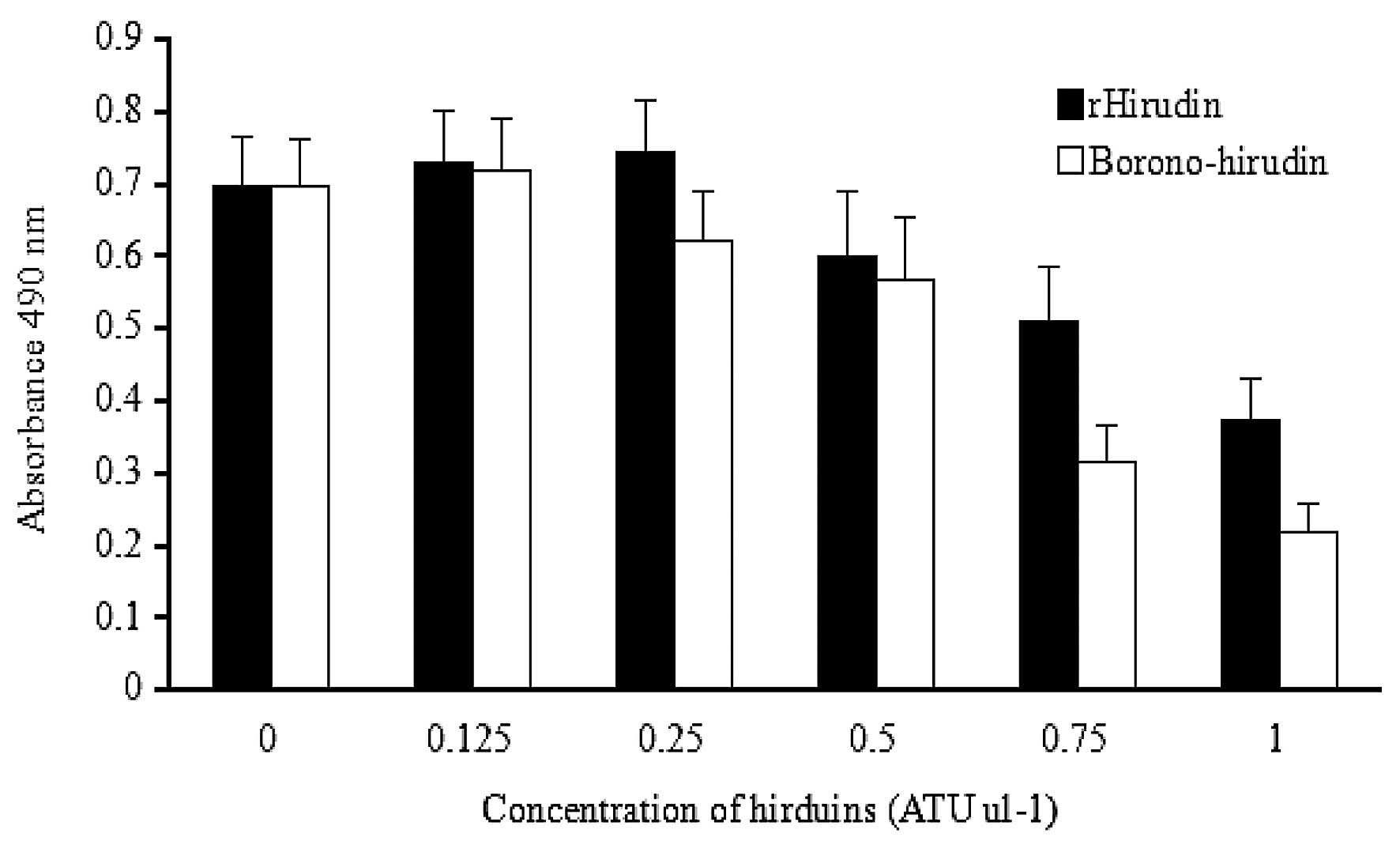
Fibroblast cell proliferation
Both hirudin solutions were demonstrated to have an
inhibitory effect on the proliferation of L929 cells when the
concentration was 0.5 U μl−1, and this proliferation
inhibition of L929 cells was dose-dependent. The proliferation rate
of boronophenylalanine-modified hirudin-treated L929 cells at 0.75
and 1 U μl−1 was significantly lower than that of
rHirudin-treated L929 cells (Fig.
6). The inhibition rate of the fibroblasts was increased by
38.4 and 40.9% following treatment with
boronophenylalanine-modified hirudin solution at 0.75 and 1 U
μl−1, respectively, compared with the inhibition by
rHirudin. At the lower concentration of the hirudin solution
(<0.5 U μl−1), the growth of the fibroblasts was
marginally stimulated, possibly due to the increased nitrogen
source from the low-dose hirudin peptides. However, the
borono-hirudin solution demonstrated a higher proliferation
inhibition effect on the L929 cells than rHirudin at any of the
concentrations.
Discussion
Using the evolved tRNA/aminoacyl-tRNA synthetase
(aaRS)/tRNA orthogonal pairs, which are able to recognize and carry
boronophenylalanine to the amber stop codon TAG, rHirudin was
successfully modified at the 63 site in the E. coli
expression system and its antithrombin activity was improved.
To avoid the degradation of the cytoplasm and
improve the production of hirudin, the gIII signal peptide was
fused to the N-terminus of hirudin, which led to the secretion of
the proteins of interest into the culture medium efficiently and
successfully. A 6× histidine sequence at the C-terminus of the
hirudin simplified the purification procedure, as only the
full-length proteins carried this histidine tag and bound to the
Ni-NTA column. Besides, the hexapeptides at the C-terminus had no
evident effect on its anticoagulant activity (6,778 ATU
mg−1 protein) compared with previous studies, in which
no His-tag was used (7, 229.13 ATU mg−1 protein)
(18). Following Ni-NTA affinity
chromatography and gel filtration, the purity was up to 95%, as
detected by SDS-PAGE.
The boronophenylalanine-modified hirudin had an
antithrombin activity of 8,639 ATU mg−1 protein, which
was higher than that of rHirudin (6,778 ATU mg−1
protein). A number of studies demonstrated that boronophenylalanine
is able to be used as a negatively charged amino acid, and the
boronic acids of it may form reversible covalent complexes with
peptides or proteins (20). The
side chain had a good spatial overlap with the positively charged
arginine residues of the interactive proteins and afforded
effective activity to the modified proteins. The boronic acids have
the ability of binding with diols or reactive serine residues, and
it was suggested that this unique feature of boronate-containing
proteins may specifically recognize and covalently bind various
glycoproteins or proteases (21).
It may also be possible to form intramolecular serine-boronate
crosslinks in proteins and enhance the combining stability between
the complexes as previously described (21). Therefore, the boronophenylalanine
side chain may be one of the possible reasons for the enhanced
antithrombin activity of the modified hirudins, in a similar
fashion to the effect of the sulfotyrosine-modified hirudin.
Sulfotyrosine was recognized and incorporated specifically into
target proteins when an amber nonsense codon with an orthogonal
aminoacyl-tRNA synthetase/tRNA pair was employed in the E.
coli expression system, and the affinity of the resulting
sulfotyrosinyl hirudin for human thrombin was enhanced by 10-fold
(9). According to a crystal
structure study of a sulfo-hirudin - thrombin complex, it was
demonstrated that the side chain of the sulfate group on
sulfotyrosine at the 63 site formed a strong salt bridge with the
positively charged Lys 81 of thrombin, and thus improved the
affinity of the interaction between sulfo-hirudin and human
thrombin (22).
As described in previous studies, the repressed
expression of plasminogen activator inhititor-1 (PAI-1), the type I
collagen α-chain (COL1A1) and a member the of interleukin family
all resulted in the inhibition of proliferation of fibroblasts
(23). The modified hirudin may
have had a stronger effect on the expression of one or all of the
proliferation factors mentioned above. Further studies are required
to determine the mechanisms underlying this effect.
In the present study, it was identified that the
incorporation efficiency of the boronophenylalanine amber
suppressor MjBTyrRS/tRNATyr cua pair
was higher than that of the reported sulfotyrosine incorporation
amber suppressor aminoacyl-tRNA synthetase/tRNA orthogonal pair. A
total of ~52.6% full length proteins was produced and the yield was
10 mg l−1 in a 500-ml flask containing 200 ml culture
medium, while only 25% full length protein was generated in the
sulfotyrosine incorporation system, due to the low permeability of
the anionic sulfotyrosine into E. coli cells as described
previously (11).
The improved bioactivity and protein yield of the
boronophenylalanine-modified hirudin suggested a possible clinical
application of the boronophenylalanine recognition and
incorporation of orthogonal pairs for producing site-specifically
modified hirudins. The enhanced bioactivity of the
site-specifically modified hirudin suggested a higher potential for
therapeutic application over the prevailing rHirudin. Furthermore,
boronic acids do not naturally occur in polypeptides and are
introduced either as posttranslational modifications or as
co-factors. Therefore, the successful incorporation of this amino
acid into proteins allowed for a notably more selective chemical
reaction on the modified protein surface, including oxidation,
reduction, Suzuki coupling reactions or the forming of covalent
boronic esters with other polyhydroxylated compounds, and may be
used to purify native protein sequences in a one-step affinity
purification procedure.
In conclusion, the present study reported a feasible
method to improve the biological activity of hirudin and provided a
new way to access novel hirudin derivatives that may have marked
clinical utility.
Acknowledgements
The authors are grateful to Dr K.Odoi of A&M
University for reviewing the manuscript. This study was supported
by the Fundamental Research Funds for the Central Universities of
China (nos. WF1114045 and 222201314027) and the Open Project for
the State Key Laboratory of Bioreactor Engineering (no.
2060204).
References
|
1
|
Sohn JH, Kang HA, Rao KJ, Kim CH, et al:
Current status of the anticoagulant hirudin: its biotechnological
production and clinical practice. Appl Microbiol Biotechnol.
57:606–613. 2001. View Article : Google Scholar
|
|
2
|
Markwardt F: Hirudin as alternative
anticoagulant - a historical review. Semin Thromb Hemost.
28:405–414. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stone SR and Hofsteenge J: Kinetics of the
inhibition of thrombin by hirudin. Biochemistry. 25:4622–4628.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee DH, Park JB, Seo JH, et al: Expression
of hirudin in fed-batch cultures of recombinant Saccharomyces
cerevisiae. Biotechnol Lett. 16:667–670. 1994. View Article : Google Scholar
|
|
5
|
Liu CC, Brustad E, Liu WS and Schultz PG:
Crystal Structure of a Biosynthetic sulfo-hirudin complexed to
thrombin. J Am Chem Soc. 129:10648–10649. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thurieau C, Simonet S, Paladino J, et al:
New N alpha-guanidinobenzoyl derivatives of hirudin-54–65
containing stabilized carboxyl or phosphoryl groups on the side
chain of phenylalanine-63. J Med Chem. 37:625–629. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Myung J, Kim KB and Crews CM: The
ubiquitin-proteasome pathway and proteasome inhibitors. Med Res
Rev. 21:245–273. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang W, Gao X and Wang B: Boronic
acid-based sensors for carbohydrates. Curr Org Chem. 6:1285–1317.
2002. View Article : Google Scholar
|
|
9
|
Suzuki A: Recent advances in the
cross-coupling reactions of organoboron derivatives with organic
electrophiles, 1995–1998. J Organomet Chem. 576:147–168. 1999.
View Article : Google Scholar
|
|
10
|
Huang SW, Wang B, Shan ZX and Zhao DJ:
Asymmetric reduction of acetophenone with borane catalyzed by
chiral oxazaborolidinones derived from L-α-amino acids. Synthetic
Commun. 30:2423–2429. 2000. View Article : Google Scholar
|
|
11
|
Liu CC and Schultz PG: Recombinant
expression of selectively sulfated proteins in Escherichia coli.
Nat Biotechnol. 24:1436–1440. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brustad E, Bushey ML, Lee JW, et al: A
genetically encoded boronate-containing amino acid. Angew Chem Int
Ed Engl. 47:8220–8223. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu DE, Li X, Zhang GY, Niu ZG, Yi CG, Jia
BY, Xia W and Guo SZ: Experimental study on effect of hirudin in
inhibiting hyperplastic scar fibroblasts. Zhonghua Shao Shang Za
Zhi. 25:265–267. 2009.(In Chinese). PubMed/NCBI
|
|
14
|
Jääger K and Neuman T: Human dermal
fibroblasts exhibit delayed adipogenic differentiation compared
with mesenchymal stem cells. Stem Cells Dev. 20:1327–1336. 2011.
View Article : Google Scholar
|
|
15
|
Zheng L, Baumann U and Reymond JL: An
efficient one-step site-directed and site-saturation mutagenesis
protocol. Nucleic Acids Res. 32:e1152004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schägger H: Tricine-SDS-PAGE. Nat Protoc.
1:16–22. 2006. View Article : Google Scholar
|
|
17
|
Tan S, Wu W, Liu J, et al: Efficient
expression and secretion of recombinant hirudin III in E. coli
using the L-asparaginase II signal sequence. Protein Expr Purif.
25:430–436. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ferrari M, Fornasiero MC and Isetta AM:
MTT colorimetric assay for testing macrophage cytotoxic activity in
vitro. J Immunol Methods. 131:165–172. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lv J, Huang C, Zhang X and Tan S:
Extracellular secretion of anticoagulant peptide hirudin in
Lactococcus lactis using SP310mut2 signal peptide. Biotechnol Lett.
34:61–65. 2012. View Article : Google Scholar
|
|
20
|
Mohler LK and Czarnik AW: α-Amino acid
chelative complexation by an arylboronic acid. J Am Chem Soc.
116:22331994. View Article : Google Scholar
|
|
21
|
Yang W, Gao X and Wang B: Boronic acid
compounds as potential pharmaceutical agents. Med Res Rev.
23:346–368. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rydel TJ, Ravichandran KG, Tulinsky A, et
al: The structure of a complex of recombinant hirudin and human
alpha-thrombin. Science. 249:277–280. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matsui T, Ito C, Oda M, et al: Lapachol
suppresses cell proliferation and secretion of interleukin-6 and
plasminogen activator inhibitor-1 of fibroblasts derived from
hypertrophic scars. J Pharm Pharmacol. 63:960–966. 2011. View Article : Google Scholar : PubMed/NCBI
|