1. Introduction
Neural precursor cell expressed, developmentally
downregulated 9 (NEDD9), a gene exclusively expressed in the brain
during embryonic stages but not in the brains of mature mice, was
firstly identified in 1992 by Kumar et al (1) by subtractive cloning technology. In
1996, Law et al (2) was the
first to ascribe a biological function to NEDD9. They screened a
number of genes that can induce the sprouting of filamentous yeast
and found a number of proteins that was able to regulate human cell
polarity and the cell cycle. Among these proteins, human enhancer
of filamentation 1 (HEF1) is expressed in a variety of human cell
lines and effectively regulates yeast-cell polarity due to its RecQ
C-terminal domain (2). Also in
1996, Minegishi et al (3)
identified Crk-associated substrate-related protein lymphocyte type
(Cas-L) as they studied a tyrosine hyper-phosphorylated protein
under the activation of T cell β1-binding; Cas-L was shown to be
identical to NEDD9/HEF1 by sequence alignment (3). Therefore, NEDD9, HEF1 and Cas-L are
three different names for the same gene and are used
interchangeably.
To date, no evidence has indicated that NEDD, as a
cytoskeletal protein, has enzyme activity; however, several
structural domains interacting with various proteins have been
identified. In vertebrates, the two proteins p130Cas/breast cancer
anti-estrogen resistance protein (BCAR)1 (4) and embryonal Fyn-associated substrate
(EFS)/Src interacting or signal integrating protein (SIN) (5–7) in
NEDD9 and Cas families share quite similar structural domains.
p130Cas, as the first identified protein of the Cas family, is
expressed in most tissue types and cell lines and contributes to
cell adhesion and migration (8).
According to sequence analysis, p130Cas and NEDD9 display a high
degree of homology. Thus, it was once thought that p130Cas and
NEDD9 had similar functions in the regulation of cell adhesion and
migration. The present review focused on further in-depth study of
NEDD9.
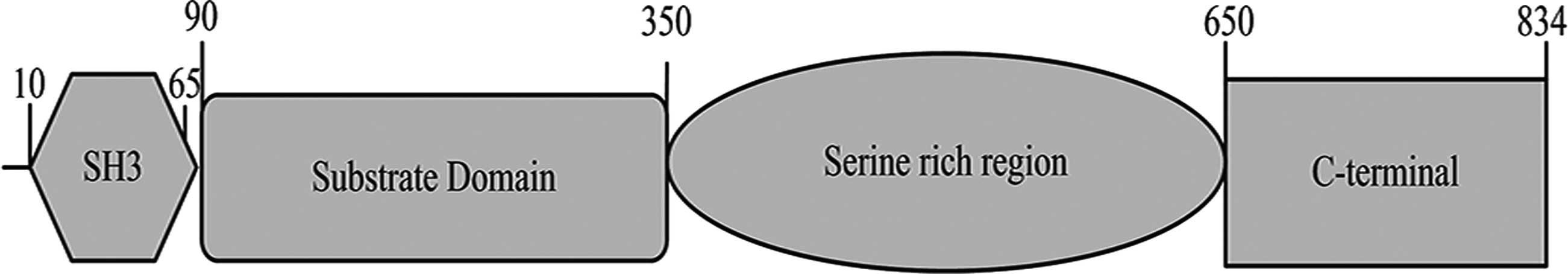
The NEDD9 gene is located in the human chromosome
6p25-24 locus and the overall length of its mRNA is 2505 nt, coding
a total of 843 amino acids (Fig.
1), where 10–65 amino acids encode the SH3 structural domain
(9). At least 15 SH2 structural
domains containing 90–350 amino acids, known as the substrate
domain (10), interact with
proteins containing the SH2 structural domain. The 350–650 amino
acids identified by bioinformatics analysis, rich in serine and
containing four helical structures (11), are likely to be protein binding
sites. The c-terminus of NEDD9 is a highly evolutionarily conserved
domain that binds with a group of spiral-loop-spiral proteins to
form dimers and heterodimers in the Cas protein (12).
2. Regulation of gene expression by
NEDD9
The regulation of NEDD9 expression is a dynamic
complex process, including phosphorylation, transcriptional
activation and proteolysis, which exerts a direct or indirect
influence on various biological processes. Normally, SDS-PAGE
analysis of NEDD9 protein displays two electrophoretic bands at
1,055 and 115 kDa (13), which are
above its molecular weight of 93 kDa, illustrating that the high
degree of phosphorylation has a key role in the regulation of gene
expression by NEDD9.
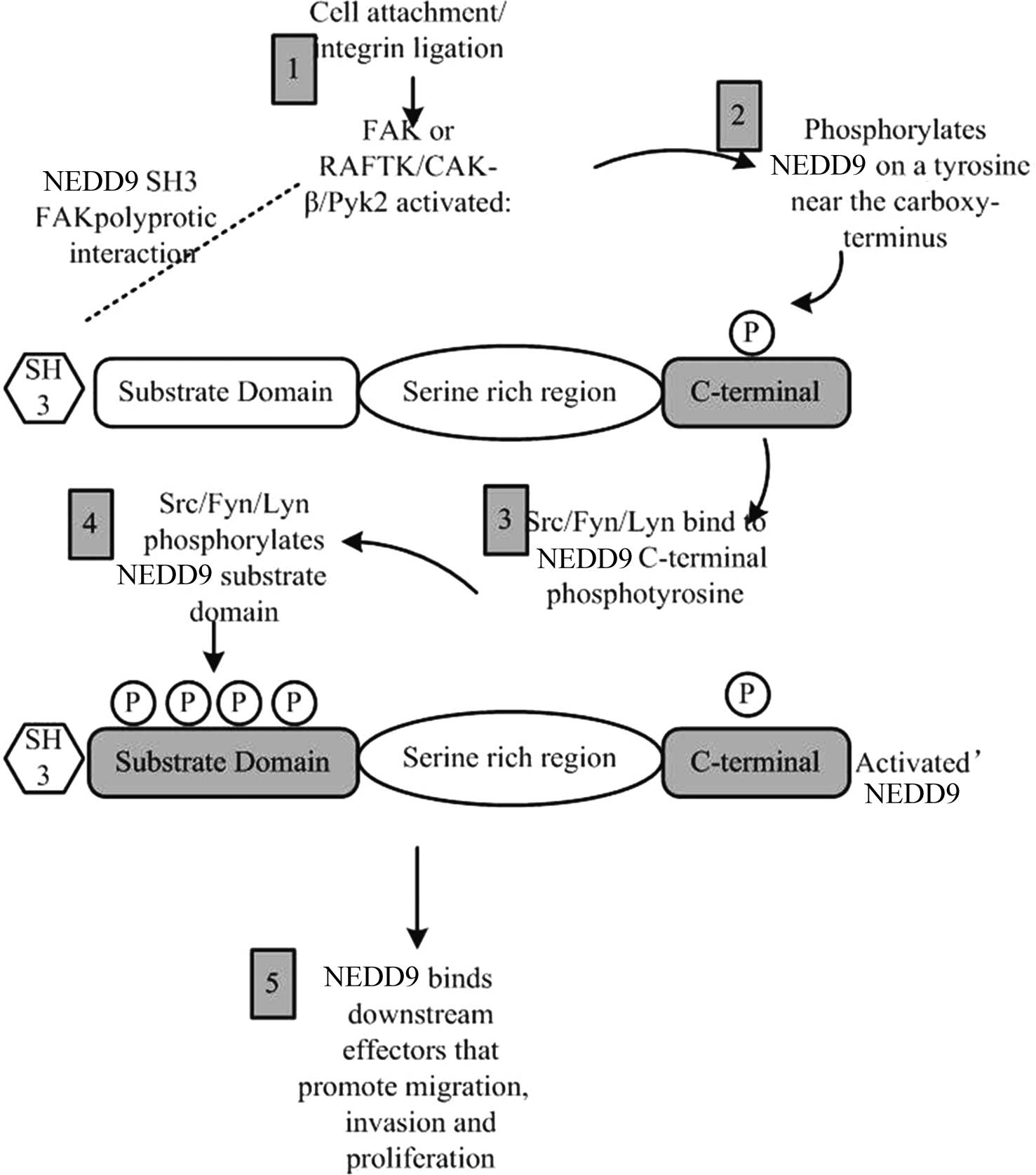
Focal adhesion kinase (FAK) and Src protein families
were the first proteins identified to be involved in the regulation
of NEDD9 phosphorylation (14,15).
In the cell adhesion process, integrin firstly activates FAK; then,
the activated FAK generates Src binding sites by tyrosine
phosphorylation near the NEDD9 C-terminal domain, and finally, more
extensive phosphorylation of the NEDD9 substrate structural domain
is generated (Fig. 2).
The phosphorylation of NEDD9 enables it to bind with
effector molecules correlated with cell migration, cell invasion
and proliferation signaling pathways. In certain types of cancer
cell, even without activation by integrin, NEDD9 phosphorylation
can be undertaken only through overexpression or activation of FAK
and Src (16). As another member
of the Cas family, EFS/SIN activates Srs with p130Cas and causes a
similar activation effect to that of NEDD9. However, their binding
domains with Srs may be different to those of NEDD9 (17). A study showed that FAK acts as an
activation agent of NEDD9 (18).
NEDD9 phosphorylation is affected by cytoskeleton actin integrity.
Actin fracture caused by pharmaceutical substances can result in
NEDD9 dephosphorylation. A study by Bargon et al (19) indicated that NEDD9
dephosphorylation brings about a change of Rho kinase activity and
a change in the hardness of cytoskeleton actin.
The expression levels of NEDD9 are low in resting
cells; however, they sharply increase when cells enter cell cycle
(13). Although the regulation of
NEDD9 expression has not been thoroughly elucidated, certain
inducible factors were found to regulate NEDD9 expression.
Transforming growth factor (TGF)-β was identified to upregulate
NEDD9 mRNA levels and enhance protein expression (20). In two different retinoblastoma cell
lines, the metabolite of vitamin A, all-cis retinoic acid (asRA),
induced NEDD9 expression, illustrating that NEDD9 expression is
associated with nerve-cell development (21,22).
In a rat model of cerebral ischemia, NEDD9 was shown to be highly
expressed in cerebral cortex and Hippocampal neurons (23). Studies on ovarian cancer and
melanoma cells indicated that enhancement of NEDD9 expression is an
important process in promoting cancer metastasis (18,24).
(Sex determining region Y)-box 2 and NANOG were also found to
combine with the promoter site of NEDD9 (25). However, the functions of these two
proteins and their association with cancer are required to be
confirmed by further studies.
Negative regulation of NEDD9 levels, including
proteolysis or degradation, occurs after gene transcription and
results in corresponding decreases in biological function. At the
telophase of mitosis, the amount of NEDD9 decreases due to
degradation caused by proteolytic enzymes (13,26).
During cell apoptosis, the specific DLVD and DDYD cleavage sites
for caspase are incised into several short fragments, which
negatively regulates the NEDD9 signaling pathway (27).
TGF-β signaling pathways also have a role in NEDD9
proteolysis. NEDD9 directly interacts with ubiquitin ligase, Smad
proteins and certain factors correlated with target protein
degradation or proteolytic cleavage, finally resulting in NEDD9
proteolysis. Furthermore, NEDD9 can regulate the activity of Smad
protein as well as inhibit the TGF-β signaling pathway (28–32).
The close association with TGF-β indicates that NEDD9 has an
important role in tumor metastasis.
3. Roles of NEDD9 in cell migration,
adhesion and invasion
Cell migration is a complex process including the
change of cell polarity, formation of microfilament and
microtubules, and finally more complex regulation, involving cell
membrane dynamics and adhesion plaque formation. NEDD9 is located
in the cell adhesion plaque and influences cell migration through
regulating the interaction of significant molecules that induce
cell migration (33). As a normal
physiological process of the body, cell migration has a positive
impact on embryonic development and the inflammatory response.
However, cell migration is abnormally activated in a large
proportion of malignant cancer cells, which is attributed to the
abnormal regulation of normal cells' non-pathological migration
mechanism in metastatic carcinoma. It was also found that changes
in NEDD9 expression have an important role in the non-pathological
movement of hematopoietic system cells (34), as well as in the migration
processes of melanoma (18),
breast cancer (35) and glioma
(36). The C-terminus of peptides
in NEDD9-overexpressing cells can induce the cells to become round
and more extended (37), followed
by loss of inter-cellular junction adhesion (38). These studies demonstrated that
NEDD9, similar to p130Cas (39),
directly regulates the formation and dissociation of focal
adhesion. In vitro migration and in vivo invasion
assays showed that the interaction of NEDD9 and FAK is a crucial
initial event during cell migration and invasion processes
(40–42). After phosphorylation by Src and
FAK, NEDD9 directly interacts with adaptor molecule Crk (15). Studies on p130Cas of the Cas family
showed that Crk can interact with p130Cas, recruit exchange factor
dedicator of cytokinesis 180, activate guanosine triphosphatase
(GTPase) Ras-related C3 botulinum toxin substrate (RAC) (43) and finally cause the cell membrane
to fluctuate and extend through polymerization of actin-related
protein 2/3 (44,45) and activation of p21-activated
kinase (46).
In addition, Crk was able to activate C3G and
another migration pathway through GTPase Ras-related protein 1
(Rap1) (47). Even though, the
activity of NEDD9 appears similar to that of p130Cas, the
mechanisms of the cell migratory pathways stimulated by the two
factors require further study. It is noteworthy that the substrate
binding site domain of NEDD9 has a binding area that can bind with
Crk as well, whereas it remains elusive whether it can cause the
activation of Rac and Rap. NEDD9 can also interact with signaling
proteins, including BCAR3/AND-34/SHEP2/Nsp2 and CHAT-H/SHEP1, which
activate downstream effector molecules through regulating the
activity of GTP enzyme (48–51)
and finally promote cell migration and invasion.
In vitro experiments showed that NEDD9
overexpression in various cell types can promote cell migration,
including the speed of random migration and its tropism (37,40–42),
while downregulation of NEDD9 expression can decrease cell
chemotaxis (34). It was suggested
that the roles of NEDD9 and p130Cas in cell migration are not
identical but associated with tissue specificity. For instance,
Natarajan et al (36) found
that NEDD9 can promote cell migration and invasion in glioma, while
p130Cas does not have this function. Another study showed that
p130Cas cannot replace the role of NEDD9 in lymphocyte migration in
NEDD9 knockout rat models (34).
Inhibition of the expression of Pho kinase,
decreased expression of FAK or dominant negative mutation
inhibition of FAK can reverse cell migration caused by NEDD9
expression (18,19,50).
Of note, in epithelial cells in which Rho expression is inhibited,
NEDD9 can induce the formation of neurite-like extensions (19), which indicates that a variety of
downstream extension factors may exist. The overexpression of NEDD9
can also activate downstream factors, including mitogen-activated
protein kinase, extracellular signal-regulated kinase 1/2 and INK,
though the specific function of these factors in the cell migration
pathway regulated by NEDD9 has not been elucidated (37). The overexpression of NEDD9 can also
activate certain genes with roles in cell migration, including
matrix metalloproteinases (MMPs), myosin light-chain kinase,
depolymerization-associated genes, Rho kinase, Nck-interacting
kinase, receptors of TGF and ErbB2/Her2/Neu receptors (37). Although the precise effects of
these cell factors have not been fully elucidated, it was shown
that factors including MMPs and depolymerization-associated
proteins promote cell migration and invasion (52).
4. Roles of NEDD9 in cell apoptosis
Normal cell apoptosis is initiated by caspase
cleavage, which is thereby activated and cleaves other proteins and
cell components. Apoptosis is accompanied by morphological
alterations, including round cell shape, cell membrane
protuberances and adhesion plaque decomposition. Cleavage of cell
components executed by caspases can cause target molecule
activation or inactivation during cell apoptosis. NEDD9 and p130Cas
are target proteins for caspases-mediated cleavage (53,54).
Cleavage of NEDD9 can inhibit integrin activity, which means that
NEDD9 serves as a sensor in the formation of adhesion plaque.
Accordingly, inhibition of integrin can cause the activation of
caspases (55). In MCF-7 breast
cancer cells and other cancer cell types (27), overexpression of the NEDD9
c-terminal 28 kD peptide can induce cell apoptosis. Furthermore,
overexpression of whole NEDD9 protein can induce apoptosis as well.
It has been suggested that low-level cleavage of NEDD9 generates a
small amount of p28 that causes adhesion plaque decomposition and
cell apoptosis (56). In MCF-7
cells, NEDD9 initially promotes cell migration and finally causes
apoptosis, indicating that the role of NEDD9 is cell
cycle-dependent (57). However,
studies on Jurkat cells, glioma cells and melanoma cells showed
that overexpression of NEDD9 does not result in apoptosis; in
contrast to MCF-7 breast cancer cells, which have a relatively low
tumor formation ability, these cells are all highly metastatic and
invasive (58). A possible
explanation for this observation is that cell survival pathway
activation is indispensable to the conversion process of the
metastatic phenotype in cancer cells. NEDD9 can only promote cancer
cell metastasis on the premise of previous survival-pathway
activation.
5. Roles of NEDD9 in cell cycle control
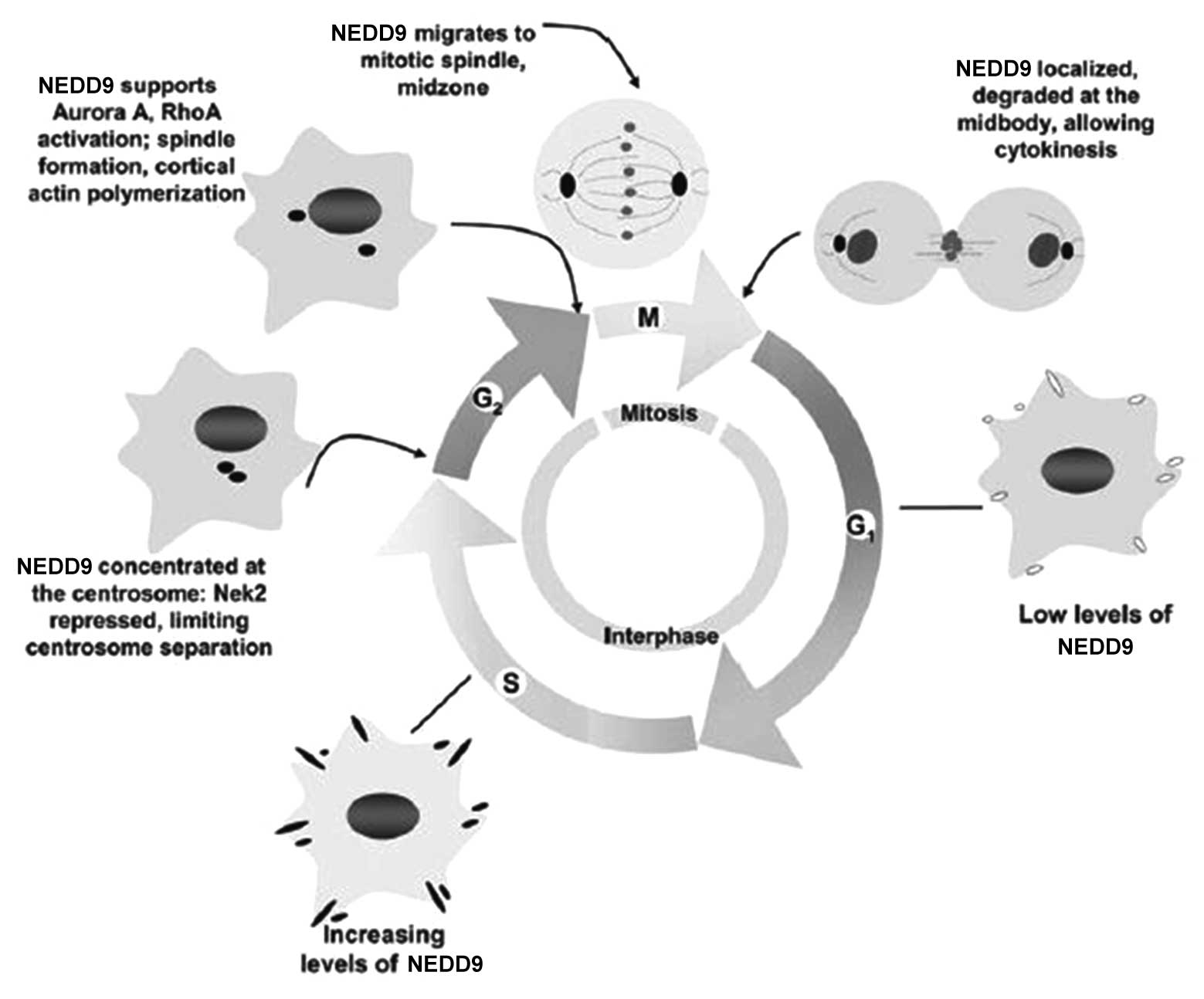
In the quiescent stage and G1 stage of the cell
cycle, the expression of NEDD9 is low. It gradually increases in S
stage and reaches a peak in G2/M stage (13). Pugacheva and Golemis (59) found that NEDD9 mainly lies in the
centrosome of mitosis in G2 stage; when mitosis commences, NEDD9
moves to the intermediate zone of mitosis along the spindle; when
the cytoplasm divides, NEDD9 is present in the midbody.
Overexpression of NEDD9 can increase the number of spindles and
centrosomes in mitosis phase, which leads to the failure of
cytoplasmic separation (60).
However, low expression of NEDD9 can cause pre-mature centrosome
separation and a lack of tubulin activation during the separation
process, which generates single-stage or asymmetric spindles and
leads to cell division failure. Cells with abnormal expression of
NEDD9 remain in G1 stage, which is consistent with the view that
NEDD9 triggers cells to enter mitosis, and due to this cell cycle
arrest, cells finally enter apoptosis.
To date, although the precise mechanism of the
regulation of the cell cycle by NEDD9 has not been fully
elucidated, certain mechanisms have been preliminarily confirmed
(Fig. 4). Initially, prior to cell
mitosis, NEDD9 combines with centrosome kinase Nek2 to induce Nek2
activation, which results in centrosome separation. In
NEDD9-negative cells, NeK2 is prematurely activated and causes the
premature separation of the centrosome. Furthermore, during the
progression from G2 to M stage, NEDD9 activates aurora A and the
timely activation of aurora A has a crucial role in the process of
mitosis. If aurora A cannot be activated in time for cell division,
cells present with the same phenotype as that of NEDD9-negative
cells (60). Finally, NEDD9 can
interact with epithelial cell transforming 2, which can
specifically activate RhoA during mitosis (61). The activation of RhoA can regulate
several key steps of mitosis. Therefore, overexpression of NEDD9
causes abnormal increases of RhoA activity and cells stay in
mitosis phase.
6. Roles of NEDD9 in development
NEDD9 also has an important role in signal
transduction of developmental cells and non-cancerous cells. Using
gene knockout technology, Seo et al (34) found that NEDD9-deficient mice
gained enhanced survival and fertility without any obvious tissue
abnormalities, while p130Cas-knockout mice died on the eleventh day
of the embryonic period (62),
which means that the function of NEDD9 can be completely replaced
by p130 or other proteins. A great number of studies showed that
the deficiency of NEDD9 leads to disturbances in development.
Studies on the normal differentiation of nerve cells and brain
development showed that NEDD9 has a significant role during this
process. Merrill et al (21,22)
found that NEDD9 has an important role in brain development by
screening all-trans retinoic acid (atRA) in a cDNA-subtractive
library, which represents the gene sequence that is expressed in
target cells, but not expressed in second cells (different types or
cell under different conditions). In the brain developmental
process, atRA, as an important regulatory factor, can promote the
extension and development of neurites. Upregulation of NEDD9
expression may be a crucial approach to active atRA. Furthermore,
NEDD9 also can interact with molecule interacting with Casl, which
can regulate the activity of plexin A and activate the semaphorin
3A signaling pathway to regulate the development of the nervous
system. Studies of gonadal differentiation in mice also found that
NEDD9 is a gender-specific gene; however, the roles of NEDD9 in
sexual development require further investigation.
7. NEDD9 as a target for cancer therapy
As a cytoskeletal protein, NEDD9 serves as a link in
signal transduction processes. NEDD9 contributes to the cell cycle
and expression or activation of numerous regulatory proteins. Due
to the 'router' function in cells, NEDD9 has a wide-ranging
influence on cell proliferation. In the early phase of normal cells
and cancer cells, the increased expression of NEDD9 can enhance the
migration and invasion abilities of cells. Furthermore, NEDD9 can
initiate post-mitotic defects associated with the failure of
cytoplasm movement. Once NEDD9 is fragmented into segments, it
causes cell adhesion and apoptosis. NEDD9-overexpressing malignant
tumors commonly feature a wide range of pre-cancerous lesions,
including the inhibition of p16Ink4, activation of Ras, translation
of human T-lymphotropic virus 1 and the BCR-ABL generated by
trans-location (18,42,63).
Hence, cancer cell invasion, apoptosis and cell division can be
inhibited through modification of these signaling pathways, which
provides novel approaches for inhibiting NEDD9-overexpressing
metastatic tumors.
NEDD9 has various roles in tumorigenesis depending
on the tumor type; this should be taken into account by clinicians
in the interpretation of NEDD9 overexpression in various tumor cell
types. For example, in solid tumors, overexpression of NEDD9 may
have different biological functions from those in hematopoietic
tissue tumors. It is hard to tell whether NEDD9 overexpression is
the key factor for tumor invasion in hemocytes, since it is normal
for hemocytes to invade other tissues. However, in epithelial
cells, NEDD9 can be identified as a biomarker of invasive solid
tumors (64).
Previous studies of NEDD9 focused on the first step
of cell invasion: Tumor cells escaping from the settlement site
(65,66). Therefore, it is likely that
activation of NEDD9 may lead to cell migration. The overexpression
of NEDD9, a significant biomarker in metastatic melanoma, can
promote lung metastases in malignant tumors; however, the processes
NEDD9 is involved in are more complex. For example, NEDD9
expression was decreased in the highly metastatic breast cancer
cell line MDA-MB231, further indicating that NEDD9 has different
roles depending on the tumor type (57).
NEDD9 is a tumor metastasis-promoting gene; however,
p130Cas does not have the same function. In normal cells, the
expression levels of NEDD9 are maintained in a dynamic balance
through transcriptional and proteolytic enzyme degradation
regulation (13). In normal cells,
various biological effects, including changes of the cell cycle,
apoptosis and cell migration, can be achieved by strict regulation
of NEDD9 expression.
To date, a profound understanding of the role of
NEDD9 in the change of benign to invasive and malignant tumors has
been acquired. At the same time, NEDD9 provides a novel target for
cancer therapy, particularly that of invasive tumors. However,
since NEDD9 does not have any obvious catalytic activity, it
remains difficult to target NEDD9 with drugs. However, by blocking
the interaction of NEDD9 with other proteins through drugs, or by
using RNA interference technology to reduce NEDD9 expression levels
may inhibit tumor metastasis.
NEDD9 is probably not an essential protein, as NEDD9
knockout mice can survive, which implies that treating tumors with
drugs or through inhibiting NEDD9 expression is feasible. The
overexpression of NEDD9 leads to the activation of Ras; therefore,
inhibition of the BRC-ABL signaling pathway or Ras function by
drugs may produce therapeutic effects against invasive and
malignant tumors caused by NEDD9-overexpression (67,68).
There remains a large amount of unanswered questions
regarding NEDD9, including the pathways via which it regulates cell
migration, its distinctive functions in different tumor stages and
its association with other diseases. Further study of NEDD9 may
provide a more profound understanding of the development of
invasive tumors. NEDD9 may serve as a potential novel target for
tumor therapy, therefore having a positive significance.
Acknowledgments
The present review was supported by the Key
Technology Research Project of Henan province (grant no.
132102310391).
References
|
1
|
Kumar S, Tomooka Y and Noda M:
Identification of a set of genes with developmentally
down-regulated expression in the mouse brain. Biochem Biophys Res
Commun. 185:1155–1161. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Law SF, Estojak J, Wang B, Mysliwiec T,
Kruh G and Golemis EA: Human enhancer of filamentation a novel
p130cas-like docking protein, associates with focal adhesion kinase
and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol
Cell Biol. 16:3327–3337. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Minegishi M, Tachibana K, Sato T, Iwata S,
Nojima Y and Morimoto C: Structure and function of Cas-L, a 105-kD
Crk-associated substrate-related protein that is involved in beta-1
integrin-mediated signaling in lymphocytes. J Exp Med.
184:1365–1375. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim M, Gans JD, Nogueira C, Wang A, Paik
JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al:
Comparative oncogenomics identifies NEDD9 as a melanoma metastasis
gene. Cell. 125:1269–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sakai R, Iwamatsu A, Hirano N, Ogawa S,
Tanaka T, Mano H, Yazaki Y and Hirai H: A novel signaling molecule,
p130, forms stable complexes in vivo with v-Crk and v-Src in a
tyrosine phosphorylation dependent manner. EMBO J. 13:3748–3756.
1994.PubMed/NCBI
|
|
6
|
Alexandropoulos K and Baltimore D:
Coordinate activation of c-Src by SH3- and SH2-binding sites on
anovel, p130Cas-related protein, Sin. Genes Dev. 10:1341. 1995.
View Article : Google Scholar
|
|
7
|
Alexandropoulos K, Donlin LT, Xing L and
Regelmann AG: Sin: Good or bad? A T lymphocyte perspective. Immunol
Rev. 192:181–195. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ishino M, Ohba T, Sasaki H and Sasaki T:
Molecular cloning of a cDNA encoding a phosphoprotein, Efs, which
contains a Src homology 3 domain and associates with Fyn. Oncogene.
11:2331–2338. 1995.PubMed/NCBI
|
|
9
|
Abassi YA, Rehn M, Ekman N, Alitalo K and
Vuori K: p130Cas Couples the tyrosine kinase Bmx/Etk with
regulation of the actin cytoskeleton and cell migration. J Biol
Chem. 278:35636–35643. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li SS: Specificity and versatility of SH3
and other proline-recognition domains: Structural basis and
implications for cellular signal transduction. Biochem J.
390:641–653. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Machida K and Mayer BJ: The SH2 domain:
Versatile signaling module and pharmaceutical target. Biochim
Biophys Acta. 1747:1–25. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Canutescu AA and Dunbrack RL Jr: MollDE: A
homology modeling framework you can click with. Bioinformatics.
21:2914–2916. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Law SF, Zhang YZ, Fashena S, Toby G,
Estojak J and Golemis EA: Dimerization of the docking/adaptor
protein HEF1/NEDD9/CAS-L via a carboxy-terminal helix-loop-helix
domain. Exp Cell Res. 252:224–235. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Law SF, Zhang YZ, Klein-Szanto AJ and
Golemis EA: Cell-cycle regulated processing of HEF1 to multiple
protein forms differentially targeted to multiple subcellular
compartments. Mol Cell Biol. 18:3540–3551. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Law SF, Estojak J, Wang B, Mysliwiec T,
Kruh G and Golemis EA: Human Enhancer of Filamentation 1 a novel
p130cas-like docking protein, associates withfocal adhesion kinase
and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol
Cell Biol. 16:3327–3337. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sima N, Cheng X, Ye F, Ma D, Xie X and Lü
W: The overexpression of scaffolding protein NEDD9 promotes
migration and invasion in cervical cancer via tyrosine
phosphorylated FAK and SRC. PLoS One. 8:e745942013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ruest PJ, Shin NY, Polte TR, Zhang X and
Hanks SK: Mechanisms of CAS substrate domain tyrosine
phosphorylation by FAK and Src. Mol Cell Biol. 21:7641–7652. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim M, Gans JD, Nogueira C, Wang A, Paik
JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al:
Comparative oncogenomics identifies NEDD9 as a melanoma metastasis
gene. Cell. 125:1269–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bargon SD, Gunning PW and O'Neill GM: The
Cas family docking protein, HEF1, promotes the formation of
neurite-like membrane extensions. Biochim Biophys Acta.
1746:143–154. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng M and McKeown-Longo PJ: Regulation
of HEF1/NEDD9/CAS-L expression and phosphorylation by TGF-beta 1
and cell adhesion. J Biol Chem. 277:39599–39608. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Merrill RA, See AW, Wertheim ML and
Clagett-Dame M: Crk-associated substrate (Cas) family member,
NEDD9, is regulated in human neuroblastoma cells and in the
embryonic hindbrain by all-trans retinoic acid. Dev Dyn.
231:564–575. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Merrill RA, Ahrens JM, Kaiser ME,
Federhart KS, Poon VY and Clagett-Dame M: All-trans retinoic
acid-responsive genes identified in the human SH-SY5Y neuroblastoma
cell line and their regulated expression in the nervous system of
early embryos. Biol Chem. 385:605–614. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sasaki T, Iwata S, Okano HJ, Urasaki Y,
Hamada J, Tanaka H, Dang NH, Okano H and Morimoto C: Nedd9 protein,
a Cas-L homologue, is upregulated after transient global ischemia
in rats. Possible involvement of Nedd9 in the differentiation of
neurons after ischemia. Stroke. 36:2457–2462. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Donninger H, Bonome T, Radonovich M,
Pise-Masison CA, Brady J, Shih JH, Barrett JC and Birrer MJ: Whole
genome expression profiling of advance stage papillary serous
ovarian cancer reveals activated pathways. Oncogene. 23:8065–8077.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boyer LA, Lee TI, Cole MF, Johnstone SE,
Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG,
et al: Core transcriptional regulatory circuitry in human embryonic
stem cells. Cell. 122:947–956. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pugacheva EN and Golemis EA: The focal
adhesion scaffolding protein HEF1 regulates activation of the
Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol.
7:937–946. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Law SF, O'Neill GM, Fashena SJ, Einarson
MB and Golemis EA: The docking protein HEF1 is an apoptotic
mediator at focal adhesion sites. Mol Cell Biol. 20:5184–5195.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nourry C, Maksumova L, Pang M, Liu X and
Wang T: Direct interaction between Smad3, APC10, CDH1 and HEF1 in
proteasomal Degradation of HEF1. BMC Cell Biol. 5:202004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu X, Elia AE, Law SF, Golemis EA, Farley
J and Wang T: A novel ability of Smad3 to regulate proteasomal
degradation of a cas family member, HEF1. EMBO J. 19:6759–6769.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng L, Guedes S and Wang T:
Atrophin-1-interacting protein 4/human Itch is a ubiquitin E3
ligase for human enhancer of filamentation 1 in transforming growth
factor-beta signaling pathways. J Biol Chem. 279:29681–29690. 2009.
View Article : Google Scholar
|
|
31
|
Inamoto S, Iwata S, Inamoto T, Nomura S,
Sasaki T, Urasaki Y, Hosono O, Kawasaki H, Tanaka H, Dang NH, et
al: Crk-associated substrate lymphocyte type regulates transforming
growth factor-beta signaling by inhibiting Smad6 and Smad7.
Oncogene. 26:893–904. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang T: The 26S proteasome system in the
signaling pathways of TGF-beta superfamily. Front Biosci.
8:1109–1127. 2003. View
Article : Google Scholar
|
|
33
|
Wozniak MA, Modzelewska K, Kwong L and
Keely PJ: Focal adhesion regulation of cell behavior. Biochim
Biophys Acta. 1692:103–119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seo S, Asai T, Saito T, Suzuki T,
Morishita Y, Nakamoto T, Ichikawa M, Yamamoto G, Kawazu M, Yamagata
T, et al: Crk-associated substrate lymphocyte type is required for
lymphocyte trafficking and marginal zone B cell maintenance. J
Immunol. 175:3492–3501. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu
W, Giri DD, Viale A, Olshen AB, Gerald WL and Massagué J: Genes
that mediate breast cancer metastasis to lung. Nature. 436:518–524.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Natarajan M, Stewart JE, Golemis EA,
Pugacheva EN, Alexandropoulos K, Cox BD, Wang W, Grammer JR and
Gladson CL: HEF1 is a necessary and specific downstream effector of
FAK that promotes the migration of glioblastoma cells. Oncogene.
25:1721–1732. 2006. View Article : Google Scholar
|
|
37
|
Fashena SJ, Einarson MB, O'Neill GM,
Patriotis C and Golemis EA: Dissection of HEF1-dependent functions
in motility and transcriptional regulation. J Cell Sci. 115:99–111.
2002.PubMed/NCBI
|
|
38
|
O'Neill GM and Golemis EA: Proteolysis of
the docking protein HEF1 and implications for focal adhesion
dynamics. Mol Cell Biol. 21:5094–5108. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Webb DJ, Donais K, Whitmore LA, Thomas SM,
Turner CE, Parsons JT and Horwitz AF: FAK-Src signalling through
paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell
Biol. 6:154–161. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Van Seventer GA, Salman HJ, Law SF, et al:
Focal adhesion kinase regulates beta1 integrin dependent migration
through an HEF1/NEDD9/CAS-L effector pathway. Eur J Imm.
31:1417–1427. 2001. View Article : Google Scholar
|
|
41
|
Ohashi Y, Iwata S, Kamiguchi K and
Morimoto C: Tyrosine phosphorylation of Crk-associated substrate
lymphocyte-type is a critical element in TCR- and beta1
integrin-induced T lymphocyte migration. J Immunol. 163:3727–3734.
1999.PubMed/NCBI
|
|
42
|
Iwata S, Souta-Kuribara A, Yamakawa A,
Sasaki T, Shimizu T, Hosono O, Kawasaki H, Tanaka H, Dang NH,
Watanabe T, et al: HTLV-I Tax induces and associates with
Crk-associated substrate lymphocyte type (Cas-L). Oncogene.
24:1262–1271. 2005. View Article : Google Scholar
|
|
43
|
Klemke RL, Leng J, Molander R, Brooks PC,
Vuori K and Cheresh DA: CAS/Crk coupling serves as a 'molecular
switch' for induction of cell migration. J Cell Biol. 140:961–972.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ridley AJ: Rho proteins: Linking signaling
with membrane trafficking. Traffic. 2:303–310. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Smith LG and Li R: Actin polymerization:
Riding the wave. Curr Biol. 14:R109–R111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cai D, Iyer A, Felekkis KN, Near RI, Luo
Z, Chernoff J, Albanese C, Pestell RG and Lerner A: AND-34/BCAR3, a
GDP exchange factor whose overexpression confers anti estrogen
resistance, activates Rac, PAK1 and the cyclin D1 promoter. Cancer
Res. 63:6802–6808. 2003.PubMed/NCBI
|
|
47
|
Tamada M, Sheetz MP and Sawada Y:
Activation of a signaling cascade by cytoskeleton stretch. Dev
Cell. 7:709–718. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cai D, Felekkis KN, Near RI, O'Neill GM,
van Seventer JM, Golemis EA and Lerner A: The GDP exchange factor
AND-34 is expressed in B cells, associates with HEF1 and activates
Cdc42. J Immunol. 170:969–978. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gotoh T, Cai D, Tian X, Feig LA and Lerner
A: p130Cas regulates the activity of AND-34, a novel Ral, Rap1 and
R-Ras guanine nucleotide exchange factor. J Biol Chem.
275:30118–30123. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sakakibara A, Hattori S, Nakamura S and
Katagiri T: A novel hematopoietic adaptor protein, Chat-H,
positively regulates T cell receptor-mediated interleukin-2
production by Jurkat cells. J Biol Chem. 278:6012–6017. 2003.
View Article : Google Scholar
|
|
51
|
Sakakibara A and Hattori S: Chat, a
Cas/HEF1-associated adaptor protein that integrates multiple
signaling pathways. J Biol Chem. 275:6404–6410. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lucas JT Jr, Salimath BP, Slomiany MG and
Rosenzweig SA: Regulation of invasive behavior by vascular
endothelial growth factor is HEF1-dependent. Oncogene.
29:4449–4459. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gervais FG, Thornberry NA, Ruffolo SC,
Nicholson DW and Roy S: Caspases cleave focal adhesion kinase
during apoptosis to generate a FRNK-like polypeptide. J Biol Chem.
273:17102–17108. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kook S, Shim SR, Choi SJ, Ahnn J, Kim JI,
Eom SH, Jung YK, Paik SG and Song WK: Caspase-mediated cleavage of
p130Cas in etoposide-induced apoptotic Rat-1 cells. Mol Biol Cell.
11:929–939. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Stupack DG, Puente XS, Boutsaboualoy S,
Storgard CM and Cheresh DA: Apoptosis of adherent cells by
recruitment of caspase-8 to unligated integrins. J Cell Biol.
155:459–470. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Frisch SM: Anoikis. Methods Enzymol.
322:472–429. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chang JX, Gao F, Zhao GQ and Zhang GJ:
Role of NEDD9 in invasion and metastasis of lung adenocarcinoma.
Exp Ther Med. 4:795–800. 2012.PubMed/NCBI
|
|
58
|
Biscardi JS, Belsches AP and Parsons SJ:
Characterization of human epidermal growth factor receptor and
c-Src interactions in human breast tumor cells. Mol Carcinog.
21:261–272. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Pugacheva EN and Golemis EA: HEF1-aurora A
interactions: Points of dialog between the cell cycle and cell
attachment signaling networks. Cell Cycle. 5:384–391. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dadke D, Jarnik M, Pugacheva EN, Singh MK
and Golemis EA: Deregulation of HEF1 impairs M-phase progression by
disrupting the RhoA activation cycle. Mol Biol Cell. 17:1204–1217.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fritz G and Kaina B: Rho GTPases:
Promising cellular targets for novel anticancer drugs. Curr Cancer
Drug Targets. 6:1–14. 2006.PubMed/NCBI
|
|
62
|
Honda H, Oda H, Nakamoto T, Honda Z, Sakai
R, Suzuki T, Saito T, Nakamura K, Nakao K, Ishikawa T, et al:
Cardiovascular anomaly, impaired actin bundling and resistance to
Src-induced transformation in mice lacking p130Cas. Nat Genet.
19:361–365. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
63
|
Dail M, Kalo MS, Seddon JA, Côté JF, Vuori
K and Pasquale EB: SHEP1 function in cell migration is impaired by
a single amino acid mutation that disrupts association with the
scaffolding protein cas but not with Ras GTPases. J Biol Chem.
279:41892–41902. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Speranza MC, Frattini V, Pisati F, Kapetis
D, Porrati P, Eoli M, Pellegatta S and Finocchiaro G: NEDD9, a
novel target of miR-145, increases the invasiveness of
glioblastoma. Oncotarget. 3:723–734. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chang JX, Gao F, Zhao GQ and Zhang GJ:
Expression and clinical significance of NEDD9 in lung tissues. Med
Oncol. 29:2654–2660. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu Y, Wang D, Zhao KL, Zhu JW, Yin HB,
Wei YZ, Wu ZJ, Cheng GJ, Wang F, Ni F, et al: NEDD9 overexpression
correlates with poor prognosis in gastric cancer. Tumour Biol.
35:6351–6356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kato-Stankiewicz J, Hakimi I, Zhi G, Zhang
J, Serebriiskii I, Guo L, Edamatsu H, Koide H, Menon S, Eckl R, et
al: Inhibitors of Ras/Raf-1 interaction identified by two-hybrid
screening revert Ras-dependent transformation phenotypes in human
cancer cells. Proc Natl Acad Sci USA. 99:14398–14403. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Druker BJ: Perspectives on the development
of a molecularly targeted agent. Cancer Cell. 1:31–36. 2002.
View Article : Google Scholar : PubMed/NCBI
|