Introduction
Chronic hepatitis B (CHB) is a major global health
problem, with ~350 million individuals affected worldwide.
Infection with the hepatitis B virus (HBV) is a major cause of the
development of cirrhosis, decompensated liver disease and
hepatocellular carcinoma (1).
Therefore, therapies which effectively inhibit HBV replication and
prevent the progression of HBV-associated liver diseases are
urgently required (2,3). Nucleoside/nucleotide analogs (NUCs)
provide one of the currently available therapies for the management
of CHB, including lamivudine (LAM), adefovir (ADV), telbivudine,
entecavir (ETV) and tenofovir (TDF) (4). NUCs are widely used for treating CHB
due to a number of advantages, including the ease of oral
administration, good tolerance and rapid viral suppression;
however, long-term therapies with NUCs, particularly early approved
NUCs, results in the emergence of viral mutations, which are
responsible for virological, and subse-quently, biochemical
breakthrough, followed by a worsening of the liver disease
(5). Virological breakthrough is
the first manifestation of the disease, predominantly caused by
resistance in patients treated with NUCs (6). Therefore, developing a further
understanding of the underlying mechanism of virological
breakthrough during treatment with NUCs may hold promise for the
development of optimal strategies for NUC therapies, and the
management of drug resistance. Currently, techniques which are
available to investigate mutations in HBV are limited. Among them,
Sanger sequencing is widely used for analyzing DNA sequences,
although its usefulness is limited by its low sensitivity and the
long duration required, in addition to an inability to perform
haplotype analysis, which renders the method unsuited for
investigating the mechanism that underlies the evolution of HBV
quasispecies during treatment with NUCs (7). However, technological advances are
improving the situation, including the development of next
generation sequencing techniques, which are capable of detecting
minor and longitudinal drug-associated mutations (8). Ultra-deep pyrosequencing (UDPS) is
based on the 454 sequencing technology, and it is useful for
detecting thousands of clonally amplified sequences. UDPS has
previously been applied to the investigation of HIV (9,10)
and hepatitis C virus (HCV) (11–13),
and noteworthy results were generated in these early studies.
However, the data which have been obtained from the application of
UDPS to HBV are limited, particularly regarding longitudinal
studies of the dynamic pattern of viral evolution during antiviral
therapy.
In the present study, the underlying mechanism of
virological breakthrough in patients with CHB receiving NUCs was
investigated using UDPS in the reverse transcriptase (RT) region of
HBV, and the dynamic viral evolution pattern during virological
breakthrough was further investigated.
Materials and methods
Enrollment of the patients and the study
design
Patients with CHB, together with compensated
cirrhosis, were selected from a prospective study, based on a
treatment regimen over a 96 week period, which comprised
combination therapy with LAM and ADV (14). Briefly, the patients were treated
with LAM monotherapy for the first 24 weeks. At week 24, a decision
was taken to switch to combination therapy or to continue with the
monotherapy, according to the response of the patient to the
treatment. Combination therapy was performed for all patients
starting from week 48, and the treatment ended at week 96. For each
patient, measurements of the DNA level of HBV were serially
recorded at weeks 0, 12, 24, 36, 48, 72 and 96. Plasma samples were
collected at weeks 0, 24, 36, 48 and 96, and were subsequently
stored at −80°C prior to the UDPS analysis. A partial virological
response was defined as a decrease in the level of HBV DNA of >1
log10 IU/ml, but with detectable levels of HBV DNA
remaining following 6 months of therapy in compliant patients
(15). Virological breakthrough
was defined as an increase of >1 log10 IU/ml in the
level of the HBV DNA from nadir during the treatment (15). To investigate the mechanism
underlying the virological breakthrough, patients were enrolled in
the present study who experienced viral breakthrough and were
receiving ADV add-on as a rescue therapy. The present study (ref.
no. 07-05) was approved by the ethics committee of Jing'an District
Central Hospital (Shanghai, China).
Polymerase chain reaction (PCR)
amplification and 454 sequencing
The DNA from serum samples was extracted using a
Qiagen® DNeasy Blood & Tissue kit (Qiagen, Inc.,
Valencia, CA, USA), according to a previously published procedure
(16). Briefly, the DNA was
isolated and purified through a spin column, according to the
manufacturer's instructions. The DNA was initially adsorbed onto
the silica of the column, followed by several washes with wash
buffer, containing 70% ethanol. The DNA was subsequently eluted
from the column, prior to the PCR amplification of the RT region
for the identification of mutations by 454 sequencing. The RT
region sequence was divided into three overlapping fragments for
PCR amplification. The template-specific sequences of the primer
pairs used in the present study are listed in Table I. The full forward primers
consisted of a directional GS FLX Titanium Primer A sequence
(synthesized by Sangon Biotech Co., Ltd., Shanghai, China). A 10
base pair barcode sequence upstream of the template-specific
forward sequence was used for the identification of the samples.
The reverse primers consisted of a GS FLX Titanium Primer B
sequence barcode, in addition to the template-specific reverse
sequence. The PCR cycling conditions were 94°C for 5 min, followed
by 30 cycles of denaturation at 94°C for 30 sec, annealing at 63°C
for 30 sec and extension at 72°C for 40 sec, using the Ex Taq™ DNA
polymerase (Takara Bio, Inc., Dalian, China). The fragments were
purified using a MinElute gel extraction kit (Qiagen, Inc.). The
DNA concentration in the purified PCR products was measured using a
PicoGreen quantification assay, according to the manufacturer's
instructions (Invitrogen Life Technologies, Carlsbad, CA, USA). The
barcoded samples were pooled with the identical quantity of DNA
from each sample, and the pooled PCR products were subjected to
standard 454 DNA sequencing, according to the manufacturer's
instructions (GS FLX system; 454 Life Sciences, Branford, CT,
USA).
 | Table IOligonucleotide primers used for
polymerase chain reaction amplification and deep sequencing of HBV
reverse transcriptase genes. |
Table I
Oligonucleotide primers used for
polymerase chain reaction amplification and deep sequencing of HBV
reverse transcriptase genes.
| Primer | Sequence |
|---|
| HBV-F1 |
5′-CTGCTGGTGGCTCCAGTT-3′ |
| HBV-R1 |
5′-GCAGGTCTTGCATGGTCCCGT-3′ |
| HBV-F2 |
5′-ATGTTGCCCGTTTGTCCTC-3′ |
| HBV-R2 |
5′-CCCAACTTCCAATTACATA-3′ |
| HBV-F3 |
5′-TAATAAAACCAAACGTTGGGGC-3′ |
| HBV-R3 |
5′-AGGAGTTCCGCAGTATGGAT-3′ |
Data and statistical analysis
The 454-generated FASTA (.fna) and quality score
(.qual) files were obtained as raw sequence data. The multiplexed
reads were split and assigned to samples on the basis of their
unique nucleotide barcode. The sequences were screened, trimmed and
filtered using programs written by Encode Genomics (Suzhou, China),
and the qualified sequence fragments were subsequently used for
Basic Local Alignment Search Tool (BLAST) analysis to identify the
mutations, by comparing with the sequence of the HBV RT region as
the reference from the National Center for Biotechnology
Information database (http://www.ncbi.nlm.nih.gov). The perl script for
formatting the data for BLAST analysis was designed in-house.
Sequences which were too short (<50 bp) and sequences with a
low-quality score were removed. After having calculated that the
mutation rate for the clean data was <1, a cut-off of 1% was
used to differentiate an authentic variant from a possible
artificial error, therefore, only a mutation rate >1 was
considered to indicate the real existence of the mutation in the RT
region (17). All data were
analyzed using the SPSS 19.0 statistical software package (IBM
SPSS, Chicago, IL, USA), and all continuous variables are expressed
as the median (range). All P values were two-sided and P<0.05
was considered to indicate a statistically significant
difference.
Results
Baseline characteristics of the
patients
A total of 12 patients (patients A-L) with CHB were
enrolled in the present study. The patients were initially treated
with LAM monotherapy, and ADV was added to the regimen as a rescue
therapy once the patients experienced virological breakthrough
(with the exception of patient C, who received ADV prior to the
virological breakthrough due to only a partial response to LAM).
The combination therapy lasted until week 96. The baseline
characteristics of the patients enrolled in this study are
summarized in Table II. A total
of 10/12 patients were HBeAg positive prior to the treatment. Among
these patients, only patient K experienced a HBeAg seroconversion
during the treatment with LAM. The mean age of the 12 patients was
47±10.03 years, and the mean baseline levels of alanine transminase
and aspartate transaminase levels were 67.31±30.83 and 49.87±16.79
IU/L respectively.
 | Table IIBaseline characteristics of the 12
patients enrolled in the present study. |
Table II
Baseline characteristics of the 12
patients enrolled in the present study.
| Patient no. | Age (year) | Gender | HBV e antigen
status | HBV DNA
level
(log10 IU/ml) | ALT (IU/L) | HBsAg (IU/ml) |
|---|
| A | 48 | M | + | 6.50 | 82 | 2,118.03 |
| B | 36 | M | + | 6.15 | 54 | |
| C | 48 | F | − | 8.04 | 96 | 8,938.29 |
| D | 60 | F | + | 8.00 | 107 | 118.50 |
| E | 55 | M | − | 5.13 | 52 | 1,587.81 |
| F | 43 | M | + | 6.39 | 114 | 1,933.33 |
| G | 43 | M | + | 6.99 | 69 | 1,239.75 |
| H | 59 | M | + | 7.33 | 77 | 3,443.97 |
| I | 30 | M | + | 6.11 | 35 | 2,558.67 |
| J | 61 | F | + | 8.04 | 30 | 4,053.79 |
| K | 39 | M | + | 6.05 | 76 | 998.37 |
| L | 42 | F | + | 5.40 | 16 | 156.86 |
Mutation of the HBV RT region detected by
UDPS at different time points during the treatment
UDPS was performed to investigate the emergence of
any drug resistance-associated mutations in the 12 patients at
weeks 0, 24, 36, 48 and 96 during the therapy, representing a total
of 54 serial samples (six of the serum samples were too low to
perform a UDPS assay). A total of ~345,025 sequences were
generated, with 6,389±916 sequences per sample and a mean length of
380±117 nucleotides, following the elimination of unqualified and
excessively short sequences (<50 bp).
The whole RT region was evaluated for each patient,
and the rates of the mutation sites for the 12 patients at the
different time points are summarized in Table III. Although all 12 patients
experienced viral breakthrough during the treatment with LAM, low
rates of drug resistance-associated mutations were observed at week
0 prior to the treatment, with the exception of patient D, who
exhibited a rate of 94.03 for M204I, and patient G, who exhibited a
rate of 97.59 for A181V. Upon the initiation of the LAM
monotherapy, which was accompanied by a decrease in the DNA level
of HBV, the HBV DNA evolved on an individual basis, although
several common characteristics were observed. The mutation rates of
LAM-resistance-associated sites, including rt204, rt180 and rt80,
slowly increased following the initiation of the LAM monotherapy,
and the combination therapy resulted in an increase in the mutation
rates at ADV-resistance-associated sites, including rt181 and rt236
(Table III). Notably, the
mutation rate of rtM204I/V was markedly increased at the time of
virological breakthrough in eight patients, and this was
accompanied by an increased mutation rate of rtL180M and/or
rtL80I/V. In addition, different HBV evolution profiles were
observed during the ADV add-on therapy.
 | Table IIIMutation rates detected using
ultra-deep pyrosequencing analysis in the 12 patients at different
time points during the treatment. |
Table III
Mutation rates detected using
ultra-deep pyrosequencing analysis in the 12 patients at different
time points during the treatment.
Patterns of viral evolution in the
patients with LAM-induced virological breakthrough
Since the above results suggested that the marked
mutation rate observed at the rt204 site was the predominant cause
of the virological breakthrough, the 12 patients were further
divided into three groups on the basis of the type of rt204
mutation. Notably, common characteristics were observed in the
three types of rt204 mutation-associated virological
breakthrough.
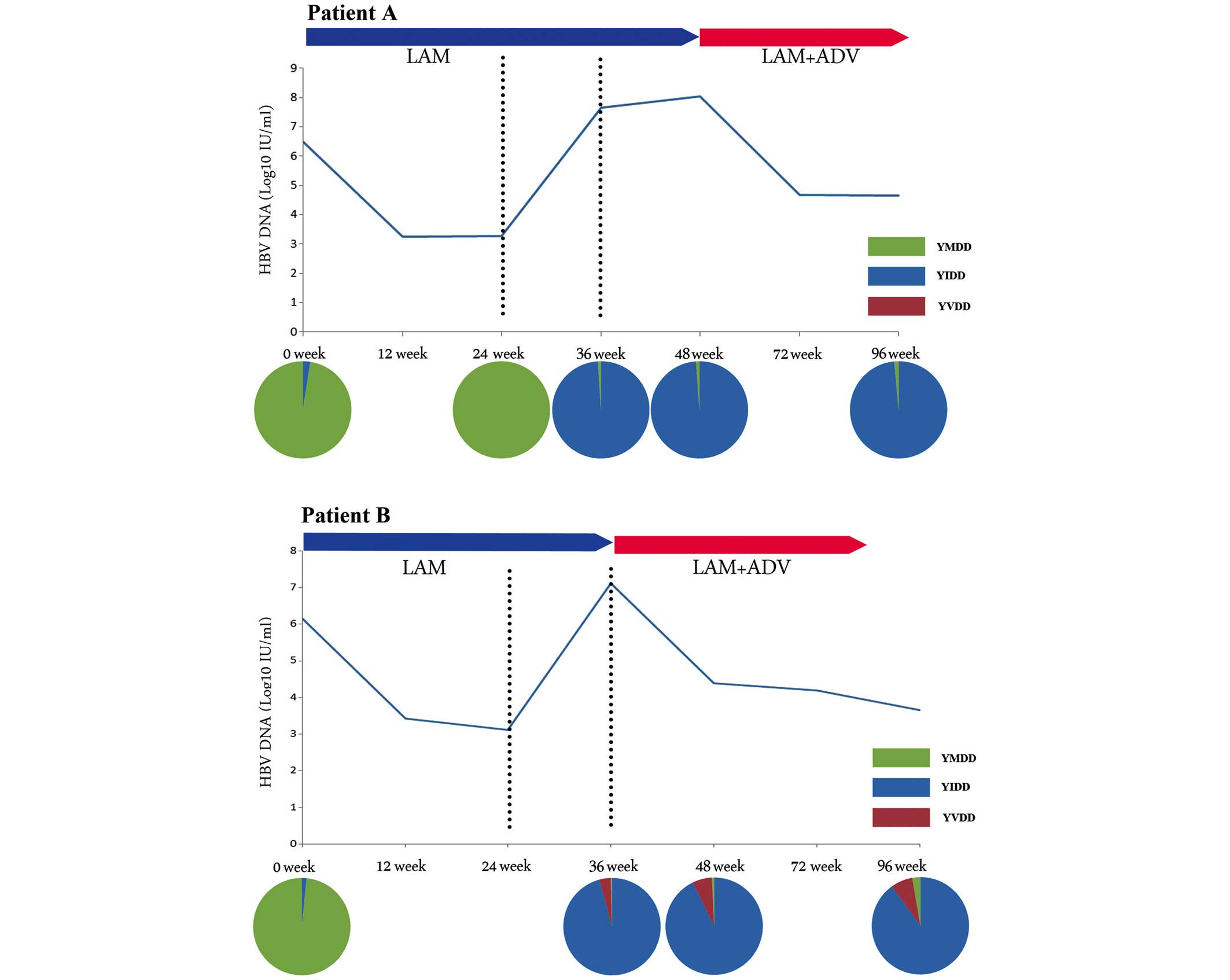
YIDD-motif-variant-dominated virological
breakthrough
Virological breakthrough in patients A, B, C and D
was dominated by the YIDD motif variant, since their virological
breakthrough was caused by a markedly increased mutation rate of
M204I during the LAM treatment. As shown in Fig. 1, the characteristics in common of
these four patients were that the viral breakthrough was caused by
the high rate of M204I mutation during the treatment with LAM, and
that the domi nation of the M204I mutation continued following the
ADV add-on therapy. Furthermore, the increased mutation rate of
rt180 or rt80 was accompanied by the occurrence of the high
mutation rate of M204I during the virological breakthrough
(Fig. 1 and Table III).
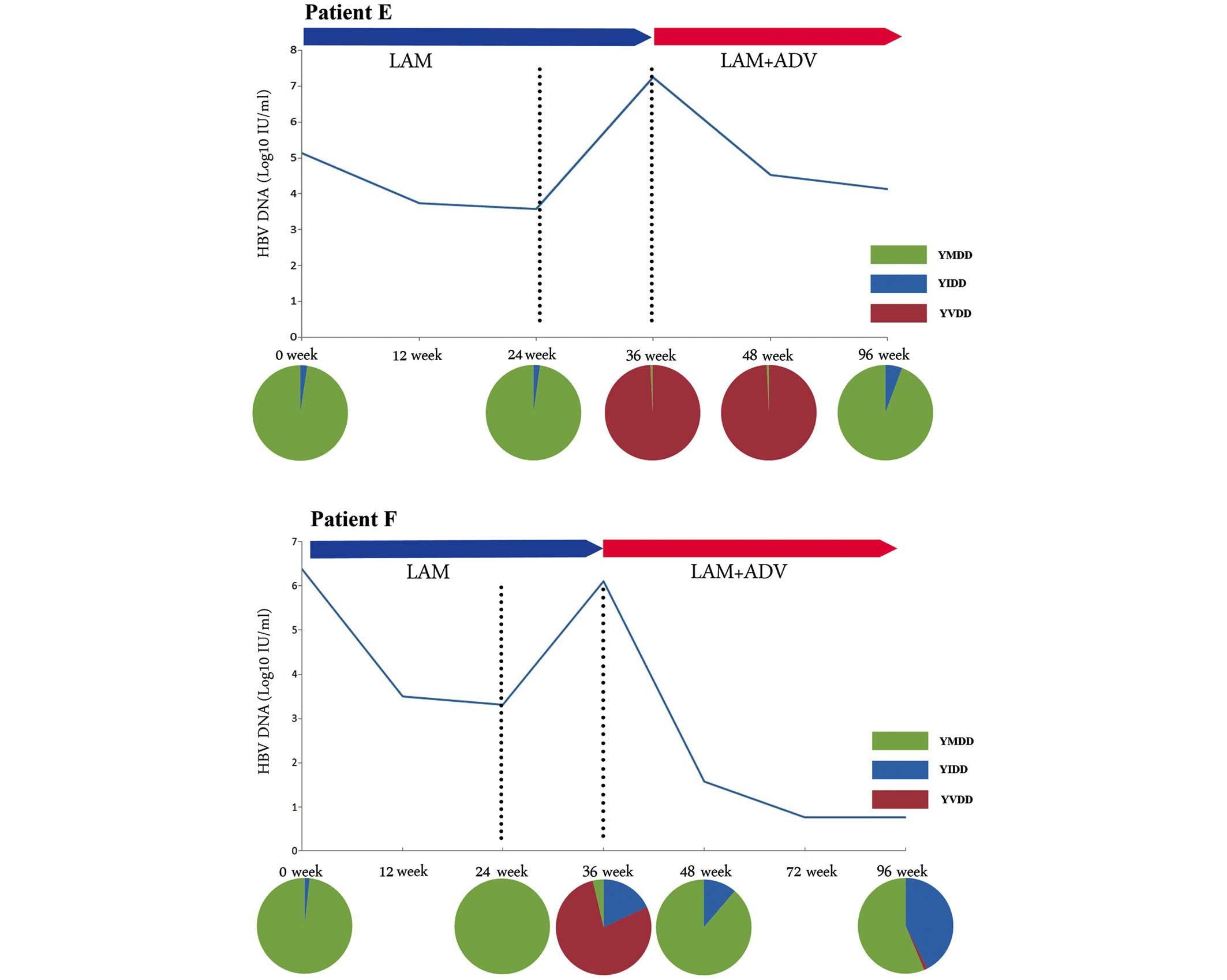
YVDD-motif-variant-dominated virological
breakthrough
This group included patients E, F, G and H, and
their viral breakthroughs were predominantly caused by the YVDD
motif variant. As shown in Fig. 2,
the M204V mutation in patients E and F eventually disappeared
following the ADV add-on therapy, which also occurred in the other
two patients. In all four patients, the M204V mutation was
accompanied by the L180M mutation, and an identical trend was
observed in the two mutation sites (Table III).
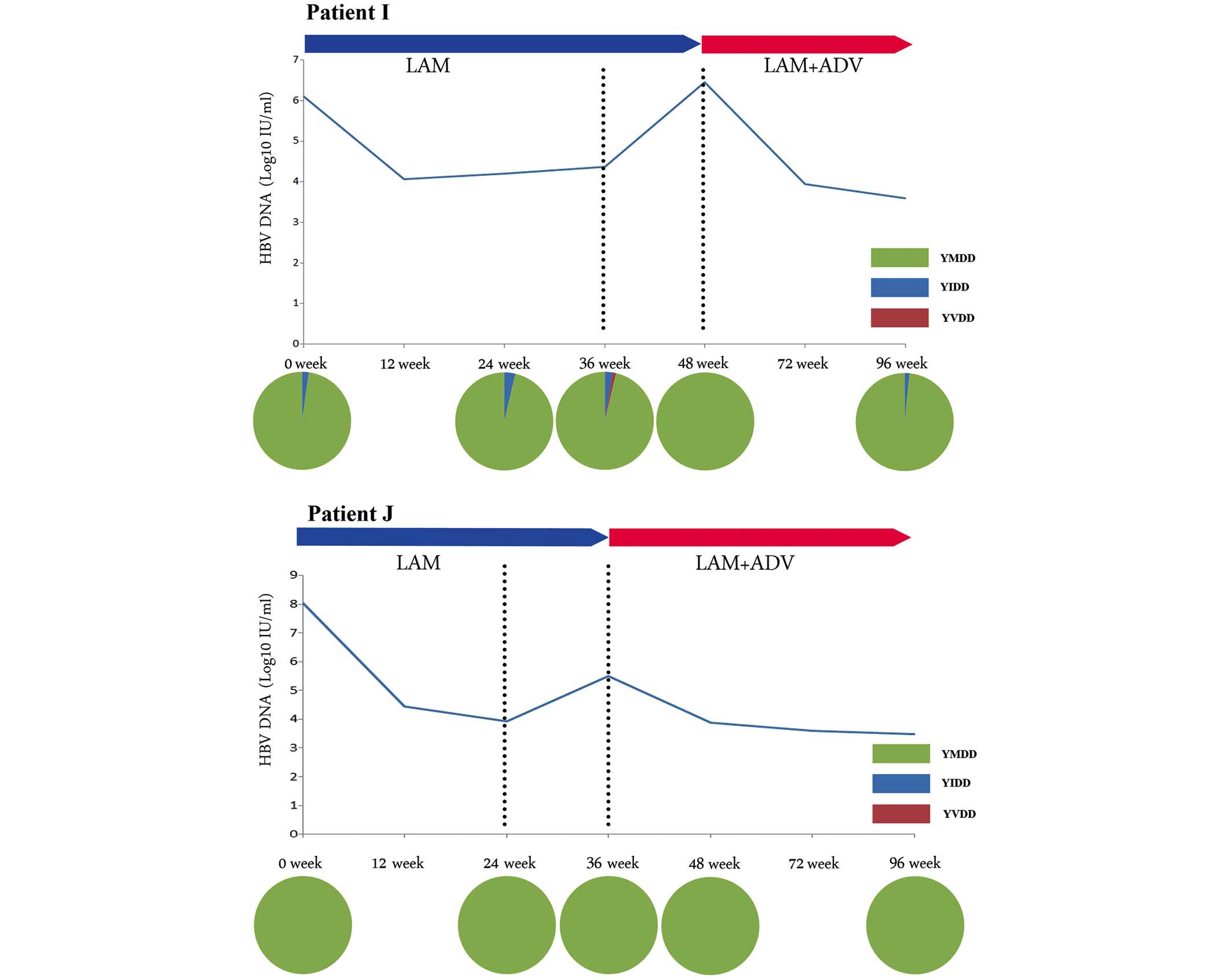
YMDD-wild-type-dominated virological
breakthrough
YMDD-wild-type-dominated virological breakthrough
was identified in patients H, I, J and K. This type of virological
breakthrough was observed in the patients lacking any mutation of
the YMDD motif (Fig. 3). The level
of HBV DNA in these four patients declined following the ADV add-on
therapy. The YMDD wild-type motif persisted during the treatment
with LAM monotherapy or the LAM + ADV combination therapy.
Discussion
Virological breakthrough is a clinical manifestation
in the treatment of CHB, which is defined as an increase of ≥1
log10 IU/ml compared with nadir during treatment in
patients with good adherence (15). Virological breakthrough during the
treatment of CHB was predominantly caused by drug resistance,
although its underlying mechanism remains to be fully elucidated
(18,19). The mutation rate identified for the
recently approved NUCs, including ETV and TDF, is very low. ETV and
TDF accounted for 1.5 and 0 of the identified drug-resistance rate,
respectively, in patients who had received 5 years of treatment
(15). However, the situation is
less optimistic as far as the early approved NUCs is concerned,
including LAM, which continues to be widely used in developing
countries (20). It was reported
that LAM, with a low genetic barrier to resistance, resulted in
drug resistant mutations in 70% of the patients following 5 years
of treatment (15). The risk of
drug resistance is associated with high baseline levels of HBV DNA,
the administration of a low-genetic-barrier drug and a gradual
decrease in the level of HBV DNA during treatment, which creates an
environment in which viral evolution may occur (21). However, the extent of mutation
which is required to give rise to a virological breakthrough and
the predominant dynamic pattern of viral evolution during
virological breakthrough remain to be fully elucidated, largely as
a consequence of the limitations of the current technology for
detecting drug resistance.
Previous studies focused on using UDPS to reveal the
underlying mechanisms associated with HCV and HIV. An increasing
number of studies are recruiting UDPS to detect minor mutations in
HBV, and these studies have demonstrated that UDPS is more
sensitive as a technique for detecting minor mutations compared
with previous technologies (22,23).
Notably, longitudinal studies focused on the dynamic evolution of
the HBV DNA during treatment are limited, which may offer further
insights into the mechanism underlying drug associated resistance.
UDPS was used previously to investigate the dynamic evolution
pattern of HBV variants in patients with CHB who developed
resistance to ADV, and the viral loads were identified to consist
of different types of HBV variants; furthermore, the absolute
levels of these variants varied during the treatment with ADV
(24). In the present study, it
was demonstrated that the YMDD wild-type motif of HBV was almost
entirely replaced by the LAM-resistant variants in eight patients
following viral breakthrough, indicating that virological
breakthrough was caused by the rapid replication of novel resistant
variants. The present study also revealed that the NUC-associated
mutations existed in treatment naive patients, for example, patient
D, who exhibited a high mutation rate of M204I at the baseline.
In the present study, the LAM-induced virological
breakthrough was further subdivided into three types, according to
which variant dominated during viral breakthrough, and common
characteristics were identified among the three types. The findings
of the present study provided novel insights into the mechanism
underlying the drug resistance and the virological breakthrough,
however, this study also gives rise to further questions. Firstly,
it was identified that the implementation of ADV add-on therapy
resulted in an eventual reversion of the YVDD variant into
wild-type YMDD, although the YIDD variant remained dominant. It is
an open question whether patients who experienced
YVDD-variant-dominated viral breakthrough benefitted more from ADV
add-on therapy compared with those who experienced the
YIDD-variant-dominated viral breakthrough. Secondly, a previous
study revealed that ~40% of the virological breakthroughs in
patients receiving NUCs were not associated with drug resistance
during clinical practice, which the authors of the study suggested
was caused by medication adherence (25). However, the adherence profile of
the patients in the present study was examined during the
treatment, and virological breakthrough was revealed to be caused
by YMDD wild-type-dominated variants. Therefore, LAM resistance
associated mutation sites remain to be identified, and the
mechanism underlying the phenomenon remains to be elucidated.
Thirdly, it was revealed that the mutation sites, rt80 and rt180,
were able to compensate for rt204 in patients who developed LAM
resistance (26), a result which
was further confirmed in the present study. Additionally, the
present study revealed that YVDD-variant-dominated virological
breakthrough was closely associated with the L180M mutation, and an
identical trend in the changes of the M204V and L180M mutations was
observed, which raised the question of the specific mechanism of
rt204 and its supplementary sites. Taken together, the present
study provided novel insights into the mechanism underlying
virological breakthrough and the evolution pattern of HBV DNA
during antiviral therapy. However, the present study was limited by
the small number of enrolled patients, and therefore a larger
cohort study in the future will be useful to substantiate these
results.
In 2007, 'the roadmap concept' was recommended for
the treatment of patients with CHB, which entailed using early
virological responses to optimize long-term outcomes for patients
with CHB (27). Previous studies
demonstrated that patients may benefit more from treatment
strategies with NUCs which are guided by the roadmap concept
(28–30). Notably, the present study revealed
that virological breakthrough may be caused by the rapid
replication of the drug resistant variants selected by the
treatment with NUCs, including YIDD- and YVDD-variant-dominated
viral breakthroughs. Therefore, more frequent and earlier
monitoring of the level of the HBV DNA level and the viral mutation
rate are urgently required in patients who experience suboptimal
treatment responses during the treatment with NUCs. Taken together,
the results from the present study may assist in improving current
understanding of HBV evolution during NUC treatment and contribute
to the development of novel treatment strategies.
Acknowledgments
The present study was partly supported by the Key
Medical Specialties Found of Shanghai Municipal Health Bureau (no.
05II011 2-1). The authors would like to thank Dr Keguang Chen
(Department of Otorhinolaryngology-Head and Neck Surgery, Eye and
ENT Hospital, Fudan University (Shanghai, China) for editing the
figures in this paper.
References
|
1
|
Lavanchy D: Hepatitis B virus
epidemiology, disease burden, treatment and current and emerging
prevention and control measures. J Viral Hepat. 11:97–107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Trépo C, Chan HL and Lok A: Hepatitis B
virus infection. Lancet. 384:2053–2063. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tujios SR and Lee WM: Update in the
management of chronic hepatitis B. Curr Opin Gastroenterol.
29:250–256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma H and Jia J: Why do I treat
HBeAg-positive chronic hepatitis B patients with a nucleoside
analogue. Liver Int. 33(Suppl 1): S133–S136. 2013. View Article : Google Scholar
|
|
5
|
Fung J, Lai CL, Seto WK and Yuen MF:
Nucleoside/nucleotide analogues in the treatment of chronic
hepatitis B. J Antimicrob Chemother. 66:2715–2725. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chotiyaputta W and Lok AS: Hepatitis B
virus variants. Nat Rev Gastroenterol Hepatol. 6:453–462. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rodriguez-Frias F, Buti M, Tabernero D and
Homs M: Quasispecies structure, cornerstone of hepatitis B virus
infection: Mass sequencing approach. World J Gastroenterol.
19:6995–7023. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Capobianchi MR, Giombini E and Rozera G:
Next-generation sequencing technology in clinical virology. Clin
Microbiol Infect. 19:15–22. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abbate I, Rozera G, Tommasi C, Bruselles
A, Bartolini B, Chillemi G, Nicastri E, Narciso P, Ippolito G and
Capobianchi MR: Analysis of co-receptor usage of circulating viral
and proviral HIV genome quasispecies by ultra-deep pyrosequencing
in patients who are candidates for CCR5 antagonist treatment. Clin
Microbiol Infect. 17:725–731. 2011. View Article : Google Scholar
|
|
10
|
Armenia D, Vandenbroucke I, Fabeni L, Van
Marck H, Cento V, D'Arrigo R, Van Wesenbeeck L, Scopelliti F,
Micheli V, Bruzzone B, et al: Study of genotypic and phenotypic
HIV-1 dynamics of integrase mutations during raltegravir treatment:
A refined analysis by ultra-deep 454 pyrosequencing. J Infect Dis.
205:557–567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Trimoulet P, Pinson P, Papuchon J, Foucher
J, Vergniol J, Chermak F, Wittkop L, Castaing N, Merrouche W,
Reigadas S, et al: Dynamic and rapid changes in viral quasispecies
by UDPS in chronic hepatitis C patients receiving telaprevir-based
therapy. Antivir Ther. 18:723–727. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Applegate TL, Gaudieri S, Plauzolles A,
Chopra A, Grebely J, Lucas M, Hellard M, Luciani F, Dore GJ and
Matthews GV: Naturally occurring dominant drug resistance mutations
occur infrequently in the setting of recently acquired hepatitis C.
Antivir Ther. 20:199–208. 2015. View
Article : Google Scholar :
|
|
13
|
Cortes KC, Zagordi O, Perlejewski K,
Laskus T, Maroszek K, Bukowska-Ośko I, Pawełczyk A, Płoski R, Berak
H, Horban A and Radkowski M: Deep sequencing of hepatitis C virus
hypervariable region 1 reveals no correlation between genetic
heterogeneity and antiviral treatment outcome. BMC Infect Dis.
14:3892014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Ji YY, Yao GB, Ma XY, Xie Q, Pang
HY, Wu SM, Li J, Chen CW, Xu XW and Gu EL: Two years efficiency of
lamivudine and adefovir dipivoxil combined therapy in chronic
hepatitis B patients. Eur Rev Med Pharmacol Sci. 17:636–643.
2013.PubMed/NCBI
|
|
15
|
European Association For The Study Of The
Liver: EASL clinical practice guidelines: Management of chronic
hepatitis B virus infection. J Hepatol. 57:167–185. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu J, Liu W, He L, Huang F, Chen J, Cui P,
Shen Y, Zhao J, Wang W, Zhang Y, et al: Sputum microbiota
associated with new, recurrent and treatment failure tuberculosis.
PLoS One. 8:e834452013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hamady M and Knight R: Microbial community
profiling for human microbiome projects: Tools, techniques, and
challenges. Genome Res. 19:1141–1152. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zoulim F and Locarnini S: Hepatitis B
virus resistance to nucleos(t)ide analogues. Gastroenterology.
137:1593–1608.e1. –e2. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rhee SY, Margeridon-Thermet S, Nguyen MH,
Liu TF, Kagan RM, Beggel B, Verheyen J, Kaiser R and Shafer RW:
Hepatitis B virus reverse transcriptase sequence variant database
for sequence analysis and mutation discovery. Antiviral Res.
88:269–275. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chan HL and Jia J: Chronic hepatitis B in
Asia-new insights from the past decade. J Gastroenterol Hepatol.
26(Suppl 1): 131–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wargo AR and Kurath G: Viral fitness:
Definitions, measurement, and current insights. Curr Opin Virol.
2:538–545. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gong L, Han Y, Chen L, Liu F, Hao P, Sheng
J, Li XH, Yu DM, Gong QM, Tian F, et al: Comparison of
next-generation sequencing and clone-based sequencing in analysis
of hepatitis B virus reverse transcriptase quasispecies
heterogeneity. J Clin Microbiol. 51:4087–4094. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ramírez C, Gregori J, Buti M, Tabernero D,
Camós S, Casillas R, Quer J, Esteban R, Homs M and Rodriguez-Frías
F: A comparative study of ultra-deep pyrosequencing and cloning to
quantitatively analyze the viral quasispecies using hepatitis B
virus infection as a model. Antiviral Res. 98:273–283. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rodriguez C, Chevaliez S, Bensadoun P and
Pawlotsky JM: Characterization of the dynamics of hepatitis B virus
resistance to adefovir by ultra-deep pyrosequencing. Hepatology.
58:890–901. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hongthanakorn C, Chotiyaputta W,
Oberhelman K, Fontana RJ, Marrero JA, Licari T and Lok AS:
Virological breakthrough and resistance in patients with chronic
hepatitis B receiving nucleos(t)ide analogues in clinical practice.
Hepatology. 53:1854–1863. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Menendez-Arias L, Alvarez M and Pacheco B:
Nucleoside/nucleotide analog inhibitors of hepatitis B virus
polymerase: mechanism of action and resistance. Curr Opin Virol.
8:1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Keeffe EB, Zeuzem S, Koff RS, Dieterich
DT, Esteban-Mur R, Gane EJ, Jacobson IM, Lim SG, Naoumov N,
Marcellin P, et al: Report of an international workshop: Roadmap
for management of patients receiving oral therapy for chronic
hepatitis B. Clin Gastroenterol Hepatol. 5:890–897. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lo AO, Wong VW, Wong GL, Chan HY, Cheung
CM and Chan HL: Efficacy of entecavir switch therapy in chronic
hepatitis B patients with incomplete virological response to
telbivudine. Antivir Ther. 18:671–679. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Piratvisuth T, Komolmit P, Tanwandee T,
Sukeepaisarnjaroen W, Chan HL, Pessôa MG, Fassio E, Ono SK, Bessone
F, Daruich J, et al: 52-week efficacy and safety of telbivudine
with conditional tenofovir intensification at week 24 in
HBeAg-positive chronic hepatitis B. PLoS One. 8:e542792013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun J, Xie Q, Tan D, Ning Q, Niu J, Bai X,
Fan R, Chen S, Cheng J, Yu Y, et al: The 104-week efficacy and
safety of telbivudine-based optimization strategy in chronic
hepatitis B patients: A randomized, controlled study. Hepatology.
59:1283–1292. 2014. View Article : Google Scholar : PubMed/NCBI
|