Introduction
Diabetes mellitus involves the metabolism of various
tissues and comprehensive signaling pathways. Type 2 diabetes is
characterized by insulin resistance accompanied by inadequate
insulin secretion (1).
Investigations into the pathogenesis of diabetes and diabetes drug
development has presented a requirement for suitable animal models.
In the past two decades, several animal models have been
established reflecting human disease pathogenesis from various
perspectives (2,3). Furthermore, animal models can assist
in improving current understanding of the nature of a disease, and
can provide a practical basis for the development of innovative
therapies for preclinical experiments (4). With innovations in medicine and
biotechnology, rodent models dominate this field. However, pigs are
more similar to humans than rodents in terms of genetics,
morphology, anatomy, physiology, dietary habits and
pharmacokinetics (5), and minipigs
are particularly useful due to their advantages of a small size and
ease of handling. Thus, minipig models have emerged as an ideal
tool in translational medicine (6).
However, minipig models are not ideal for
constructing metabolic disease models through diet or drug
induction. Certain disease models are induced through diet over a
long duration (7–10), whereas a transgenic animal offers
the potential for a shorter induction duration. Disease models
generated by drug induction, examine only one aspect, for example a
type 2 diabetes model induced by STZ, which involves pancreatic
damage, leading to insulin deficiency (11). Therefore, multi-transgenic models
enable a more comprehensive analysis of pathology. Several
multi-transgenic pigs have been developed for disease models and
animal breeding (12–15). In addition, polygenic modified
porcine models have extensive and more efficient potential,
compared with single transgenic pigs (12). Dieckhoff et al used
multi-transgenic pigs for a series of investigations on
retroviruses. These polygenic pigs were obtained by crossbreeding
single transgenic pigs (13). In
addition, Webster et al incubated sperm cells with three
marker vectors and generated multi-transgenic fluorescent pigs
(14). To simplify vector
construction procedures; increase modeling efficiency, stability
and integrated uniformity; and reduce the difficulty of
transfection, several studies have attempted to use polycistronic
vectors to load multiple genes. Deng et al adopted a single
vector with 2A peptides linking four marker genes, to prepare
multi-transgenic fluorescent pigs (12). Jeong et al used an internal
ribosome entry site (IRES)-mediated polycistronic vector to
co-express human CD59, CD55 and H-transferase in Yucatan minipig
models (15). Park et al
also used the 2A peptide to generate shTNFRI-Fc and HA-hHO-1
Yucatan transgenic (Tg) pigs (16). However, there have been no previous
reports of a multi-transgenic porcine diabetes model. Therefore,
the present study aimed to create a multi-transgenic minipig
diabetes model, which can express functional genes directly through
a foot and mouth disease virus 2A (F2A)-mediated
polycistronic system.
In pilot investigations, diabetic pig models have
been successfully manufactured by alteration of a single important
gene (3,17–19).
However, to develop a model involving the alteration of multiple
crucial genes, the present study selected three genes:
11-β-hydroxysteroid dehydrogenase 1 (11β-HSD1), which is involved
in insulin resistance (20); human
islet amyloid polypeptide (hIAPP) (3) and C/EBP homologous protein (CHOP)
(21), which can disrupt the
islets. The present study aimed to investigate whether increased
hepatic production of glucocorticoid, catalyzed by 11β-HSD1
directly, induces insulin resistance with adipose deposition, and
whether elevated expression levels of hIAPP and CHOP lead to
pancreatic cell damage in the animals. Ideally, the
multi-transgenic pig islet β-cell stress-associated apoptosis
pathways are activated (22),
which thereby enable a reduction in the number of islet β-cells,
resulting in the absolute lack of insulin secretion (23). In addition, insulin resistance is
caused by 11β-HSD1 (24) and
significantly impaired glucose tolerance, with consistently high
levels of fasting glucose in the future (20), culminating in generating the
diabetes model. The model aims to support investigations of the
mechanisms involved these two pathways (25), and may also be used for developing
novel drugs, with 11β-HSD1 as a target, for the treatment of
diabetes, Cushing's syndrome and other metabolic diseases (26), and for developing drugs to promote
insulin secretion (25,27).
Materials and methods
Experimental animals
The donor cells for use in in vitro somatic
cell nuclear transfer (SCNT) were porcine fetal fibroblasts (PFFs)
obtained 35 day fetuses from Wuzhishan miniature pigs (WZSPs). The
WZSPs used in the present study were obtained from the Germplasm
Resource Center of Chinese Experimental Minipig at the Institute of
Animal Sciences, Chinese Academy of Agricultural Sciences (Beijing,
China). The recipient animals were Landrace x Yorkshire pigs
(8-month-old females; Tianjing Yililai Breeding Co., Ltd., Tianjin,
China), and received humane care according to the criteria outlined
in the Guide for the Care and Use of Laboratory Animals, Institute
of Animal Sciences, Chinese Academy of Agricultural Sciences
(Beijing, China). The procedures were approved by the Institute of
Animal Sciences, Chinese Academy of Agricultural Sciences (Beijing,
China; permit no. ACGRCM2013-035). All animals were housed under
controlled conditions (temperature, 18–22°C; relative air humidity,
30–70%) with free access to water. The animals were sacrificed
through overdose of ketamine (100 mg/kg; cat. no. 087K1253;
Sigma-Aldrich, St. Louis, MO, USA) and xylazine (25 mg/kg; cat, no.
KH070901; Hengrui, Lianyungang, China). The tissues were
immediately frozen in liquid nitrogen and stored at −80°C for
subsequent analysis.
Construction of a multi-transgenic
tissue-specific polycistronic system
A recombinant plasmid vector (Fig. 1) containing multiple genes
(11β-HSD1, CHOP and hIAPP) was constructed based on pcDNA3.1c (+)
(cat. no. V790-20; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). This vector consisted of two expression
cassettes separated by a matrix-attachment region (MAR) insulator
(28). One cassette carried the
porcine liver-specific apolipoprotein E promoter (PapoE) (29) and porcine 11β-HSD1 cDNA (GenBank:
NM_214248.1), and the other cassette contained the porcine
pancreas-specific insulin promoter (PIP), which was cloned by
designed primers basing on the GenBank human counterpart, and the
murine CHOP (GenBank: NM_007837.3) and human IAPP (GenBank:
NM_000415.2) cDNAs. The foot and mouth disease virus 2A sequence
(F-2A fragment) following the furin digested site (5′-ATC ACG AAT
TCC AGC TGT TGA ATT TTG ACC TTC TTA AGC TTG CGG GAG ACG TCG AGT CCA
ACC CCG GGC CCG AAT TCG TCG AGACC-3′) linked CHOP with IAPP. All
sequences between PapoE and bovine growth hormone polyadenylation
signal were synthesized (Invitrogen; Thermo Fisher Scientific,
Inc.). In addition, MluI and NotI restriction sites
were designed to locate the start and end of this synthesized
fragment, respectively. The multiple cloning site of pcDNA3.1 (+)
was digested using the MluI and NotI enzymes (New
England Biolabs, Beijing, China), and the cytomegalovirus (CMV)
promoter fragment was replaced with this synthesized fragment.
Thus, the present study successfully generated the
pcDNA3.1-PapoE-HSD11B1-PIP-CHOP-IAPP recombinant vector. The
recombinant DNA molecular was digested using the endonuclease,
ScaI (New England Biolabs), and the excised fragment was
separated by agarose gel electrophoresis (Biowest Regular Agarose
G-10 Biowest, Hongkong, China). Purification (Zymoclean Gel DNA
Recorvery Kit™; cat. no. D4008; Zymo Research Corporation, Irvine,
CA, USA) and sequencing (Invitrogen; Thermo Fisher Scientific,
Inc.) was performed for verification. This strategy successfully
generated the pcDNA3.1-PapoE-11β-HSD1-PIP-CHOP-hIAPP recombinant
vector (Fig. 1).
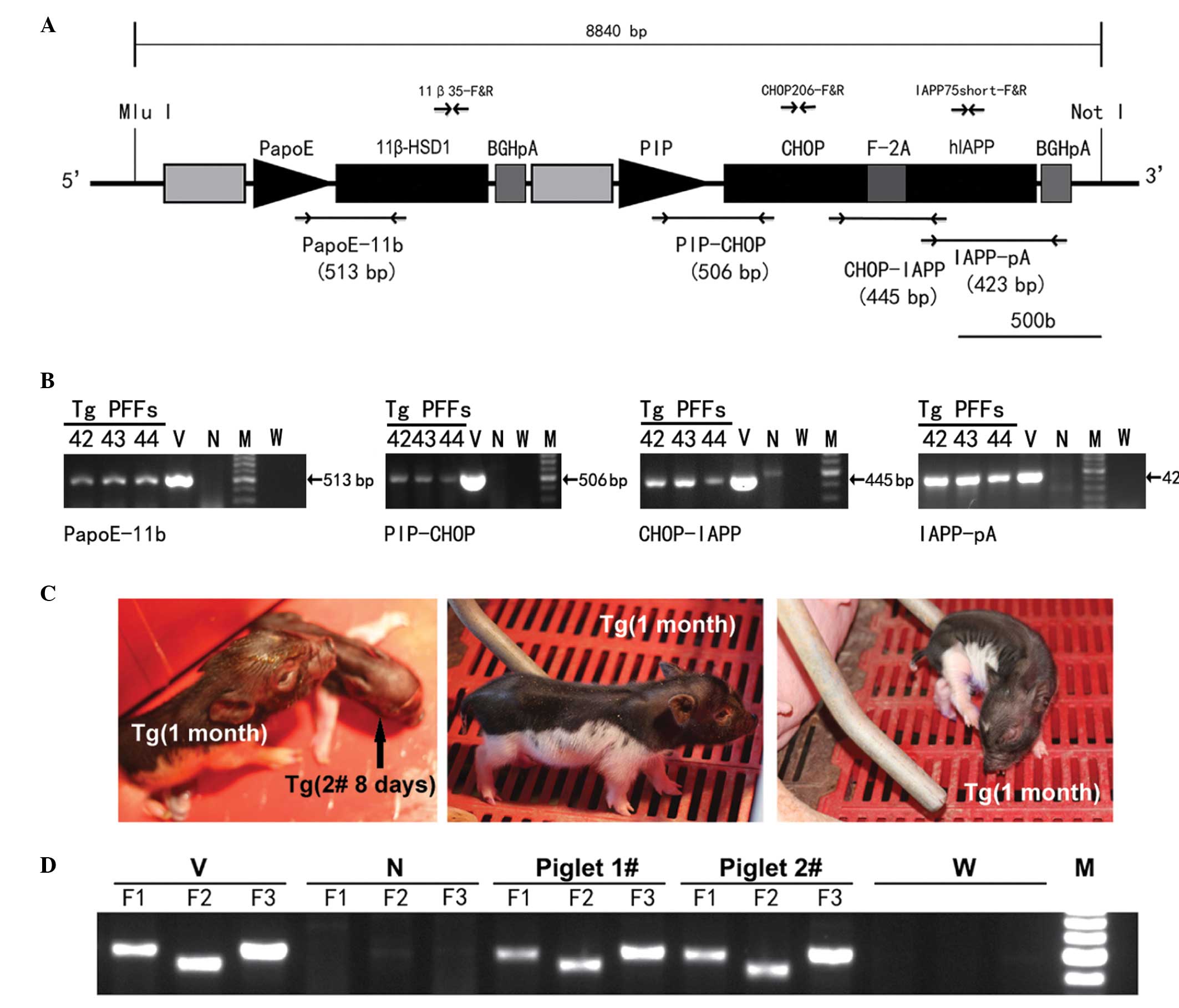 | Figure 1Generation and identification of
multi-transgenic PFFs and minipigs. (A) Schematic structure of
tissue-specific polycistronic system (8,840 bp). The head-to-head
arrows represent the primers for transgenic recognition, copy
number measurement and gene expression analysis. The fragment
between the two restriction sites comprises two cassettes isolated
by an insulator: (1) 11β-HSD1
driven by the liver-specific PapoE; (2) hIAPP and CHOP linked to the F-2A
peptide driven by the PIP. (B) PCR screening of multi-transgenic
PFFs. Amplification of the PapoE-11b, PIP-CHOP, CHOP-IAPP and
IAPP-pA fragments is shown, respectively. Lanes 42–44, three
representative PFFs transfected by the vector. (C) Multi-transgenic
piglets produced by somatic cell nuclear transfer. (D) Genomic DNA
PCR identification of piglet 1#, 2# and
negative control. F1–F3 indicate the three anticipated bands
corresponding to PapoE-11b, PIP-CHOP and CHOP-IAPP, respectively.
Tg, transgenic; 11β-HSD1, 11-β-hydroxysteroid dehydrogenase 1;
PapoE; hIAPP; human islet amyloid polypeptide; PIP, porcine
pancreas-specific insulin promoter; CHOP; C/EBP homologous protein;
PCR, polymerase chain reaction; V, positive vector; N, negative
control; M, 100 bp DNA ladder; W, ddH2O. MluI, MluI
restriction enzyme site; NotI, NotI restriction enzyme site;
PFFs, porcine fetal fibroblasts. |
Transfection of PFFs and preparation of
in vitro maturation enucleated oocytes
A linear DNA molecule was generated using the
ScaI endonuclease to digest the recombinant vector for
extraction from agar gel electrophoresis (Zymoclean Gel DNA
Recorvery Kit™; cat. no. D4008). At 35 days following the birth of
the WZSPs, PFFs were digested with 0.25% trypsin (Yaxin
Biotechnology, Co., Ltd., Shanghai, China) at 37°C and cultured in
complete culture medium, containing DMEM (cat. no. D5648;
Sigma-Aldrich), 2 g/L NaHCO3 (cat. no. S5761;
Sigma-Aldrich), 15–20% fetal bovine serum (FBS; cat. no. 16000-044;
Sigma-Aldrich), 1% penicillin-streptomycin (cat. nos. P7794 and
S1277-5G; Sigma-Aldrich). Routine steps of cell recovery, passage
and cryopreservation were adopted, following which electroporation
was performed following transfection (12,15,30).
At 2–4 days prior to transfection, the PFFs were thawed and
subcultured at 37°C in complete culture medium with 15% FBS
(1–2×106 cells per well) until the cells reached 70–90%
confluence, the cells were trypsinized and planked in cell board.
Generally, the required number of cells per well was
0.5–1×106 following primary cell counting, and 2
µg pcDNA3.1-PapoE-HSD11B1-PIP-C HOP-IAPP was used per well.
The reaction required 100 µl electroporation solution (BTX
Technologies, Inc., Hawthorne, NY, USA), comprising 82 µl
Nucleofector® solution and 18 µl supplement.
Electroporation was performed, according to the manufacturer's
instructions (Lonza Group, Basel, Switzerland) to select the
optimal transfer program (T-016). Following electroporation, the
cells were transferred to the cell incubator and incubated at 37°C
for 48 h, seeded directly into a 96-well cell plate (500–1,000
cells/well) (30). During the
first 2 days, the cells were selected using 800 ng/µl G418
(Merck Millipore, Beijing, China), and were then screened with a
concentration of 600 ng/µl in the following 10 days. In the
last 3 days, the G418 concentration was reduced to 200
ng/µl. G428 selection continued for 15 days. Following G418
screening and colony formation, positive cells were identified and
expanded in order to select optimally growing clones to perform
SCNT.
Ovaries were collected from the Yorkshire pigs in a
slaughter house (Beijing Shunyi Slaughter Company, Changping,
Beijing, China). The blood and other contaminants were removed by
washing in preheated double-antibiotic normal saline (penicillin
and streptomycin) three to four times. The cumulusoocyte complexes
(COCs) and follicular fluid were collected from 3–6 mm (diameter)
follicles. A 10 ml disposable syringe with a 12# needle
were used, and the needle opening was directed downwards when
inserted into the side tissues of the follicle. The follicular
fluid was transferred into 50 ml centrifuge tubes for 15–20 min at
25°C, following which the supernatant was discarded. Subsequently,
the precipitation was washed 2–4 times using 38°C polyvinyl
alcohol-Tyrode's lactate-Hepes (PVA-TL-HEPES) medium (Nunc,
Vedbaek, Denmark), with the supernatant carefully discarded after
standing for 10 min each time. The washed follicular fluid (~6 ml)
was transferred to 60 mm Petri dishes. Under a stereoscope
(SMZ1500; Nikon, Tokyo, Japan), oocytes were picked up using a
mouth pipette and transferred onto a 35 mm Petri dish prefilled
with PVA-TL-HEPES, then washed twice. In the washing process, COCs
surrounded by three or more layers of cumulus cells were selected
under a dissecting microscope (SZX7; Olympus Corporation, Toyko,
Japan), which had a regular shape, uniform cytoplasm and were
diaphanous. The oocytes were washed with PVA-TL-HEPES twice in a 35
mm petri dish and transferred to another dish containing balanced
free-hormone Tissue Culture Medium 199 solution (Gibco; Thermo
Fisher Scientific, Inc.) and washed twice again. Consequently,
60–80 oocytes/well were distributed into a 4-well plate, under
conditions of 38.5°C, 5% CO2 and saturated humidity for
40 h. MII oocytes with an integrated vitelline membrane, clear
perivitelline space, symmetrical cytoplasm and a conspicuous
polocyte were selected for SCNT (31).
SCNT and the generation of
multi-transgenic pigs
Positive PapoE-11β-HSD1-PIP-CHOP-hIAPP WZS PFFs
served as nuclear donor cells. Initially, the first polar body and
a section of the surrounding cytoplasm of the MII oocytes were
drawn out using a microinjection needle. Secondly, positive cells
were injected into the perivitelline space at the same location of
the oocyte. The reconstructed oocyte-donor cell complexes were
fused and activated by electric shock (CF-150B; BIological
Laboratory Equipment, Maintenance and Service, Ltd., Budapest,
Hungary). The activated complexes were placed in PZM3 culture
medium (Greiner Bio One, Frickenhausen, Germany) and cultivated at
38°C in 5% CO2 for 9 days. Finally, well-developed
embryos with the desired shapes were selected for implantation.
Caesarean surgery and eutocia were combined ~114 days later, based
on pregnancy status, to deliver the piglets.
Sample collection
Of the piglets examined, one piglet was sacrificed 8
days following birth, and another was stillborn. The ears were
removed and placed in 75% ethanol. The pancreas, left lobe of
liver, kidneys and longissimus muscles were dissected, samples of
which were rapidly frozen in liquid nitrogen. The remaining tissue
samples were fixed in 4% paraformaldehyde (cat. no. P1110;
Solarbio, Beijing, China).
RNA isolation, cDNA preparation and total
protein extraction
The liquid nitrogen-frozen tissues were triturated
in duplicate using a Precellys 24 homogenizer (Bertin Technologies,
Montigny-le-Bretonneu, France) and were used for RNA extraction and
total protein isolation. Total RNA was extracted using TRIzol
reagent (Ambion; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The remaining genomic DNA was eliminated
using DNase I (Tiangen Biotech Co., Ltd., Beijing, China), and
cDNAs were reverse-transcribed from the total RNA using a Revert
Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific,
Inc.). The conditions were as follows: 5 min at 37°C, followed by
60 min at 42°C and 10 min at 72°C. The cDNA samples were cooled at
−20°C as soon as possible, and were used to evaluate the
transcriptional expression of target genes using quantitative
polymerase chain reaction (qPCR) analysis.
Total protein was obtained from the tissues using
T-PER Tissue Protein Extraction reagent (Thermo Fisher Scientific,
Inc.). In addition, protease inhibitor containing cocktail tablets
(cat. no. 4693159001; Roche Diagnostics GmbH, Mannheim, Germany)
was used to protect protein integration. The total protein
concentration was determined using Working reagent (Thermo Fisher
Scientific, Inc.) and ELISA (Spectra Max M5; Molecular Devices;
Thermo Fisher Scientific, Inc. Following SDS degeneration, the
protein solution was stored at −20°C for subsequent western blot
analysis.
PCR and qPCR analysis
Genomic DNA was isolated from the ear clippings of
the WZS Tg and non-Tg piglets (BioTeke Corporation, Beijing,
China). PCR was used to screen the positive PFFs and to identify
multi-transgenic piglets. The PCR primers were designed using
Primer Premier 5.0 software (Premier Biosoft International, Palo
Alto, CA, USA). A total of four primer pairs (Table I) were used for these
amplifications. PCR was performed in a 20 µl system,
containing 2 µl 20X PCR buffer (Mg2+), 100
µmol/l dNTP, 1 µmol of each PCR primer, 2X U Taq DNA
polymerase (Takara Biotechnology Co., Ltd., Dalian, China) and 2
µl DNA derived from the PFF clone or WZS multi-transgenic
piglet ears. The PCR program was as follows: 95°C for 5 min
denaturation, 35 cycles of 94°C for 30 min denaturation, 60°C for
30 sec annealing, 72°C for 30 sec, followed by a final extension of
5 min at 72°C. All reactions were performed in duplicate. The
products were stained with Gel Red (Generay Biotech Co., Ltd.,
Shanghai, China) and analyzed using 1% agarose gel electrophoresis
(G-10; Biowest). qPCR was performed to determine the levels of
expression of the transcriptional genes, which was performed using
an ABI PRISM® 7500 Real-Time PCR system to determine the
transcriptional expression of the transgenes in the liver,
pancreas, muscle (longissimus dorsi) and kidney. SYBR®
Premix Ex Taq™ reagent and ROXII calibrating liquid (Takara
Biotechnology Co., Ltd.) was also used. The forward and reverse
primers are listed in Table I.
Each qPCR reactive mixture contained 10 µl SYBR®
Premix Ex Taq™ (2x), 0.8 µl forward and reverse
primers (10 µM; Table I),
0.4 µl ROXII, 2 µl DNA templates and dH2O,
to total volume of 20 µl. The qPCR analysis was performed as
a two-step procedure: Stage 1, initial denaturation of 30 sec at
95°C; stage 2, PCR reaction, comprising 40 cycles of 5 sec at 95°C
and 34 sec at 60°C; stage 3, dissociation stage comprising 15 sec
at 95°C, 1 min at 60°C and 15 sec at 95°C. All reactions were
performed in triplicate. ABI 7500 System SDS software (Version 1.4)
was used to analyze the data. Amplification plots were constructed
to reflect the gene amplification status. The specificity of the
primers with the cDNA templates were determined from the
dissociation melting curve, which ideally had only one peak.
Quantification of target gene expression levels were calculated by
the comparative threshold (Ct) value, the expression levels of all
transgenes were normalized to that of the porcine
glyceral-dehyde-3-phosphate dehydrogenase (GAPDH) mRNA Ct value and
expressed as 2−ΔΔCt (32)
 | Table IPrimers used for multi-transgenic
identification and gene expression analysis. |
Table I
Primers used for multi-transgenic
identification and gene expression analysis.
| Primer | Sequence
(5′-3′) | Temp
(°C) | Product
length (bp) | Use |
|---|
| PapoE-11b F |
GCTCCCTTTCCCCCTTAACC | 60 | 513 | Analysis of Tg PFFs
and pigs |
| PapoE-11b R |
AGGCCAAGAAGATCCCCAGA | | | |
| PIP-CHOP F |
AGGGAAATGATCCAGAAAGTGC | 57 | 506 | Analysis of Tg PFFs
and pigs |
| PIP-CHOP R | GGACGCAGGGTCAAG
AGTAGTG | | | |
| CHOP-IAPP F |
GAAACGGAAACAGAGTGG | 57 | 445 | Analysis of Tg PFFs
and pigs |
| CHOP-IAPP R |
GTTGCTGGAATGAACTAAAA | | | |
| IAPP-pA F |
TCATCAGGTGGAAAAGCGGAA | 65 | 423 | Analysis of Tg
PFFs |
| IAPP-pA R |
TAGCCAGACCATAGAGCCCA | | | |
| 11β35-F |
AGACACAGACACGGCCATGA | 59 | 62 | Analysis of copy
number and gene expression |
| 11β35-R |
TTCGGAGATGGTTGTACGTTGA | | | |
| CHOP 206-F |
CAACAGAGGTCACACGCACAT | 58 | 69 | Analysis of copy
number and gene expression |
| CHOP 206-R |
CCTGGGCCATAGAACTCTGACT | | | |
| IAPP75 short-F |
TGAAAGTCATCAGGTGGAAAAGC | 59 | 60 | Analysis of copy
number and gene expression |
| IAPP75 short-R |
AGGCGCTGCGTTGCA | | | |
| GAPDH-F |
AGGGCATCCTGGGCTACACT | 60 | 166 | Reference for gene
expression |
| GAPDH-R |
TCCACCACCCTGTTGCTGTAG | | | |
| TFRC-F |
GAGACAGAAACTTTCGAAGC | 60 | 81 | Reference for copy
number |
| TFRC-R |
GAAGTCTGTGGTATCCAATCC | | | |
Multi-transgene copy number
determination
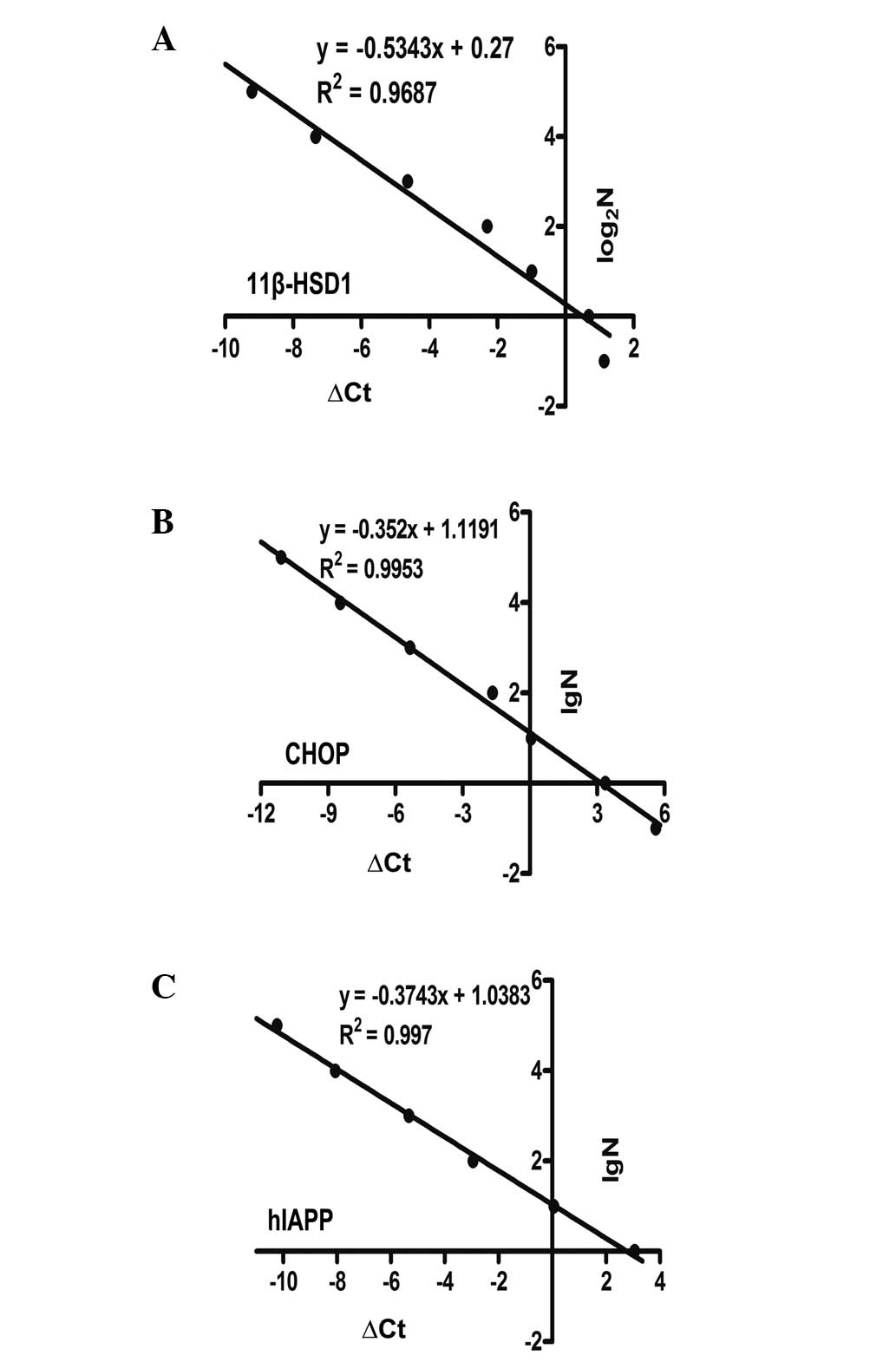
qPCR with the ΔCt-LogaN absolute standard
curve method was used to determine copy numbers (33). ΔCt was defined as Ct[transgene] -
Ct[transferrin receptor (TFRC)]. TFRC is known to exist as a single
copy in the porcine haploid genome and is used as a reference gene
(34). The primers, which were
used to amplify TFRC are listed in Table I. A mixed gradient copy number
standard was used for standardization (N=105,
104, 103, 102, 10, 1 or 64, 32,
16, 8, 4, 2, 1, 0.5), with 100 ng of wild-type genomic DNA.
Standard curves were constructed to clarify the correlation between
ΔCq, and the lgN or log2N and calculated the copy number
of target genes, respectively.
Western blot analysis
The expression levels of Tg proteins were measured
in the tissue samples from the liver and pancreas of the
multi-transgenic piglets. High-quality protein was extracted using
Tissue Protein Extraction Reagent (T-PER; Thermo Fisher Scientific,
Inc.). Protease inhibitors (Roche Diagnostics GmbH) were added to
the T-PER reagent, just prior to use. The tissues were homogenized
in this lysis buffer (cat. no. 78510; Thermo Fisher Scientific,
Inc.) using pellets and the sample was centrifuged at 10,000 ×g for
5 min at 4°C. The supernatant was collected and protein
concentration was determined using a Micro BCA Protein Assay kit
(Pierce Biotechnology, Inc., Rockford, IL, USA). An equivalent
quantity of protein (27 µg) was loaded in each lane,
separated by SDS-PAGE, and then transferred onto a nitrocellulose
membrane by electroblotting. The membrane was blocked with 5%
fat-free milk or bovine serum albumin (BSA), following which the
membranes were incubated with primary antibodies against 11β-HSD1
(polyclonal rabbit anti-pig; 1:1,000 dilution; cat. no. ab83522;
Abcam, Cambridge, MA, USA), CHOP (monoclonal rabbit anti-mouse;
1:1,000 dilution; cat. no. ab11419; Abcam), hIAPP (polyclonal
rabbit anti-human; 1:500 dilution; sc-20936; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) and β-actin (polyclonal
rabbit anti-mouse; 1:1,000 dilution; cat. no. TA-09; Zhongshan
Biotech Co., Ltd., Beijing, China). Subsequently, the membranes
were incubated with a horseradish peroxidase-conjugated goat
anti-rabbit antibody (1:5,000 dilution; cat. no. ab97051; Abcam)
for 1 h at room temperature. Following washing, the immunoblots
were detected using Super Signal West Pico Chemiluminescent
substrate (Thermo Fisher Scientific, Inc.).
Hepatic lipid biochemistry
The liver tissues (1#: 64.7 mg;
2#: 77 mg, Nc: 52 mg) were thoroughly homogenized in
1,000 µl normal saline using procedure 1 (5000-2×10-030) in
a Precellys 24 homogenizer (Bertin Technologies), and the debris
precipitate was removed by centrifugation at 12,000 rpm.
Subsequently, 800 µl of the supernatant was used to assay
the triglyceride and total cholesterol contents via the oxidase
method (17082 Hitachi Automatic Biochemical Analyzer; Hitachi,
Tokyo, Japan) (35). The
concentration of cortisol in the hepatic tissue was determined via
electrochemiluminescence using an automatic electrochemical
luminescence analyzer (Elecsys2010; Roche Diagnostics GmbH). The
final values were converted to comparable proportions.
Immunohistochemistry
The tissues were sliced (5 µm) following 4%
paraformaldehyde treatment and being embedded in paraffin. The
sections were stained with hematoxylin (5%; Solarbio) and eosin
using routine method. An anti-insulin antibody (anti-pig insulin;
1:300 dilution; Abcam) was used for the immunohisto-chemical
localization of the pancreas islets. Primary antibodies against
CHOP (1:50 dilution; Abcam) were used to validate transgene
overexpression. Pancreatic apoptosis was analyzed using blotting
with anti-caspase 3 antibody (1:400 dilution; Abcam). The
paraffin-embedded pancreas sections of the Tg and negative control
(Nc) animals were stained with the indicated antibodies. The
primary antibody was incubated for 1.5 h at room temperature,
followed by incubation with the secondary antibody (Abcam).
Diaminobenzidine (Sigma-Aldrich) served as the color development
reagent. Finally, hematoxylin was used to counterstain the
sections. An Olympus microscope (CX31; Olympus Corporation)
connected to a Pixera digital camera (Pro 120es; Pixera
Corporation, San Jose, CA USA) was used to capture images of the
sections.
Statistical analysis
The data are presented as the mean ± standard error
of mean. Two-sample unpaired two-tailed t-tests were used to
measure the statistical significance of differences (Microsoft
Office Excel 2010; Microsoft Corporation, Redmond, WA, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Transgenesis of PFFs and screening of
positive cells
The linearized
pcDNA3.1-PapoE-11β-HSD1-PIP-CHOP-hIAPP was introduced into porcine
PFFs by electroporation (Fig. 1A).
Stably transfected PFFs were selected with G418 for 15 days. PCR
was used to verify the multi-transgenic PFFs (Fig. 1B). When all target fragments
co-occurred, the PFFs were identified as Tg. Clones 1-44 had
successful integration events, and nine of these clones with
high-efficiency amplification were selected to prepare SCNT
embryos.
Development of SCNT embryos in vivo and
generation of multi-transgenic WZS miniature piglets
A total of 1,459 Tg-SCNT embryos were produced and
surgically transplanted into the fimbriae of the fallopian tubes of
three naturally estrous Landrace x Yorkshire gilts (Table II). Two recipients became
successfully pregnant, and another one returned to estrus. The
first surrogate mother delivered a live pigletby eutocia (Fig. 1C Tg; 1 month). The second gilt gave
birth to two babies by caesarean, including one stillborn piglet
(not shown, but termed piglet 1#) and one, which was
sacrificed at 8 days old due to feeding difficulty and artificial
feeding indigestion (Fig. 1C Tg
(2#; 8 days). Caesarean section was also performed in
the third recipient; in which a mass of black-brown debris was
found and surgically removed (Table
II). Unfortunately, the eldest (Tg; 1 month) did not survive as
a result of umbilical cord inflammation.
 | Table IICharacteristics of pregnant
recipients, embryos and transgenic piglets. |
Table II
Characteristics of pregnant
recipients, embryos and transgenic piglets.
| Recipient
number | Breed | Status of
receptor | Transferred embryos
(n) | Embryonic age
(days) | Grade of embryonic
development | Status of
delivery | Number of piglets
(liveborn) |
|---|
| 504 | Landrace x
Yorkshire | Surrogate gilt | 573 | 2 | B+ | Eutocia | 1 (1) |
| 597 | Landrace x
Yorkshire | Surrogate gilt | 460 | 1 | B− | Caesarean | 2 (1) |
| 525 | Landrace x
Yorkshire | Surrogate gilt | 426 | 2 | B− | Returned to
estrus | Back debris
(0) |
Genotyping of multi-transgenic pigs and
determination of transgene copy numbers
Genomic DNA was extracted from the ear clippings of
piglets 1#, 2# and the Nc. Three primer pairs
(Table I) were used to amplify
three segments to confirm the presence of the three exogenous genes
in the multi-transgenic pigs (1# and 2#) when
the three aim bands emerged at the same time (Fig. 1D). Copy number determination in
single-transgenic pigs is being increasingly reported (24,35).
However, there are few reports in multi-transgenic animals. This
approach revealed very notable and inconsistent results in the
polycistronic system-transfected pigs in the present study
(Fig. 2 and Table III).
 | Table IIIMulti-transgene copy numbers. |
Table III
Multi-transgene copy numbers.
| Gene | 1# | 2# | Negative
control |
|---|
| 11β-HSD1 | 12.16 | 10.09 | 0.41 |
| hIAPP | 40.52 | 28.01 | 0.01 |
| CHOP | 54.68 | 46.13 | <0.01 |
Overexpression of the three transgenes in
specific tissues of different individuals
Piglets 1# and 2# were used to
examine the transcription and translation of target genes. Piglet
1# was stillborn, and piglet 2# was
sacrificed at 8 days old. Secretion insufficiency and insulin
resistance-associated tissues, including the pancreas, liver,
muscle and kidney, were used for analysis. The transcriptional
expression levels of the three genes were determined by qPCR in
various individuals and organs (Fig.
3A–C). These results indicated that the transgenes were
overexpressed, and that the PapoE and PIP worked. The relative mRNA
levels of 11β-HSD1 in piglet 2# were almost two-fold
those of the piglets in the Nc group (Fig. 3A; left). As piglet 1#
was stillborn, its mRNA was more likely to be degraded, and the
gross expression of 11β-HSD1, including endogenesis and Tg
exogenesis, was low (Fig. 3A;
left). The levels of CHOP and hIAPP were high in the positive
piglets (1#, P<0.05; 2#, P<0.01;
Fig. 3A middle and right). It is
necessary to emphasize that the CHOP and hIAPP transgenes are
exogenous; the CHOP transgene is murine and the hIAPP cDNA is
human. These two genes were undetected in the control animals. The
tissue specificities of the PapoE and PIP were also almost ideal
(Fig. 3B and C). The mRNA
expression levels of 11β-HSD1 in the livers of piglets
1# and 2# were marginally higher than in the
muscle and kidneys (Fig. 3B and C;
left). In addition, the expression levels of CHOP and hIAPP in the
pancreas were higher, compared with those in the muscle and kidneys
(Fig. 3B and C; middle and right).
The nonspecific expression in the other tissues may be to the
selecting of specific promoters with their core regions for utility
reasons, and these core regions can be involved in their specific
tissues (29). These promoters may
be expressed in other undesired organs or tissues due to certain
intracellular transcription factors binding to the core region
non-specifically, driving transgene expression (36). This issue requires consideration
and investigation in the future.
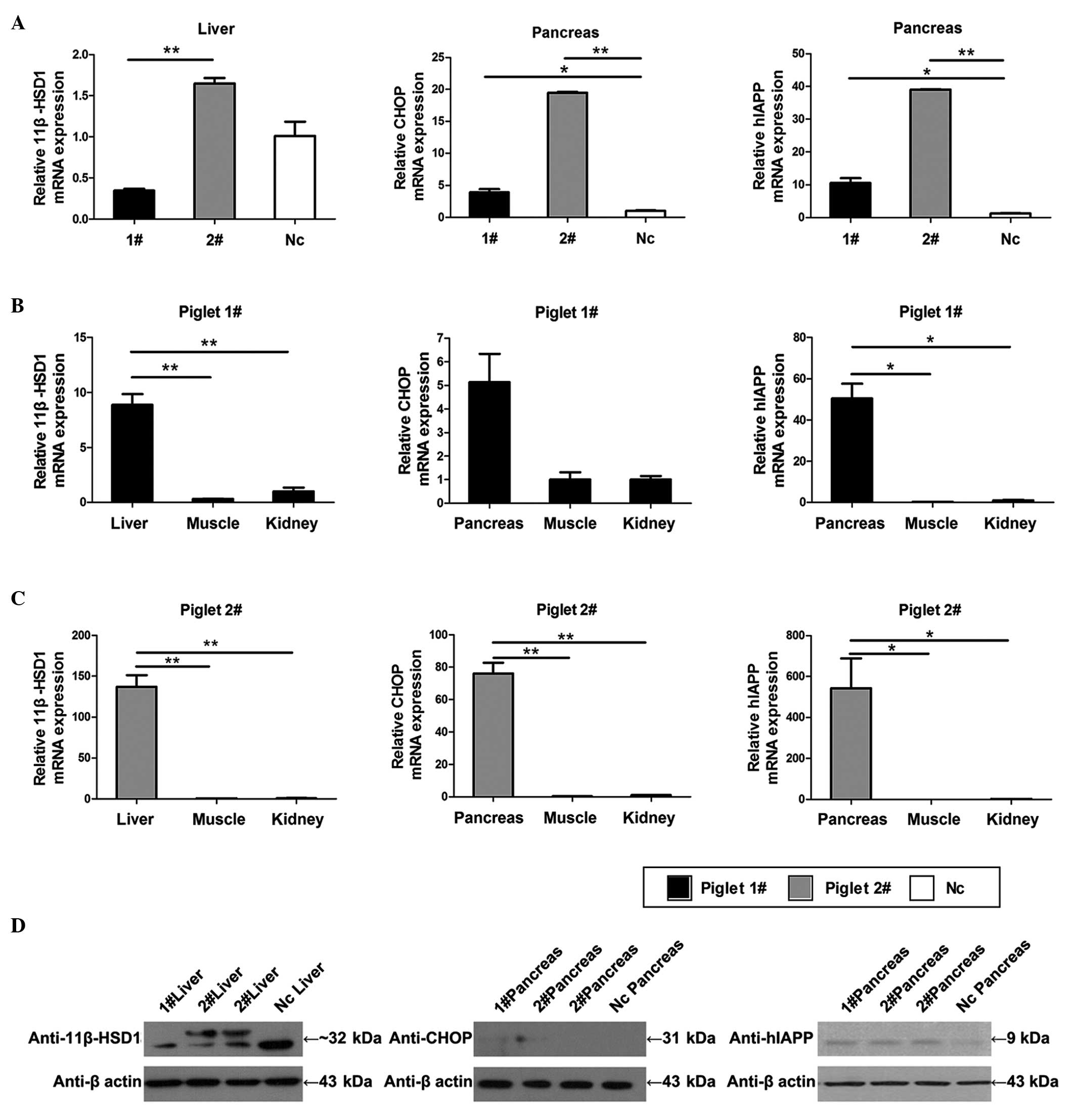 | Figure 3Protein expression and transgene
expression analyses. (A) Expression levels of 11β-HSD1 in the liver
of the 1#, 2# and Nc piglets. The mRNA of
1# liver was likely degraded, so that its relative
expression was lower than in the Nc piglet. Expression levels of
CHOP and hIAPP in the pancreas of the 1# and
2# pigs were greater than in Nc. (B and C) Specific
expression levels in the liver were compared with those in the
muscle and kidney of the same Tg piglet. The specific expression
levels of CHOP and hIAPP in the pancreas were high, compared with
the levels in the other tissues of the same Tg piglet. (D) Results
of exogenous protein expression analysis. Transgenic 11βHSD-1 was
confirmed using a porcine polyclonal anti-11β-HSD1 antibody in the
liver of piglet 2#. CHOP was detected at low levels in
piglets 1# and 2#, and was not detected in Nc
using a monoclonal mouse anti-CHOP antibody. Transgenic hIAPP was
overexpressed in the pancreas of piglets 1# and
2#, as assessed using human polyclonal anti-hIAPP
antibody. Each sample was analyzed in triplicate and the data are
expressed as the mean ± standard error of the mean (n=3, three
parts of the tissue samples). *P<0.05;
**P<0.01. Tg, transgenic; Nc, negative control;
1β-HSD1, 11-β-hydroxysteroid dehydrogenase 1; CHOP, C/EBP
homologous protein; hIAPP, human islet amyloid polypeptide. |
Western blot analysis was used to detect protein
expression levels, and to identify whether the protein specific
levels of these factors were consistent with the mRNA expression
levels (Fig. 3D). A doublet
corresponding to 11β-HSD1 was observed in piglet 2#; and
this second band may represent the Tg expression protein or a
splice variant of the Tg protein, compared with the Nc. Similar to
the mRNA expression profile, hIAPP was expressed at a high level in
the pancreas of the Tg piglets. Tg murine CHOP was observed at low
levels in the pancreas. The present study then performed
immunohistochemical analysis to validate the expression of CHOP
(Fig. 4A; middle), and the results
indicated that the hybridized intensities of CHOP in the Tg group
were higher, compared with those of the Nc group.
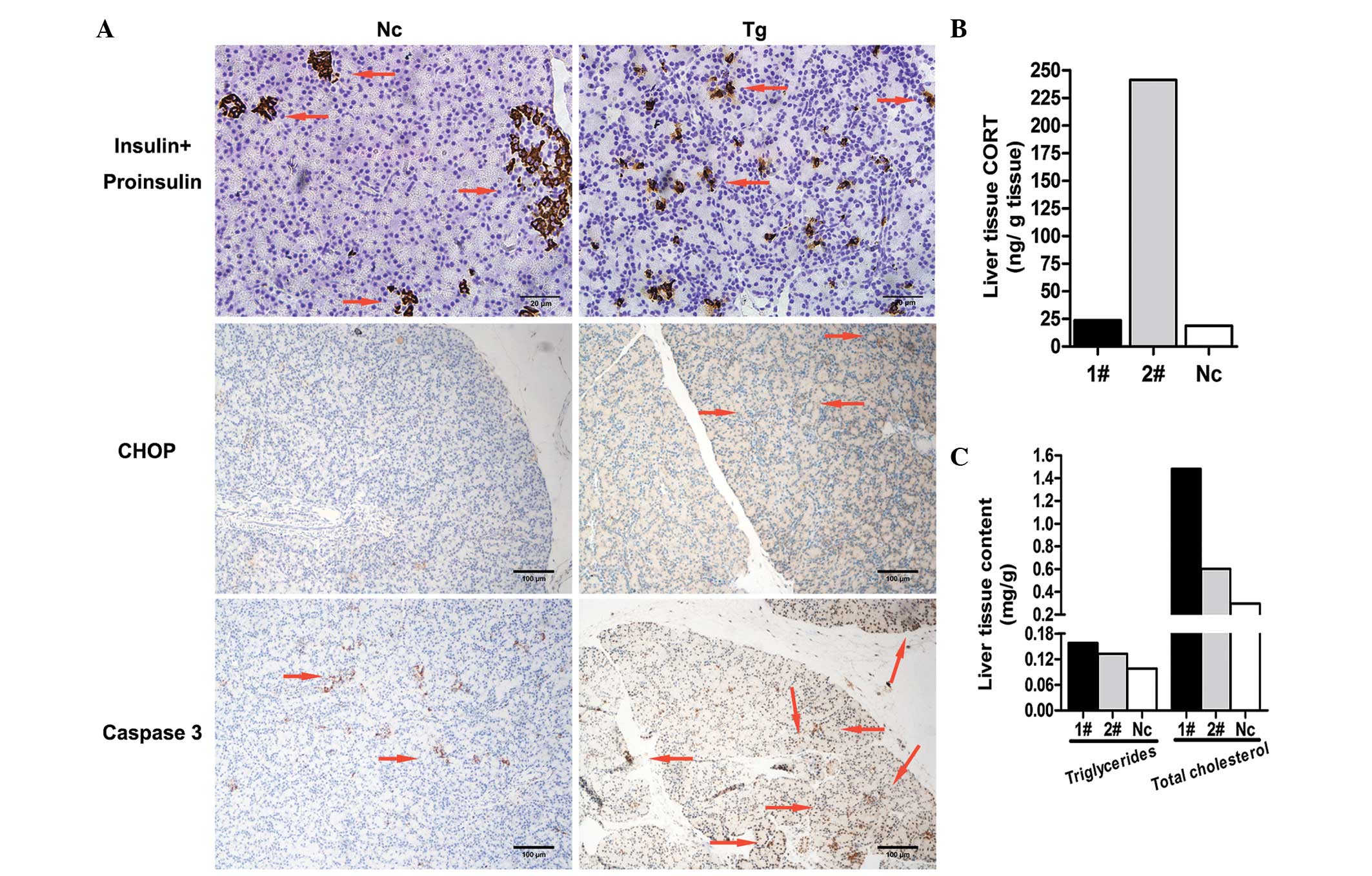 | Figure 4Phenotypic analysis. (A)
Photomicrograph of pancreas immunohistochemistry. Tg piglet
2# and an Nc were sacrificed. Top row, insulin
immunohistochemistry of the pancreases of Tg piglet 2#
and Nc shows the status of islet cell development; observing the
profile of the islet cells allows the estimation of β cell
proliferation (magnification, ×400; scale bar=20 µm). Middle
row, pancreas sections stained with anti-CHOP antibody (1:50; scale
bar=100 µm). Bottom row, positive caspase 3 signals show
apoptosis in Tg piglet 2# and Nc (scale bar=100
µm). (B) CORT, (C) triglyceride and total cholesterol
concentrations were determined in the Tg 1#, Tg
2# and Nc piglets. Expression of 11β-HSD1,
11-β-hydroxysteroid dehydrogenase 1 in the Tg piglets led to an
increase in CORT (2#). Subsequently, the hepatic lipid
contents (triglycerides and total cholesterol) increased and
exceeded those in the Nc piglet. These values were measured twice
and are expressed as the mean. Tg, transgenic; Nc, negative
control; CHOP, C/EBP homologous protein; CORT, cortisol. |
Primary identification of function and associated
phenotype. Due to the chronic nature of Type 2 diabetes, the blood
glucose level of the Tg pig was normal. The Tg pig continually
showed significant disease symptoms, and may have required an
extended period. Future investigations may involve the induction of
hyperglycemia with high-fat, high-calorie diets. However, notable
original pathological changes were observed without neonatal
hyperglycemia.
The results of the anti-insulin immunohistochemistry
confirmed that the normal shapes of the islet cells were affected,
and that their volume was reduced, compared with the normal islet
cells in the Nc piglet (Fig. 4A;
upper). The results suggested that the development of the islet of
Langerhans in the Tg piglets was also impaired. In addition, the
increase in the active form of caspase 3 is used as an indicator of
cell apoptosis (37). In the
present study caspase 3 immunohistochemistry revealed increasing
apoptosis (Fig. 4A; lower).
Consequently, it was reasonable to hypothesize that coexpression of
the two genes in the pancreas enhanced the decline in insulin
secretion (38).
The 11β-HSD1 gene expresses the enzyme,
11-β-hydroxysteroid dehydrogenase 1, which can catalyze the
activation of cortisone to an active glucocorticoid (cortisol)
(20,31). Subsequently, it can facilitate the
lipid content generation of triglycerides and total cholesterol
(26,39). 11β-HSD1 can accelerate liver
adipose tissue accumulation and lead to insulin resistance
(24,40). In the present study, piglet
2# was neonatal, and no clear adipose vacuoles were
visualized in the HE sections. Therefore, the liver tissue cortisol
(Fig. 4B), triglyceride and total
cholesterol (Fig. 4C) contents
were determined. The Tg piglets had higher levels of these factors,
compared with the Nc piglet. The cortisol level of piglet
2# was high, and the triglyceride and total cholesterol
levels of piglet 1# were marginally higher than those of
piglet 2#. Although these differences between the two Tg
piglets were observed, it is important to note that the
1# and 2# Tg piglets had higher levels of
cortisol, triglyceride and total cholesterol contents, compared
with the Nc piglet. These results also preliminarily verified the
effect of 11β-HSD1 overexpression, and suggested that 11β-HSD1 was
more likely to induce insulin resistance in the near future.
Discussion
Based on the complicated pathogenesis of type 2
diabetes, the present study hypothesized that a multi-transgenic
pig model, generated by directly altering relevant genes to cause
insulin resistance and affect insulin secretion, can be used to
provide a suitable mimic of diabetes. This was encouraged following
the success of single-transgene pig diabetes models (3,17–19).
These previous studies showed that single genes or factors affected
the molecular pathogenesis of diabetes. However, these single genes
were circumscribed, and their effectiveness was limited by
compensatory mechanisms. In particular, although the pancreas is
the pathological center of diabetes, the disease also involves a
variety of peripheral tissues, including the liver and muscles
(41). Therefore, the selection of
relevant genes in the present study was based on this
consideration. 11β-HSD1 is important in insulin resistance.
Previous studies have reported that the constitutive overexpression
of 11β-HSD1 can induce insulin resistance with metabolic syndrome
in rodents. Masuzaki et al created Tg mice overexpressing
11β-HSD1 selectively in adipose tissue. These mice developed
visceral obesity, which was exaggerated by a high-fat diet as a
result of increased adipose levels of corticosterone. In addition,
the mice also exhibited hyperphagia with hyperleptinemia, marked
insulin resistance and hyperlipidemia (20). Paterson et al generated Tg
mice selectively, showing increased activity of 11β-HSD1 in the
liver. These animals exhibited fatty liver, dyslipidemia,
transgene-dose-associated hypertension and insulin resistance, but
retained normal body weight (24).
It has also been hypothesized that the exclusive overexpression of
11β-HSD1 may be desirable for generating a porcine model of insulin
resistance (42,43). In addition, the hIAPP gene encodes
islet amyloid polypeptide, which is known to precipitate in the
islet β cells of patients with diabetes (44). This polypeptide can induce β cell
apoptosis, however, it is unclear whether hIAPP deposition and
accumulation in porcine β cells has this effect (3). Several studies have prepared hIAPP Tg
mice and demonstrated the viability of this method. For example,
Hull et al produced hIAPP Tg lines, which showed islet
amyloid formation and β-cell loss when fed high-fat diets (45), and diabetes experts have suggested
that genetically engineered hIAPP Tg pigs are suitable for
mimicking the role of islet amyloidosis in the pathogenesis of type
2 diabetes mellitus pathogenesis (3). In addition, CHOP is a direct upstream
factor, which can drive endoplasmic reticulum stress and cell
apoptosis (46). It has been
reported that, in pancreatic β-cells, endoplasmic reticulum
stress-mediated apoptosis is associated with the pathogenesis of
diabetes (47). The present study
hypothesized that co-expressing hIAPP and CHOP in the pancreas
exaggerates the β-cell apoptosis-inducing effect, and that the
multi-transgenic piglets generated in the present study using a
tissue-specific polycistronic system provide ideal animal models of
diabetes, possibly conferring insulin resistance accompanied by
declining insulin secretion. The use of multiple genes involved in
these two predominant modes of pathogeneses provides an advantage
over single-Tg diabetes pig models. In addition, the primary data
obtained in the presents study indicated that the functional gene
polycistronic system was feasible. Hepatic lipid biochemistry and
immunohistochemistry indicated that Tg hepatic lipogenesis was
facilitated, that islets of Langerhans were impaired and that
insulin secretion was likely to be affected, which may be
attributed to cooperation between CHOP and hIAPP.
Compared with previous single-vector
multi-transgenic pigs (12,15),
the primary advantage of the multi-transgenic approach is that
diabetes-associated genes involving the molecular pathogenesis of
diabetes were selected. The results of the present study verified
the hypothesis that replacing marker genes with functional genes is
achievable. The second advantage is that the present study did not
perform gene replacement only. Whereas other studies have used CMV
or CMV early enhancer/chicken β-actin promoters, which are
expressed ubiquitously in vivo, the present study used two
different tissue-specific porcine promoters, and qPCR assays
ensured that these promoters were expressed in their target organs.
Western blotting and immunohistochemistry also detected the
expression of the Tg proteins to a certain extent. In particular,
PapoE was cloned (29), and the
MAR insulator was placed between the expression cassettes to reduce
the impact of the promoters (28).
The third advantage is that, to eliminate the 2A tail, which may
affect the function of the upstream protein, a furin cleavage site
was added. Furin, an endogenous proprotein convertase, can cut 2A
tails (12), and the successful
utilization of furin was previously reported in pigs (11). 2A is a superior polycistronic
system, compard with IRES (12).
Jeong et al used an IRES-mediated vector co-expressing
xenotransplant genes in minipigs. However, hCD55 was not expressed
(15). Of note, the present study
unexpectedly observed that the copy numbers of the three genes were
not the same, and there are no previous reports regarding copy
numbers in singular polycistronic pigs. This result may be due to
random transgenes exhibiting different positional effects, or it
may indicate that the linear vector used in the present study was
fragmented, and that the integrating capacity of the segments was
different. The exact reasons for these observations require further
investigation. Therefore, copy number is a significant element that
requires consideration when manufacturing multi-transgenic pigs and
subsequently constructing their colony.
In conclusion, the present study suggested that the
Tg pig model constructed had functional transgenes and possessed
the desired diabetes-associated phenotypes. The merit of this
system was its targeted multi-transgene expression. The results
demonstrateda technique for how to co-express multiple functional
genes in specific tissues, in contrast to impractical marker genes
or ubiquitous gene expression. The original aim of the present
study was to examine comprehensive pathogenesis in multi-transgenic
pig models to develop novel strategies for how to more reasonably
mimic a human polygenic disease. Several questions remain to be
answered, however, the initial phenotypes indicated that Tg animals
with multiple functional genes are promising and have significant
potential. Prospectively, diabetes multi-transgenic models may be
suitable for diverse drug development, from peripheral insulin
resistance to central relative decreases in insulin secretion.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant. no. 31372276), the National
Science and Technology Major Project (grant. no. 2013ZX08010-003),
the Development Program of China (grant. no. 2012AA020603) and the
Agricultural Science and Technology Innovation Program (grant. nos.
ASTIP-IAS05 and ASTIP-IAS-TS-4).
References
|
1
|
American Diabetes Association: Diagnosis
and classification of diabetes mellitus. Diabetes care. 36(Suppl
1): S67–S74. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Masiello P: Animal models of type 2
diabetes with reduced pancreatic beta-cell mass. Int J Biochem Cell
Biol. 38:873–893. 2006. View Article : Google Scholar
|
|
3
|
Wolf E, Braun-Reichhart C, Streckel E and
Renner S: Genetically engineered pig models for diabetes research.
Transgenic Res. 23:27–38. 2014. View Article : Google Scholar
|
|
4
|
Rüster C and Wolf G: Models of diabetic
nephropathy. Drug discovery today: Disease models. 7:35–41.
2010.
|
|
5
|
Verma N, Rettenmeier AW and Schmitz-Spanke
S: Recent advances in the use of Sus scrofa (pig) as a model system
for proteomic studies. Proteomics. 11:776–793. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu H, Li Y, Wei Q, Liu C, Bolund L, Vajta
G, Dou H, Yang W, Xu Y, Luan J, et al: Development of transgenic
minipigs with expression of antimorphic human cryptochrome 1. PLoS
One. 8:e760982013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xia JH, Yuan J, Xin LL, Zhang Y, Kong S,
Chen Y, Yang S and Li K: Transcriptome analysis on the inflammatory
cell infiltration of nonalcoholic steatohepatitis in Bama minipigs
induced by a long-term high-fat, high-sucrose diet. Plos One.
9:e1137242014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xia JH, Zhang YY, Xin LL, Kong S, Chen Y,
Yang S and Li K: Global Transcriptomic Profiling of Cardiac
Hypertrophy and Fatty Heart Induced by Long-Term High-Energy Diet
in Bama Miniature Pigs. Plos One. 10:e01324202015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang SL, Xia JH, Zhang YY, Fan JG, Wang H,
Yuan J, Zhao ZZ, Pan Q, Mu YL, Xin LL, Chen YX and Li K:
Hyperinsulinemia shifted energy supply from glucose to ketone
bodies in early nonalcoholic steatohepatitis from high-fat
high-sucrose diet induced Bama minipigs. Sci Rep. 5:139802015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Winzell MS and Ahrén B: The high-fat
diet-fed mouse - A model for studying mechanisms and treatment of
impaired glucose tolerance and type 2 diabetes. Diabetes.
53:S215–S219. 2004. View Article : Google Scholar
|
|
11
|
Danda RS, Habiba NM, Rincon-Choles H,
Bhandari BK, Barnes JL, Abboud HE and Pergola PE: Kidney
involvement in a nongenetic rat model of type 2 diabetes. Kidney
Int. 68:2562–2571. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deng W, Yang D, Zhao B, Ouyang Z, Song J,
Fan N, Liu Z, Zhao Y, Wu Q, Nashun B, et al: Use of the 2A peptide
for generation of multi-transgenic pigs through a single round of
nuclear transfer. PLoS One. 6:e199862011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dieckhoff B, Kessler B, Jobst D, Kues W,
Petersen B, Pfeifer A, Kurth R, Niemann H, Wolf E and Denner J:
Distribution and expression of porcine endogenous retroviruses in
multi-transgenic pigs generated for xenotransplantation.
Xenotransplantation. 16:64–73. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Webster NL, Forni M, Bacci ML, Giovannoni
R, Razzini R, Fantinati P, Zannoni A, Fusetti L, Dalprà L, Bianco
MR, et al: Multi-transgenic pigs expressing three fluorescent
proteins produced with high efficiency by sperm mediated gene
transfer. Mol Reprod Dev. 72:68–76. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeong YH, Park CH, Jang GH, Jeong YI,
Hwang IS, Jeong YW, Kim YK, Shin T, Kim NH, Hyun SH, et al:
Production of multiple transgenic Yucatan miniature pigs expressing
human complement regulatory factors, human CD55, CD59 and
H-transferase genes. PLoS One. 8:e632412013. View Article : Google Scholar
|
|
16
|
Park SJ, Cho B, Koo OJ, Kim H, Kang JT,
Hurh S, Kim SJ, Yeom HJ, Moon J, Lee EM, et al: Production and
characterization of soluble human TNFRI-Fc and human HO-1 (HMOX1)
transgenic pigs by using the F2A peptide. Transgenic Res.
23:407–419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Renner S, Braun-Reichhart C, Blutke A,
Herbach N, Emrich D, Streckel E, Wünsch A, Kessler B, Kurome M,
Bähr A, et al: Permanent neonatal diabetes in INS (C94Y) transgenic
pigs. Diabetes. 62:1505–1511. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Umeyama K, Watanabe M, Saito H, Kurome M,
Tohi S, Matsunari H, Miki K and Nagashima H: Dominant-negative
mutant hepatocyte nuclear factor 1alpha induces diabetes in
transgenic-cloned pigs. Transgenic Res. 18:697–706. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Renner S, Fehlings C, Herbach N, Hofmann
A, von Waldthausen DC, Kessler B, Ulrichs K, Chodnevskaja I,
Moskalenko V, Amselgruber W, et al: Glucose intolerance and reduced
proliferation of pancreatic beta-cells in transgenic pigs with
impaired glucose-dependent insulinotropic polypeptide function.
Diabetes. 59:1228–1238. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Masuzaki H, Paterson J, Shinyama H, Morton
NM, Mullins JJ, Seckl JR and Flier JS: A transgenic model of
visceral obesity and the metabolic syndrome. Science.
294:2166–2170. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mathis D, Vence L and Benoist C: beta-cell
death during progression to diabetes. Nature. 414:792–798. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maris M, Overbergh L, Gysemans C, Waget A,
Cardozo AK, Verdrengh E, Cunha JP, Gotoh T, Cnop M, et al: Deletion
of C/EBP homologous protein (Chop) in C57Bl/6 mice dissociates
obesity from insulin resistance. Diabetologia. 55:1167–1178. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hoppener JWM, Jacobs HM, Wierup N,
Sotthewes G, Sprong M, de Vos P, Berger R, Sundler F and Ahrén B:
Human islet amyloid polypeptide transgenic mice: In vivo and ex
vivo models for the role of hIAPP in type 2 diabetes mellitus. Exp
Diabetes Res. 2008:6970352008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paterson JM, Morton NM, Fievet C, Kenyon
CJ, Holmes MC, Staels B, Seckl JR and Mullins JJ: Metabolic
syndrome without obesity: Hepatic overexpression of
11beta-hydroxysteroid dehydrogenase type 1 in transgenic mice. Proc
Natl Acad Sci USA. 101:7088–7093. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hull RL, Shen ZP, Watts MR, Kodama K, Carr
DB, Utzschneider KM, Zraika S, Wang F and Kahn SE: Long-term
treatment with rosiglitazone and metformin reduces the extent of,
but does not prevent, islet antyloid deposition in mice expressing
the gene for human islet antyloid polypeptide. Diabetes.
54:2235–2244. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Masuzaki H and Flier JS: Tissue-specific
glucocorticoid reactivating enzyme, 11 beta-hydroxysteroid
dehydrogenase type 1 (11 beta-HSD1) - a promising drug target for
the treatment of metabolic syndrome. Curr Drug Targets Immune
Endocr Metabol Disord. 3:255–262. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen ZY, Liu SN, Li CN, et al:
Atorvastatin helps preserve pancreatic beta cell function in obese
C57BL/6 J mice and the effect is related to increased pancreas
proliferation and amelioration of endoplasmic-reticulum stress.
Lipids Health Dis. 13:2014. View Article : Google Scholar
|
|
28
|
West AG, Gaszner M and Felsenfeld G:
Insulators: Many functions, many mechanisms. Genes Dev. 16:271–288.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xia J, Hu B, Mu Y, Xin L, Yang S and Li K:
Molecular cloning and characterization of the promoter region of
the porcine apolipoprotein E gene. Mol Biol Rep. 41:3211–3217.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ruan JX, Li HG, Xu K, Wu T, Wei J, Zhou R,
Liu Z, Mu Y, Yang S, Ouyang H, et al: Highly efficient
CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs.
Sci Rep. 5:142532015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lai L, Kolber-Simonds D, Park KW, Cheong
HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN, et al:
Production of alpha-1,3-galactosyltransferase knockout pigs by
nuclear transfer cloning. Science. 295:1089–1092. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(T)(-Delta Delta C) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
33
|
Bubner B and Baldwin IT: Use of real-time
PCR for determining copy number and zygosity in transgenic plants.
Plant Cell Rep. 23:263–271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luo W, Li Z, Huang Y, Han Y, Yao C, Duan
X, Ouyang H and Li L: Generation of AQP2-Cre transgenic mini-pigs
specifically expressing Cre recombinase in kidney collecting duct
cells. Transgenic Res. 23:365–375. 2014. View Article : Google Scholar
|
|
35
|
Li L, Li Q, Bao Y, Li J, Chen Z, Yu X,
Zhao Y, Tian K and Li N: RNAi-based inhibition of porcine
reproductive and respiratory syndrome virus replication in
transgenic pigs. J Biotechnol. 171:17–24. 2014. View Article : Google Scholar
|
|
36
|
Zhang Y, Wong CH, Birnbaum RY, Li G,
Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, et al: Chromatin
connectivity maps reveal dynamic promoter-enhancer long-range
associations. Nature. 504:306–310. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sadeghnia HR, Ghorbani Hesari T,
Mortazavian SM, Mousavi SH, Tayarani-Najaran Z and Ghorbani A:
Viola tricolor induces apoptosis in cancer cells and exhibits
antiangiogenic activity on chicken chorioallantoic membrane. Biomed
Res Int. 2014:6257922014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rabhi N, Salas E, Froguel P and Annicotte
JS: Role of the unfolded protein response in β cell compensation
and failure during diabetes. J Diabetes Res. 2014:7951712014.
View Article : Google Scholar
|
|
39
|
Morton NM, Paterson JM, Masuzaki H, Holmes
MC, Staels B, Fievet C, Walker BR, Flier JS, Mullins JJ and Seckl
JR: Novel adipose tissue-mediated resistance to diet-induced
visceral obesity in 11 beta-hydroxysteroid dehydrogenase type
1-deficient mice. Diabetes. 53:931–938. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Paterson JM, Holmes MC, Kenyon CJ, Carter
R, Mullins JJ and Seckl JR: Liver-selective transgene rescue of
hypothalamic-pituitary-adrenal axis dysfunction in 11
beta-hydroxysteroid dehydrogenase type 1-deficient mice.
Endocrinology. 148:961–966. 2007. View Article : Google Scholar
|
|
41
|
Lee AW and Cox RD: Use of mouse models in
studying type 2 diabetes mellitus. Expert Rev Mol Med. 13:e12011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jung EM, An BS, Kim YK, Jeong YH, Hwang WS
and Jeung EB: Generation of porcine fibroblasts overexpressing
11β-HSD1 with adipose tissue-specific aP2 promoter as a porcine
model of metabolic syndrome. Mol Med Rep. 8:751–756.
2013.PubMed/NCBI
|
|
43
|
Jeon Y, Kim Y, Yoon J, Cai L, Hwang SU,
Kim E, Lee S, Jeung EB and Hyun SH: Production of
11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) over-expressed
pigs for the study of metabolic syndrome disease. Reprod Fertil
Dev. 26:116. 2014. View Article : Google Scholar
|
|
44
|
Bram Y, Frydman-Marom A, Yanai I, Gilead
S, Shaltiel-Karyo R, Amdursky N and Gazit E: Apoptosis induced by
islet amyloid polypeptide soluble oligomers is neutralized by
diabetes-associated specific antibodies. Sci Rep. 4:42672014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hull RL, Andrikopoulos S, Verchere CB,
Vidal J, Wang F, Cnop M, Prigeon RL and Kahn SE: Increased dietary
fat promotes islet amyloid formation and beta-cell secretory
dysfunction in a transgenic mouse model of islet amyloid. Diabetes.
52:372–379. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Oyadomari S and Mori M: Roles of
CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ.
11:381–389. 2004. View Article : Google Scholar
|
|
47
|
Marchetti P, Bugliani M, Lupi R, Marselli
L, Masini M, Boggi U, Filipponi F, Weir GC, Eizirik DL and Cnop M:
The endoplasmic reticulum in pancreatic beta cells of type 2
diabetes patients. Diabetologia. 50:2486–2494. 2007. View Article : Google Scholar : PubMed/NCBI
|