Introduction
Hepatocellular carcinoma (HCC) is one of the most
common types of malignancy in humans. There is an increasing
incidence of HCC worldwide, and it is a leading cause of
cancer-associated mortality, partly because many patients are
initially diagnosed with advanced disease (1). In recent decades, great efforts have
been made to determine the molecular mechanisms underlying HCC. In
addition, dysregulated expression of oncogenes and tumor
suppressors has been reported to have a key role in the development
and progression of HCC (2).
Sprouty 2 (Spry2), which is a member of the sprouty
family, is an evolutionarily conserved inhibitor of receptor
tyrosine kinases (3). Spry2 has a
suppressive role in the regulation of mitogen-activated protein
kinase (MAPK) signaling (4). It
has previously been reported that Spry2 is frequently downregulated
in HCC, and patients with lower Spry2 expression exhibit poorer
survival and increased recurrence (5). Furthermore, downregulation of Spry2
is associated with highly malignant phenotypes, including advanced
tumor stages and vascular invasion (5); however, the regulatory mechanism
underlying Spry2 expression in HCC cells has yet to be
elucidated.
MicroRNAs (miRs) are a class of 18–25 nucleotide
long non-coding RNAs, which can induce mRNA degradation or suppress
protein translation via binding to the 3′-untranslated regions
(3′-UTRs) of specific mRNA (6).
miRs are important post-transcriptional regulators of gene
expression in normal cells, as well as in cancer cells. Therefore,
assessment of miR levels could potentially be useful for the
classification and stratification of tumors (7,8).
Dysregulated expression of specific miRs has been detected in HCC,
including miR-122, miR-124, miR-138, miR-148a and miR-203 (9–12).
Furthermore, upregulated miR-27b has been detected in five
drug-resistant HCC cell lines (13). miR-27b has been suggested to serve
as a possible marker of HCC following exposure to aflatoxins
(14). However, the detailed role
of miR-27b in mediating the metastatic potential of HCC cells
remains unclear.
The present study aimed to investigate the
association between miR-27b expression and HCC progression. In
addition, the detailed role of miR-27b in the regulation of HCC
cell migration and invasion was determined, and the underlying
molecular mechanisms involving Spry2 were discussed.
Materials and methods
Reagents
Dulbecco's modified Eagle's medium (DMEM), fetal
bovine serum (FBS), Lipofectamine 2000, TRIzol reagent and the
miRNA Reverse Transcription kit were purchased from Thermo Fisher
Scientific, Inc. (Waltham, MA, USA). miRNA quantitative polymerase
chain reaction (qPCR) Detection kit was purchased from Genecopoeia
(Rockville, MD, USA). Directed Mutagenesis kit was purchased from
Agilent Technologies, Inc. (Santa Clara, CA, USA). Rabbit
anti-human Spry2 monoclonal antibody (cat. no. ab180527), rabbit
anti-human glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
monoclonal antibody (cat. no. ab8245) and mouse anti-rabbit
secondary antibody IgG (HRP; cat. no. ab6728) were purchased from
Abcam (Cambridge, MA, USA). Enhanced chemiluminescence (ECL) kit
was purchased from Pierce Protein Biology; Thermo Fisher
Scientific, Inc. (Rockford, IL, USA). PsiCHECK 2 vector was
purchased from Promega Corporation (Madison, WI, USA). miR-27b
mimics, miR-27b inhibitor, Spry2 plasmid and Spry2-specific small
interfering (si)RNA were obtained from Guangzhou FulenGen Co., Ltd.
(Guangzhou, China).
Tissue specimens
The present study was approved by the Ethics
Committee of Yuhuangding Hospital (Yantai, China). A total of 35
primary HCC tissues and matched normal adjacent specimens were
collected. Written informed consent was obtained. The
histomorphology of all samples was confirmed by the Department of
Pathology. HCC tissues were immediately snap-frozen in liquid
nitrogen upon surgical removal.
Cell culture
The HepG2, LH86, LMH and PLHC-1 human HCC cell
lines, and the THLE-3 normal liver cell line were obtained from the
American Type Culture Collection (Manassas, VA, USA). The cells
were cultured in DMEM supplemented with 10% fetal bovine serum
(FBS) at 37°C in a humidified incubator containing 5%
CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol reagent. Tissues were homogenized prior to RNA extraction by
grinding. All samples were treated with 1 µl DNase (Thermo
Fisher Scientific, Inc.) prior to RT. The miRNA Reverse
Transcription kit was used to reverse transcribe RNA into cDNA,
according to the manufacturer's protocol. Briefly, reverse
transcription was performed at 16°C for 30 min, followed by an
incubation step at 42°C for 30 min and enzyme inactivation at 85°C
for 5 min. qPCR was performed using a miRNA qPCR Detection kit on
an ABI 7500 thermocycler (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The primers for Spry2 were:
Forward: 5′-CCTACTGTCGTCCCAAGACCT-3′ and reverse:
5′-GGGGCTCGTGCAGAAGAAT-3′ purchased from Sangon Biotech Co., Ltd.
(Shanghai, China). Briefly, the PCR cycling conditions were as
follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of
denaturation at 95°C for 15 sec and annealing/elongation at 60°C
for 60 sec. U6 was used as an internal reference gene. The
experiment was repeated 3 times. The relative expression levels
were analyzed using the 2−ΔΔCq method (15).
Western blotting
Tissues and cells were solubilized in cold
radioimmunoprecipitation lysis buffer (Beyotime Biotechnology,
Shanghai, China). Proteins were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (50 µg of
protein/lane), and transferred onto a polyvinylidene difluoride
(PVDF; Thermo Fisher Scientific, Inc.) membrane. The membrane was
incubated with phosphate-buffered saline containing 5% milk
overnight at 4°C. Subsequently, the PVDF membrane was incubated
with rabbit anti-Spry2 monoclonal antibody (1:50) and rabbit
anti-GAPDH monoclonal antibody (1:50) at room temperature for 3 h.
The membrane was then incubated with mouse anti-rabbit secondary
antibody (1:5,000) at room temperature for 1 h. An ECL kit was used
to perform chemiluminescent detection. The protein concentration
was quantified using Pierce BCA Protein Assay Kit, Thermo Fisher
Scientific, Inc. The relative protein expression levels were
analyzed using Image-Pro Plus software 6.0 (Media Cybernetics,
Inc., Rockville, MD, USA), and are presented as the density ratio
vs. GAPDH.
Transfection
Lipofectamine 2000 was used to transfect the HepG2
cells, according to the manufacturer's protocol. Briefly, cells
were cultured to 70% confluence, at a seed density of
106 and resuspended in serum-free medium. miR-27b
mimics, miR-27b inhibitor, Spry2 plasmid (Syngentech, Beijing,
China), Spry2-specific siRNA and Lipofectamine 2000 were diluted
with serum-free medium. The diluted Lipofectamine 2000 was added to
the diluted siRNA and miR-27b mimics, and the mixture was incubated
for 20 min at room temperature. Subsequently, the mixture was added
to the cell suspension. After a 6 h incubation at 37°C and 5%
CO2, the medium was replaced with the normal
serum-containing medium. Subsequently, the cells were cultured for
24 h prior to further experimentation.
Dual luciferase reporter assay
The miR-27b-predicted target sequence within the
Spry2 3′-UTR (UUUGCAACUGUGAA), and a mutant lacking complimentarity
with the miR-27b seed sequence (UUUGCACACACACC), were cloned
downstream of the luciferase gene driven by the cytomegalovirus
promoter, thus generating Luc.Spry2 and Luc.mtSpry2 vectors,
respectively. The wild type (WT) and mutant (MUT) 3′UTRs of Spry2
were obtained from Biowit Technologies (Shenzhen, China), which was
inserted into the BgIII and KpnI restriction sites 3′
to the end of the Renilla luciferase gene in the PsiCHECK vector.
HepG2 cells were transfected with Luc.Spry2 or Luc.mtSpry2 vectors
in conjunction with miR-27b mimics or scramble negative control miR
mimics using Lipofectamine 2000. A total of 24 h post-transfection,
the cells were harvested. Dual-Luciferase Reporter Assay System
(Promega Corporation) was used to determine luciferase activity.
Luciferase activity was quantified using an Lmax multiwell
luminometer (Molecular Devices, Sunnyvale, CA, USA).
Wound healing assay
Wound healing assay was performed, in order to
evaluate the cell migratory capacity of each group. Briefly, cells
were cultured to full confluence. Wounds ~1 mm wide were created in
the cell layer using a plastic scriber, and cells were washed with
phosphate-buffered saline and incubated in a serum-free medium. A
total of 24 h after the wound was generated, cells were incubated
in DMEM supplemented with 10% FBS. After further incubation for 0
and 48 h, the cells were fixed with absolute alcohol and observed
under a BX53 fluorescence microscope (Olympus Corporation, Tokyo,
Japan).
Transwell assay
Transwell assay was performed in order to examine
cell invasion. Briefly, the invasive ability of HepG2 cells was
determined in 24-well Transwell chambers, which were coated with a
layer of Matrigel. Cell suspension of DMEM (106
cells/ml) was added to the upper chamber, and DMEM supplemented
with 10% FBS was added to the lower chamber. Following a 24 h
incubation, non-invading cells as well as the Matrigel (Thermo
Fisher Scientific, Inc.) on the interior of the inserts was removed
using a cotton-tipped swab. Invasive cells on the lower surface of
the membrane were stained with gentian violet, rinsed with water
and air-dried. Five fields were randomly selected and cell number
was counted under a microscope.
Bioinformatical analysis
The target genes of miR-27 were predicted using
TargetScan (16). The following
settings were used for analysis: i) Species, human; ii) gene
symbol, Spry2 and iii) selelcted a conserved miRNA family.
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analysis was performed using SPSS 16.0 (SPSS, Inc.,
Chicago, IL, USA). The association between miR-27b expression and
clinicopathologic features in patients with HCC was analyzed using
χ2 test and Spearman rank correlation analysis. The
statistical correlation of data between the cell groups was
analyzed by one-way analysis of variance. P<0.05 was considered
to indicate a statistically significant difference.
Results
miR-27b expression pattern in HCC cell
lines and tissue samples
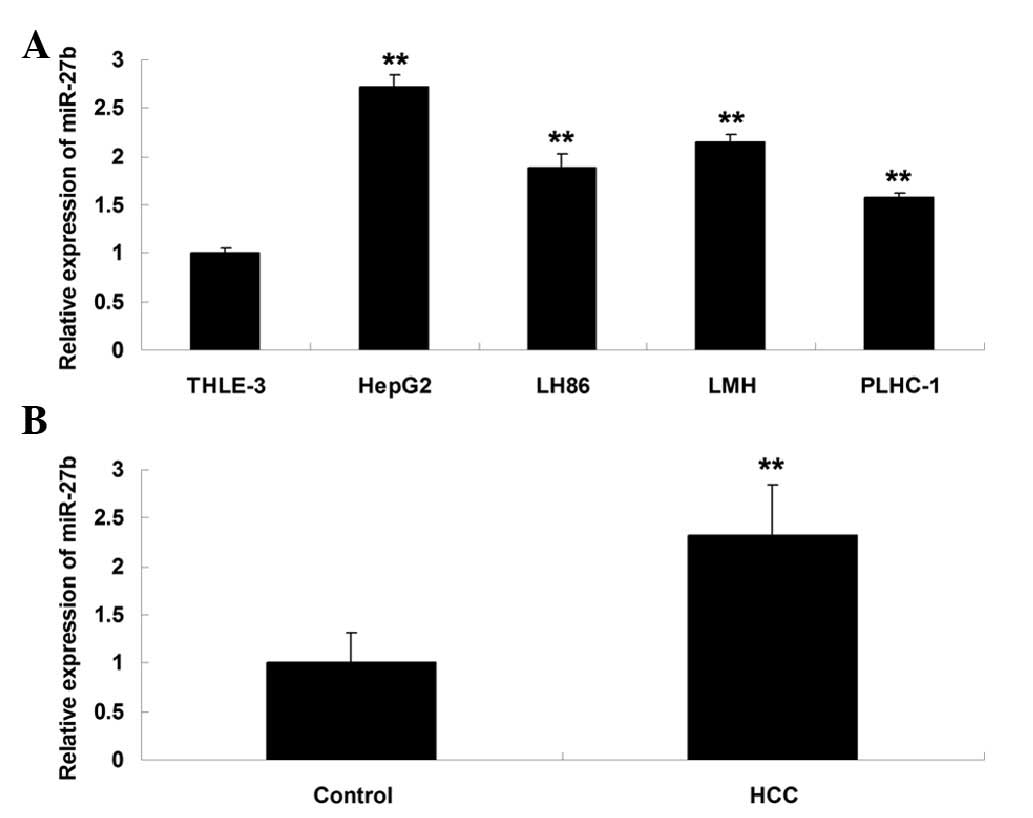
The expression levels of miR-27b were examined in
four common human HCC cell lines, HepG2, LH86, LMH and PLHC-1, as
well as in a normal liver cell line, THLE-3. HepG2 cells exhibited
a significant increase in the expression levels of miR-27b, as
compared with the normal liver THLE-3 cells (Fig. 1A), therefore this cell line was
used in further experiments. In addition, the expression levels of
miR-27b were significantly increased in the HCC tissues, as
compared with in the peritumoral noncancerous tissues (Fig. 1B). The expression levels of miR-27b
were increased in 77.1% (27/35) of the HCC tissue samples.
Correlation between miR-27b expression
and clinicopathologic features in patients with HCC
The association between miR-27b expression and
clinicopathologic features in patients with HCC was analyzed using
χ2 test. As presented in Table I, miR-27b expression had no
association with the age, gender and tumor size of the patients
(P>0.05). However, miR-27b expression was correlated with tumor
differentiation, Tumor Node Metastasis (TNM) stage and vascular
invasion (P<0.05).
 | Table ICorrelation between miR-27b expression
and clinicopathologic features in patients with hepatocellular
carcinoma. |
Table I
Correlation between miR-27b expression
and clinicopathologic features in patients with hepatocellular
carcinoma.
| Clinicopathologic
feature | Cases | miR-27b expression
| χ2 | P-value |
|---|
| Low | High |
|---|
| Age (years) | | | | 0.397 | 0.429 |
| <60 | 12 | 2 | 10 | | |
| ≥60 | 23 | 6 | 17 | | |
| Gender | | | | 0.027 | 0.602 |
| Male | 21 | 5 | 16 | | |
| Female | 14 | 3 | 11 | | |
| Tumor size (cm) | | | | 0.122 | 0.527 |
| <5 | 15 | 3 | 12 | | |
| ≥5 | 20 | 5 | 15 | | |
| Tumor
differentiation | | | | 5.402 | 0.025 |
| I+II | 18 | 7 | 11 | | |
| III+IV | 17 | 1 | 16 | | |
| TNM stage | | | | 4.610 | 0.037 |
| I+II | 19 | 7 | 12 | | |
| IIIA | 16 | 1 | 15 | | |
| Vascular
invasion | | | | 6.291 | 0.016 |
| Yes | 17 | 7 | 10 | | |
| No | 18 | 1 | 17 | | |
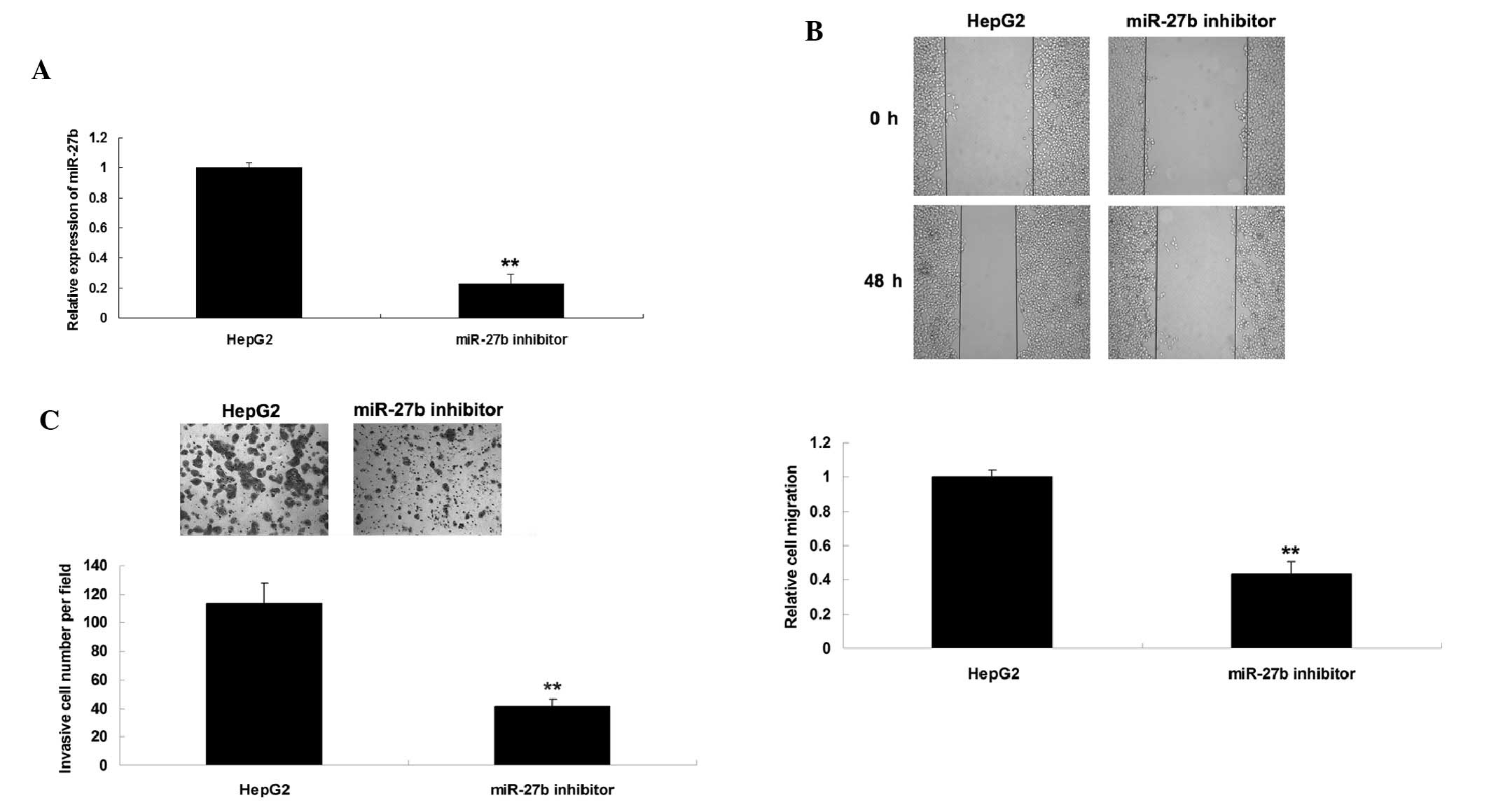
Knockdown of miR-27b inhibits HepG2 cell
migration and invasion
The present study investigated the effects of
miR-27b on the regulation of migration and invasion in HCC cells.
Since miR-27b was upregulated in HCC cells, HepG2 cells were
transfected with a miR-27b inhibitor to suppress its expression.
Post-transfection, the expression levels of miR-27b were reduced
(Fig. 2A). Wound healing and
Transwell assays were conducted to investigate the effects of
miR-27b knock-down on HCC cell migration and invasion,
respectively. As shown in Fig. 2B and
C, knockdown of miR-27b expression significantly inhibited the
migration and invasion of HepG2 cells. These results indicate that
miR-27b has a role in the regulation of HCC metastasis.
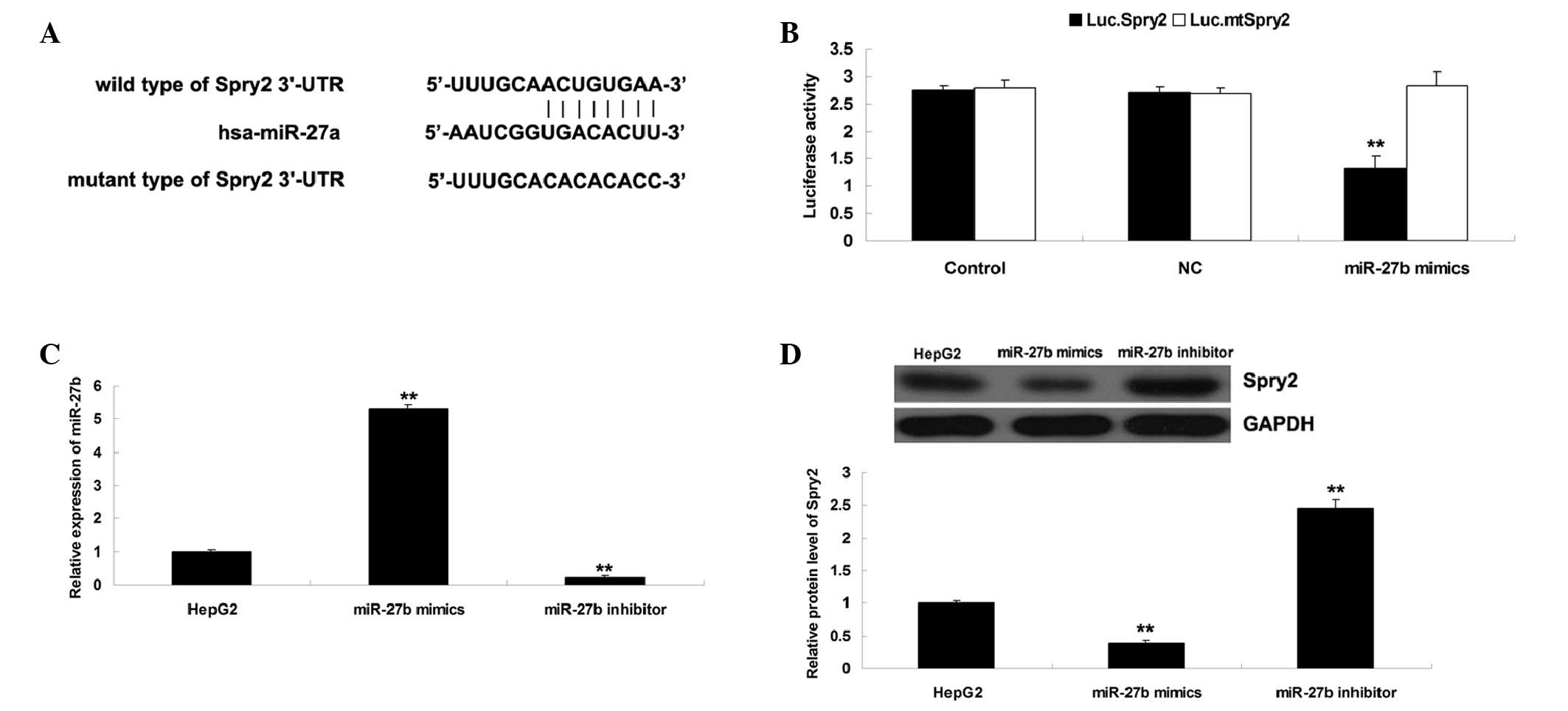
Spry2 is identified as a target gene of
miR-27b and is negatively regulated by miR-27b in HepG2 cells
Bioinformatical analysis was performed in order to
predict the target genes of miR-27b, and Spry2 was identified as a
putative target. To clarify whether Spry2 is a direct target of
miR-27b, a Luc.Spry2 vector containing the wild-type 3′-UTR of
Spry2 and a Luc.mtSpry2 vector containing a mutated form of the
3′-UTR of Spry2 were generated (Fig.
3A). Subsequently, HepG2 cells were transfected with Luc.Spry2
or Luc.mtSpry2 vector, with or without miR-27b mimics. A total of
24 h post-transfection, the luciferase activity was examined.
Compared with the control group, luciferase activity was
significantly decreased in the HepG2 cells co-transfected with the
Luc.Spry2 vector and miR-27b mimics, but was unaltered in HepG2
cells co-transfected with the Luc.mtSpry2 vector and miR-27b mimics
(Fig. 3B). These results indicate
that miR-27b directly binds to the seed sequences within the Spry2
3′-UTR in HepG2 cells.
The present study investigated the effects of
miR-27b overexpression or knockdown on the protein expression
levels of Spry2 in HepG2 cells. HepG2 cells were transfected with
miR-27b mimics or a miR-27b inhibitor. As shown in Fig. 3C, transfection with miR-27b mimics
upregulated the miR-27b expression levels; however, transfection
with the miR-27b inhibitor downregulated miR-27b expression levels.
Subsequently, western blotting was performed to examine the protein
expression levels of Spry2. As shown in Fig. 3D, upregulation of miR-27b
significantly inhibited the protein expression levels of Spry2;
however, knockdown of miR-27b expression enhanced the protein
expression levels of Spry2 in HepG2 cells. Accordingly, these
results suggest that miR-27b may negatively regulate the protein
expression levels of Spry2 in HCC HepG2 cells via directly
targeting its mRNA.
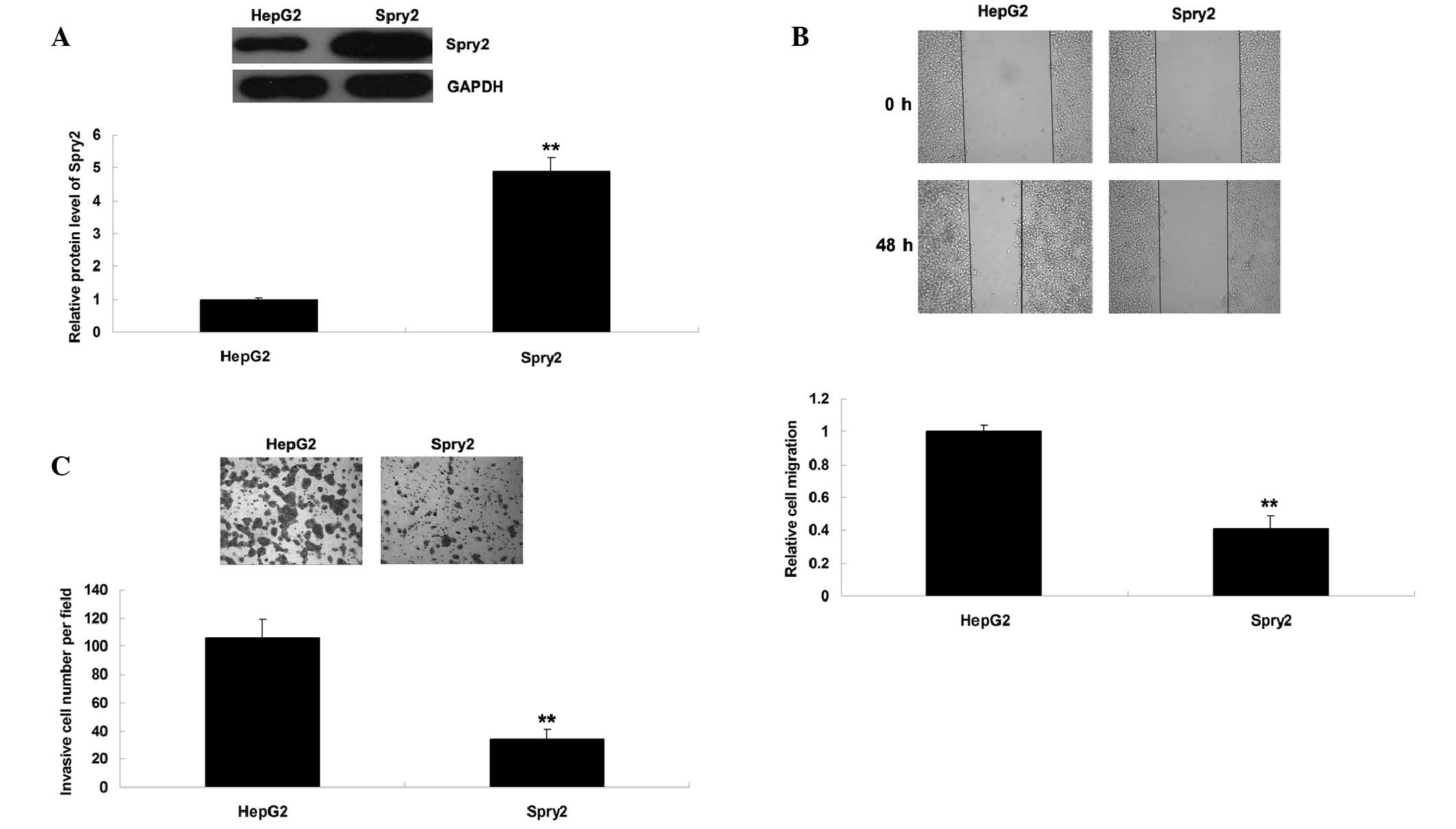
Spry2 is involved in the miR-27b-mediated
migration and invasion of HepG2 cells
Since knockdown of miR-27b upregulated Spry2
expression and suppressed HCC cell migration and invasion, the
present study hypothesized that Spry2 may be involved in
miR-27b-mediated migration and invasion of HCC cells. To verify
this hypothesis, HepG2 cells were transfected with a Spry2 plasmid.
Post-transfection, the protein expression levels of Spry2 were
significantly increased (Fig. 4A).
Consistent with miR-27b knockdown, overexpression of Spry2
suppressed HCC cell migration and invasion (Fig. 4B and C).
To further clarify whether the effects of miR-27b on
HCC cell migration and invasion were mediated by inhibition of
Spry2 expression, HepG2 cells were transfected with a miR-27b
inhibitor, or co-transfected with a miR-27b inhibitor and Spry2
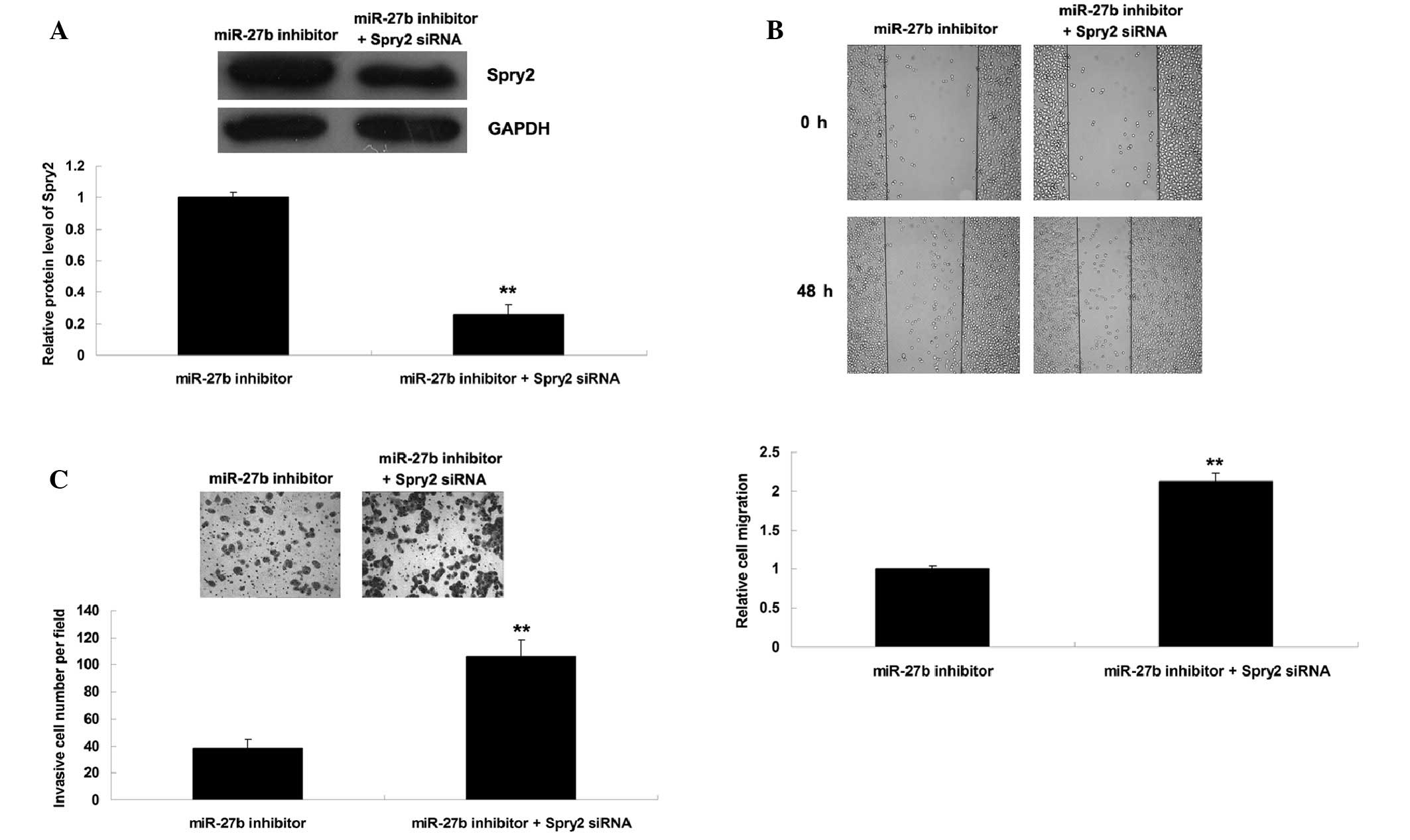
siRNA. Post-transfection, the protein expression levels of Spry2
were reduced in HepG2 cells co-transfected with a miR-27b inhibitor
and Spry2 siRNA, as compared with the HepG2 cells transfected with
a miR-27b inhibitor only (Fig.
5A). In addition, the migratory and invasive capacity was
increased in HepG2 cells co-transfected with a miR-27b inhibitor
and Spry2 siRNA, as compared with the HepG2 cells transfected with
a miR-27b inhibitor only (Fig. 5B and
C). These results indicate that knockdown of Spry2 expression
may reverse the suppressive effects of miR-27b inhibition on HCC
cell migration and invasion. Based on these findings, miR-27b may
have a promoting role in the regulation of HCC cell migration and
invasion, at least partially via directly targeting Spry2
expression.
Discussion
Dysregulated expression of miR-27b has previously
been implicated in HCC (13);
however, the detailed role of miR-27b in HCC remains largely
unknown. The results of the present study demonstrated that the
expression levels of miR-27b were increased in HCC cell lines and
tissues samples. Furthermore, elevated miR-27b expression was
significantly correlated with histological grade, depth of invasion
and TNM stage. Further investigation revealed that miR-27b had a
promoting role in the regulation of HCC cell migration and
invasion, at least partially via directly targeting Spry2
expression.
Dysregulated miR expression has been shown to be
tightly associated with the development and progression of HCC
(1,17). Dysregulation of miR-148a
discriminates not only the overall survival and recurrence free
survival rates of HCC, but also the vascular invasion (18). miR-27b has previously been reported
to be involved in hepatic lipid metabolism (19). In addition, miR-27b has been
suggested to be involved in HCC (13,14).
Zhuo et al (13)
investigated the association between miRs and multidrug resistance
using the HCC Huh-7 cell line, which was treated with adramycin,
cisplatin, carboplatin, mitomycin C or vincristine at increasing
concentrations, in order to develop drug-resistant sublines.
miR-27b was significantly upregulated in the drug-resistant HCC
cell lines, suggesting that miR-27b may have a role in drug
resistance in HCC (13).
Furthermore, miR-27b has been suggested to serve as a possible
marker of HCC following exposure to aflatoxins (14). The present study is the first, to
the best of our knowledge, to demonstrate that miR-27b was
frequently upregulated in HCC, and elevated miR-27b expression
levels were significantly correlated with tumor differentiation,
TNM stage and vascular invasion. These findings suggested that
miR-27b may act as an oncogenic miR in HCC. Further studies should
expand the sample size, and focus on the association between
miR-27b expression and the prognosis of patients with HCC.
The role of miR-27b has also been demonstrated in
other types of cancer, with the majority of studies suggesting that
miR-27b may act as a tumor suppressor in human cancer (20,21).
Lee et al (22)
demonstrated that miR-27b inhibited growth, progression and the
inflammatory response in neuroblastoma cells by targeting
peroxisome proliferator-activated receptor γ. In addition, miR-27b
was downregulated in non-small cell lung cancer (NSCLC), and
overexpression of miR-27b significantly suppressed the
proliferation and invasion of NSCLC cells (21). However, in several types of human
malignancy, miR-27b acts as an oncogene (23). miR-27b was upregulated in glioma
tissues and cells, and knockdown of miR-27b was able to trigger
growth inhibition, induce apoptosis and inhibit invasion of glioma
cells, potentially via direct or indirect inhibition of signal
transducer and activator of transcription 3, c-myc and cyclin D1
(24). The present study
demonstrated that miR-27b had an oncogenic role in the regulation
of HCC cell migration and invasion.
Since miRs have roles in human cancer via mediating
the protein expression of their target genes (25), the present study focused on the
target genes of miR-27b in HCC cells. Spry2 was identified as a
direct target gene of miR-27b in HCC cells. A previous study
reported that dysregulated Spry2 expression has been detected in
HCC (5). Song et al
(5) demonstrated that 86.3% (207
of 240) of patients with HCC exhibited downregulated Spry2
expression. Patients negative for Spry2 exhibited poorer survival
and increased recurrence. Multivariate analysis further established
Spry2 as an independent predictor of postoperative recurrence in
patients with HCC. Furthermore, downregulation of Spry2 was
associated with highly malignant phenotypes, and was positively
correlated with the metastatic potential of HCC cell lines
(5). Lee et al (26) investigated the potential role of
Spry2 in HCC by expressing dominant negative Spry2 and activated
β-catenin in the mouse liver; the results demonstrated that tumor
cells exhibited high expression levels of extracellular
signal-regulated kinases, as well as dysregulation of genes
associated with cell proliferation, apoptosis and angiogenesis. In
addition, Wang et al (27)
reported that inactivation of Spry2 enhanced AKT-driven
hepatocarcinogenesis via activation of MAPK and pyruvate kinase
muscle pathways. The present study demonstrated that overexpression
of Spry2 suppressed HCC cell migration and invasion, suggesting
that Spry2 has a suppressive role in HCC metastasis. Furthermore,
knockdown of Spry2 reversed the suppressive effects of miR-27b
inhibition on HCC cell migration and invasion, thus suggesting that
Spry2 is involved in miR-27b-meditated HCC cell migration and
invasion.
In conclusion, the present study demonstrated that
upregulation of miR-27b may be associated with HCC progression, and
suggested that miR-27b promotes the migration and invasion of HCC
cells, at least partially via suppressing Spry2 expression.
Therefore, the miR-27b/Spry2 axis may be considered a potential
therapeutic target for HCC.
References
|
1
|
Zhu AX: Molecularly targeted therapy for
advanced hepatocellular carcinoma in 2012: Current status and
future perspectives. Semin Oncol. 39:493–502. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Psyrri A, Arkadopoulos N, Vassilakopoulou
M, Smyrniotis V and Dimitriadis G: Pathways and targets in
hepatocellular carcinoma. Expert Rev Anticancer Ther. 12:1347–1357.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li P, Tao L, Yang J, Cai H, Ju X, Li J,
Shao P, Cao Q, Qin C, Meng X and Yin C: Sprouty2 is associated with
prognosis and suppresses cell proliferation and invasion in renal
cell carcinoma. Urology. 82:253e1–e7. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mei Y, Bian C, Li J, Du Z, Zhou H, Yang Z
and Zhao RC: miR-21 modulates the ERK-MAPK signaling pathway by
regulating SPRY2 expression during human mesenchymal stem cell
differentiation. J Cell Biochem. 114:1374–1384. 2013. View Article : Google Scholar
|
|
5
|
Song K, Gao Q, Zhou J, Qiu SJ, Huang XW,
Wang XY and Fan J: Prognostic significance and clinical relevance
of Sprouty 2 protein expression in human hepatocellular carcinoma.
Hepatobiliary Pancreat Dis Int. 11:177–184. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Coulouarn C, Factor VM, Andersen JB,
Durkin ME and Thorgeirsson SS: Loss of miR-122 expression in liver
cancer correlates with suppression of the hepatic phenotype and
gain of metastatic properties. Oncogene. 28:3526–3536. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar
|
|
11
|
Wang W, Zhao LJ, Tan YX, Ren H and Qi ZT:
MiR-138 induces cell cycle arrest by targeting cyclin D3 in
hepatocellular carcinoma. Carcinogenesis. 33:1113–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yin W, Zhao Y, Ji YJ, Tong LP, Liu Y, He
SX and Wang AQ: Serum/plasma microRNAs as biomarkers for
HBV-related hepatocellular carcinoma in China. Biomed Res Int.
2015:9651852015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhuo L, Liu J, Wang B, Gao M and Huang A:
Differential miRNA expression profiles in hepatocellular carcinoma
cells and drug-resistant sublines. Oncol Rep. 29:555–562. 2013.
|
|
14
|
Valencia-Quintana R, Sánchez-Alarcón J,
Tenorio-Arvide MG, Deng Y, Montiel-González JM, Gómez-Arroyo S,
Villalobos-Pietrini R, Cortés-Eslava J, Flores-Márquez AR and
Arenas-Huertero F: The microRNAs as potential biomarkers for
predicting the onset of aflatoxin exposure in human beings: A
review. Front Microbiol. 5:1022014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu
C, Zou L, Li Z, Zhao J and Lin N: MicroRNA-101 suppresses
SOX9-dependent tumorigenicity and promotes favorable prognosis of
human hepatocellular carcinoma. FEBS Lett. 586:4362–4370. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Heo MJ, Kim YM, Koo JH, Yang YM, An J, Lee
SK, Lee SJ, Kim KM, Park JW and Kim SG: microRNA-148a dysregulation
discriminates poor prognosis of hepatocellular carcinoma in
association with USP4 overexpression. Oncotarget. 5:2792–2806.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Her GM, Hsu CC, Hong JR, Lai CY, Hsu MC,
Pang HW, Chan SK and Pai WY: Overexpression of gankyrin induces
liver steatosis in zebrafish (Danio rerio). Biochim Biophys Acta.
1811:536–548. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ishteiwy RA, Ward TM, Dykxhoorn DM and
Burnstein KL: The microRNA −23b/−27b cluster suppresses the
metastatic phenotype of castration-resistant prostate cancer cells.
PLoS One. 7:e521062012. View Article : Google Scholar
|
|
21
|
Wan L, Zhang L, Fan K and Wang J: MiR-27b
targets LIMK1 to inhibit growth and invasion of NSCLC cells. Mol
Cell Biochem. 390:85–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee JJ, Drakaki A, Iliopoulos D and Struhl
K: MiR-27b targets PPARγ to inhibit growth, tumor progression and
the inflammatory response in neuroblastoma cells. Oncogene.
31:3818–3825. 2012. View Article : Google Scholar :
|
|
23
|
Jin L, Wessely O, Marcusson EG, Ivan C,
Calin GA and Alahari SK: Prooncogenic factors miR-23b- and miR-27b
are regulated by Her2/Neu, EGF, and TNF-α in breast cancer. Cancer
Res. 73:2884–2896. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen L, Li H, Han L, Zhang K, Wang G, Wang
Y, Liu Y, Zheng Y, Jiang T, Pu P, et al: Expression and function of
miR-27b in human glioma. Oncol Rep. 26:1617–1621. 2011.PubMed/NCBI
|
|
25
|
Yoshitaka T, Kawai A, Miyaki S, Numoto K,
Kikuta K, Ozaki T, Lotz M and Asahara H: Analysis of microRNAs
expressions in chondrosarcoma. J Orthop Res. 31:1992–1998. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee SA, Ho C, Roy R, Kosinski C, Patil MA,
Tward AD, Fridlyand J and Chen X: Integration of genomic analysis
and in vivo transfection to identify sprouty 2 as a candidate tumor
suppressor in liver cancer. Hepatology. 47:1200–1210. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang C, Delogu S, Ho C, Lee SA, Gui B,
Jiang L, Ladu S, Cigliano A, Dombrowski F, Evert M, et al:
Inactivation of Spry2 accelerates AKT-driven hepatocarcinogenesis
via activation of MAPK and PKM2 pathways. J Hepatol. 57:577–583.
2012. View Article : Google Scholar : PubMed/NCBI
|