Introduction
Renal cell carcinoma (RCC) tumors originate in the
renal cortex, and account for ~3% of adult malignancies and almost
90% of all renal neoplasms (1,2).
Previously estimated cancer statistics showed that the incidence
and mortality rates of RCC were among the 10 leading types of
cancer in the USA, with >65,150 new cases and a mortality rate
of >8,780 recorded in 2013 (3).
RCC is a relatively asymptomatic disease, and ~30% of patients with
RCC develop invasive disease, commonly metastasizing to the bone,
lungs, brain and liver (4,5). Patients with metastatic RCC face a
poor prognosis and have limited therapeutic options. Therefore,
increased understanding of the molecular mechanisms involved in the
progression of RCC, including its recurrence, metastasis or drug
resistance, is required to provide a rationale for effective
therapeutic methods for the treatment of RCC.
Long non-coding RNAs (lncRNAs), are transcripts of
>200 bp in length with no protein-coding function (6), and represent a class of non-coding
RNAs, which has received less attention in investigations. Several
studies have demonstrated that lncRNAs are crucial for the
regulation of chromatin structure, gene expression and
translational control (7,8). There is evidence to suggest that
lncRNAs may regulate key cancer pathways at the transcriptional,
post-transcriptional and epigenetic levels (9). Estimates suggest that the number of
human lncRNAs rivals that of protein-coding genes, ranging between
10,000 and 20,000 (10). Despite
these numbers, only a small number of lncRNAs have been
characterized. In previous years, due to the successful application
of different novel approaches, including genome-wide gene
expression screening, genome-wide association studies,
region-targeted association assays and conventional linkage
screening, designed LncRNA arrays, RIP-RNA sequencing, transgenic
expression, and gene knockdown or knockout, the functions of
LncRNAs in cancer are being increasingly characterized.
Accumulating data show that several identified LncRNAs are crucial
in tissue carcinogenesis, invasion and metastasis (11–15).
Previous sequencing for the expression of LncRNAs
showed TRIM52-AS1 was downregulated in RCC (16,17).
The present study was undertaken to verify the expression of
TRIM52-AS1 in RCC, and to assess the impact of TRIM52-AS1 on cell
proliferation, invasion and migration. The results of these
investigations may elucidate whether TRIM52-AS1 functions as a
tumor suppressor in RCC, and may lay the foundation for further
studies regarding the pathogenesis of RCC.
Materials and methods
Sample collection and RNA isolation
The present study was approved by the ethics
committee of the First Affiliated Hospital of Harbin Medical
University (Harbin, China). All RCC tissues and paired adjacent
normal tissues used in the present study were collected from The
Department of Urology of The First Affiliated Hospital of Harbin
Medical University. All specimens were obtained on the basis of
their availability for investigation purpose and under a protocol
approved by the local medical ethics committee. Written informed
consent was obtained from all patients involved in the present
study. All tissue samples were reviewed and classified using
hematoxylin and eosin staining and the 2009 American Joint
Committee on Cancer staging system (18). The clinicopathological information
of the patients is presented in Table
I. The people were all renal cell (RCC) carcinoma patients.
They could improve prognosis after nephrectomy. A total of 60 fresh
RCC and adjacent normal tissue samples, located 2.0 cm from the
visible RCC lesions, were obtained from patients with RCC by
nephrectomy. The samples were immersed in RNAlater (Qiagen, Hilden,
Germany) for 30 min and subsequently stored at −80°C. The total RNA
of each sample was extracted using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. All isolated RNA was quantified using a
Nano-Drop® spectrophotometer. Only RNA samples with
260/280 ratios of 1.8–2.0 were used for further investigation.
 | Table IClinical and pathologic
characteristics of all analyzed samples. |
Table I
Clinical and pathologic
characteristics of all analyzed samples.
| Factor | Number |
|---|
| Mean age (range),
years | 55 (25–83) |
| Gender
(male/female) | 35/25 |
| Pathology | |
| Clear cell RCC | 32 |
| Papillary RCC | 15 |
| Chromophobe RCC | 13 |
| Stage | |
| T1a | 34 |
| T1b | 16 |
| T2 | 7 |
| T3 | 2 |
| T4 | 1 |
| Fuhrman grade | |
| G1 | 5 (T1a), 4 (T1b) |
| G2 | 23 (T1a), 10 (T1b), 3
(T2) |
| G3 | 6 (T1a), 2 (T1b), 4
(T2), 2 (T3), 1 (T4) |
| G4 | 0 |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
CAKI- 2, 769- P, ACHN and 786-O renal carcinoma cell
lines were obtained from the laboratory of the Institute of Urology
of Shenzhen PKU-HKUST Medical Center (Shenzhen, China). Human
embryonic kidney 293T cells were purchased from the Shanghai
Institutes for Biological Sciences, Chinese Academy of Sciences
(Shanghai, China). The cells were maintained in Dulbecco's modified
Eagle's medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented
with 10% fetal bovine serum at 37°C in a humidified atmosphere with
5% CO2. First-strand cDNA was synthesized using oligo-dT
primers (cat. no. K1622; Fermentas, Waltham, MA, USA). The primer
sequences specific for human TRIM52-AS1 were as follows: Forward
5′-AGA GCA AGG ACT GTA TGT GTTC-3′ and reverse 5′-CTG GAG TGG CAG
AAG TAAGG-3′; with human GAPDH as an internal control, the primer
sequences for GAPDH were forward 5′-AGT GGC AAA GTG GAG ATT -3′ and
reverse 5′-GTG GAG TCA TAC TGG AACA-3′. RT-qPCR was performed using
a SYBR® Premix EX Taq™ II PCR kit (cat. no. RR820A;
Takara Biotechnology Co., Ltd., Dalian, China), according to the
manufacturer's protocol, on a Roche Light-cycler 480 Real-Time PCR
System (Roche Diagnostics). Data were calculated according to the
Applied Biosystems comparative quantification method (19).
Cell culture and transfection
Cells of the human renal carcinoma cell lines, ACHN
and 786-O, were purchased from the American Type Culture Collection
(Manassas, VA, USA). TRIM52-AS1 small hairpin shRNA (target
sequence, 5′-GCA CAG AGC AAG GAC UGU AUG UGU U-3′) were purchased
from Genechem (Shanghai, China). The TRIM52-AS1 overexpression
plasmid, TRIM52-AS1-pcDNA3.1+, was synthesized by Invitrogen
(Thermo Fisher Scientific, Inc.). Approximately 400,000 renal
cancer cells were cultured in six- well plates. The ACHN and 786-O
cells were transfected with the TRIM52-AS1 shRNA (200 pmol/well) or
TRIM52-AS1-pcDNA3.1+ (4 µg/well) vector using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Cell proliferation assay
Cell proliferation was determined using a
3-(4,5-dimethylthiazol-2yl)-2,5-diphenyl tetrazolium bromide (MTT;
Sigma-Aldrich) assay, performed according to the manufacturer's
protocol. Briefly, the cells (~5×103 cells) were seeded
into a 96-well culture plate 24 h prior to trans-fection with the
TRIM52-AS1 shRNA (200 pmol/well) or TRIM52-AS1-pcDNA3.1+ (4
µg/well)vector. At 0, 24, 48 or 72 h post-transfection, 20
µl MTT (5 mg/ml) was added to each well, and the plates were
incubated for 4 h at 37°C. Subsequently, the MTT medium mixtures
were discarded, and 150 µl dimethyl sulfoxide was added to
each well, and agitated for 10 min at room temperature to
solubilize the crystals. The results were measured at a wavelength
of 490 nm (with 630 nm as the reference wavelength) using an ELISA
microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Assays were repeated at least three times.
Cell migration assay
A wound scratch assay was used to assess the
migratory ability of the 786-O and ACHN RCC cells in vitro.
For the assay, ~5×106 cells were seeded per 6-well dish
and were transfected with TRIM52-AS1 shRNA (100 pmol) or
TRIM52-AS1-pcDNA3.1+ (4 µg) after 24 h using Lipofectamine
2000. At 6 h post-transfection, a vertical horizontal wound was
made in the cell layer using a sterile 10 µl pipette tip,
and markers were included to allow observation of cells at the same
point. The cells were then rinsed with phosphate-buffered saline
(PBS) and cultured in an incubator at 37°C. Images of the wounds
were captured with a digital camera system (C3040-AD6; Olympus
Corporation, Tokyo, Japan) 0 and 24 h following creation of the
wounds at the same points. The wound widths (lm) were measured
using a standard caliper (Caliper; PerkinElmer, Inc., Waltham, MA,
USA). The experiments were performed in triplicate and repeated at
least three times.
Cell apoptosis assay
The extent of apoptosis was evaluated using an
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI)
detection kit (Invitrogen; Thermo Fisher Scientific, Inc.).
Following transfection of the ACHN and 786-O cells with the
TRIM52-AS1 shRNA (200 pmol/well) or TRIM52-AS1-pcDNA3.1+ (4
µg/well) vector, the cells were collected, washed twice with
pre-chilled PBS and re-suspended in 1X binding buffer (pH 7.4; 10
mmol/l HEPES, 140 mmol/l NaCl, 5 mmol/l CaCl2;
Invitrogen; Thermo Fisher Scientific, Inc.), 48 h post-treatment.
The aliquots were mixed with 10 µl annexin V-FITC and 10
µl PI at room temperature for 15 min. The apoptosis assay
was performed using a flow cytometer (EPICS Xl-4, Beckman, CA,
USA). Each experiment was performed at least three times.
Statistical analysis
Statistical analysis was performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). Statistical significance
was determined using Student's t-test. For comparison of the
expression levels of TRIM52-AS1 in matched tumor, vs. normal
samples, a paired t-test was used. Data are expressed as the
mean ± standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
TRIM52-AS1 is downregulated in RCC
tissues and cell lines
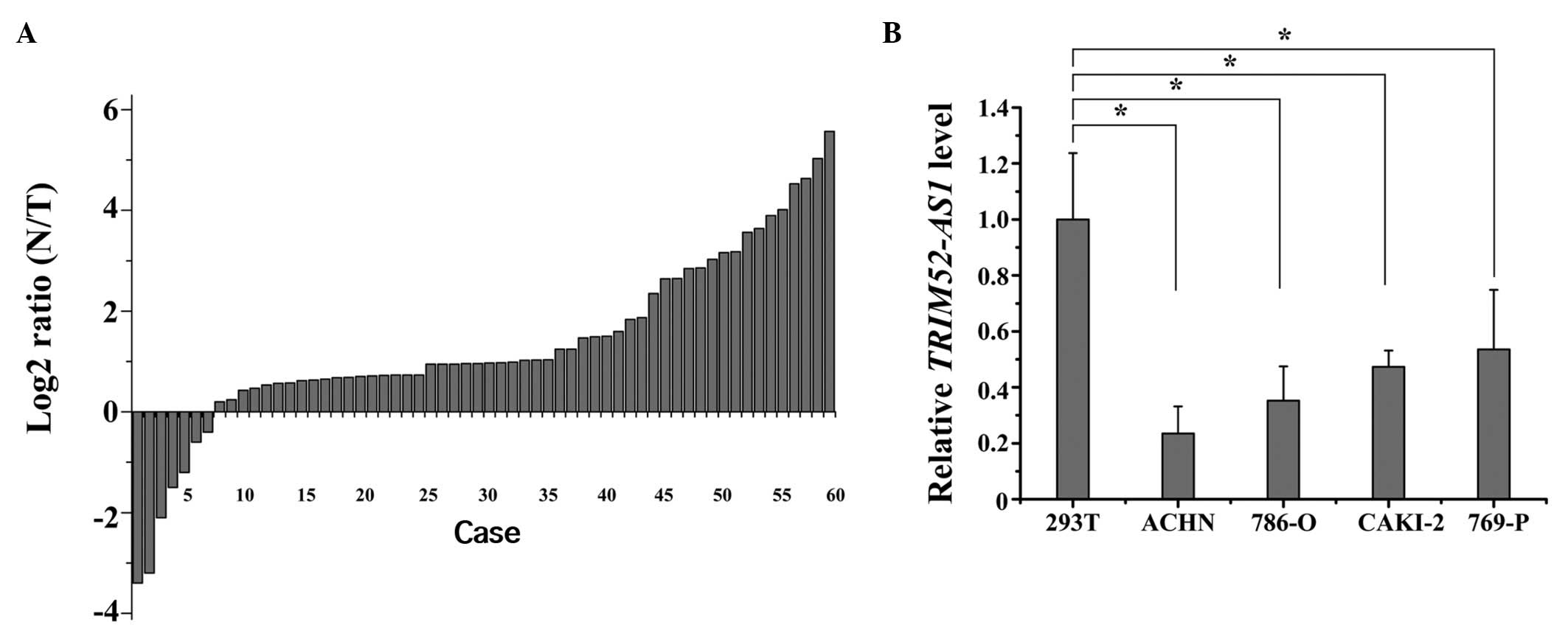
A previous study showed TRIM52-AS1 to be
downregulated in RCC tissues, as determined by lncRNAs expression
profiling (16,17). To confirm the result of sequencing,
RT-qPCR was used to quantify the expression levels of TRIM52-AS1 in
60 matched RCC tissue samples and adjacent normal tissue samples.
The relative expression of TRIM52-AS1 [log2 (N/T)] is shown in
Fig. 1A. The expression of
TRIM52-AS1 in the RCC tissues was significantly lower, compared
with that in the adjacent normal tissues. Furthermore, compared
with normal kidney 293T cells, TRIM52-AS1 was also downregulated in
the 786-O and ACHN cell lines (P<0.05; Fig. 1B). These results suggested that
TRIM52-AS1 may act as a tumor suppressor gene in RCC.
Downregulation of TRIM52-AS1 inhibits
cell proliferation in vitro
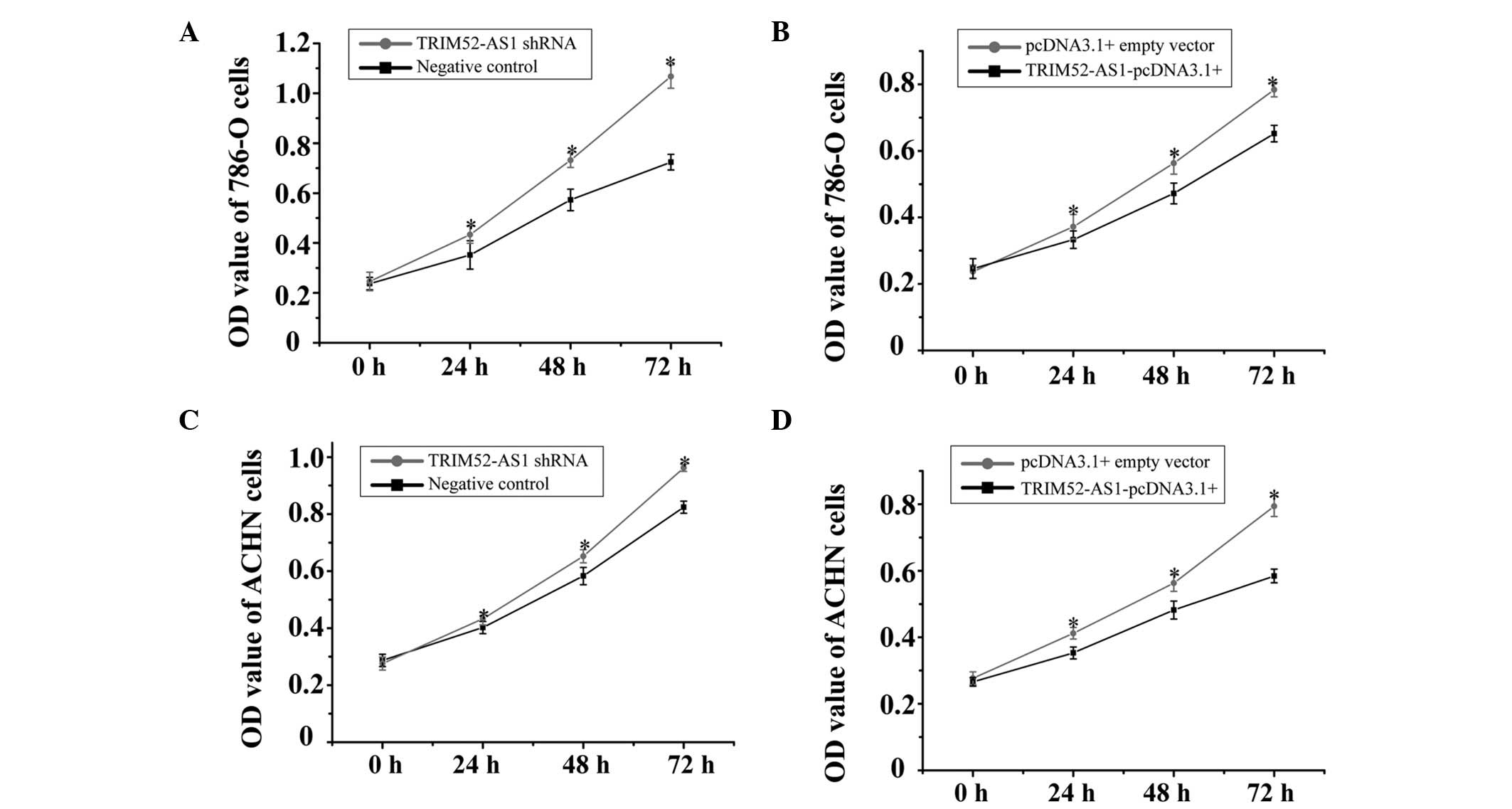
To analyze the function of TRIM52-AS1 in RCC using
an MTT assay, either TRIM52-AS1 shRNA or the TRIM52-AS1-pcDNA3.1+
vector were transfected into the 786-O and ACHN cell lines. The
optical density (OD) values of the TRIM52-AS1 shRNA or the
TRIM52-AS1-pcDNA3.1+ vector-treated groups were measured at a
wavelength of 490 nm (620 nm reference wavelength) using an ELISA
microplate reader at 0, 24, 48 and 72 h post-transfection. The
results demonstrated that the relative proliferation rates of the
TRIM52-AS1 shRNA-transfected 786-O cell line were significantly
increased by 42.8% (24 h), 31.2% (48 h) and 55.2% (72 h), whereas
proliferation rates in the TRIM52-AS1-pcDNA3.1+ vector-transfected
group were significantly decreased by 16.8% (24 h), 15.2% (48 h)
and 20.4% (72 h; Fig. 2A and B);
The relative cell proliferation rates in the TRIM52-AS1
shRNA-transfected ACHN cell line were significantly increased, by
14.8% (24 h), 17.3% (48 h) and 29.2% (72 h), whereas proliferation
rates in the TRIM52-AS1-pcDNA3.1+ vector-transfected group were
significantly decreased by 21.5% (24 h), 30.6% (48 h) and 54.7% (72
h; Fig. 2C and D). These results
indicated that the downregulation of TRIM52-AS1 had a negative
effect on cellular proliferation in RCC.
Downregulation of TRIM52-AS1 inhibits RCC
cell migration in vitro
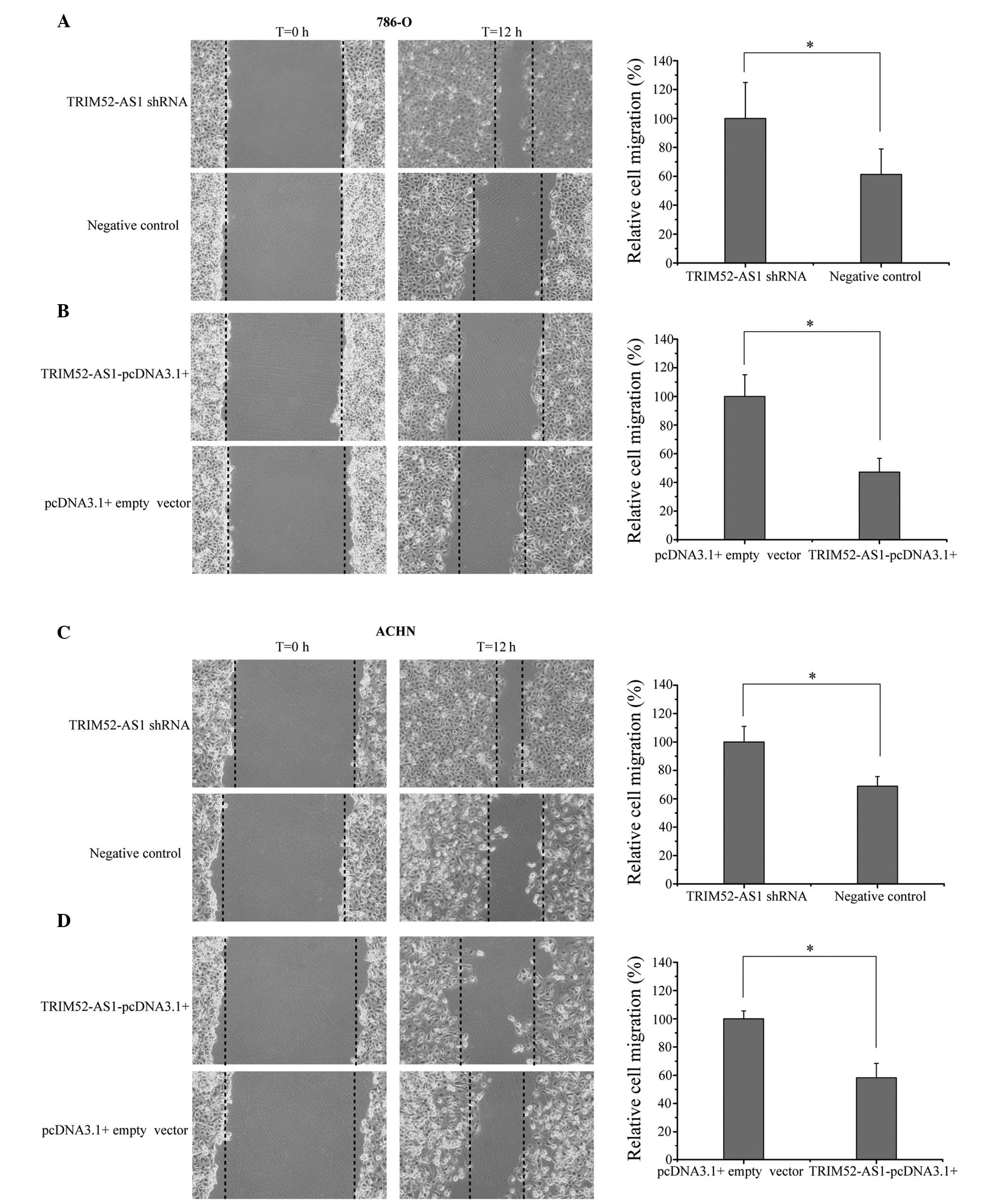
The effects of TRIM52-AS1 on cellular migration in
RCC cells were observed using a wound scratch assay. As shown in
Fig. 3, interference of the
expression of TRIM52-AS1 increased the rate of migration, whereas
the overexpression of TRIM52-AS1 decreased migration rate
(P<0.05). The results demonstrated that the wound widths in the
group of cells transfected with the TRIM52-AS1-pcDNA3.1+ vector
were markedly wider than those in the TRIM52-AS1 shRNA group, which
indicated that downregulation of TRIM52-AS1 inhibited the migration
of RCC cells.
Downregulation of TRIM52-AS1 promotes RCC
cell apoptosis in vitro
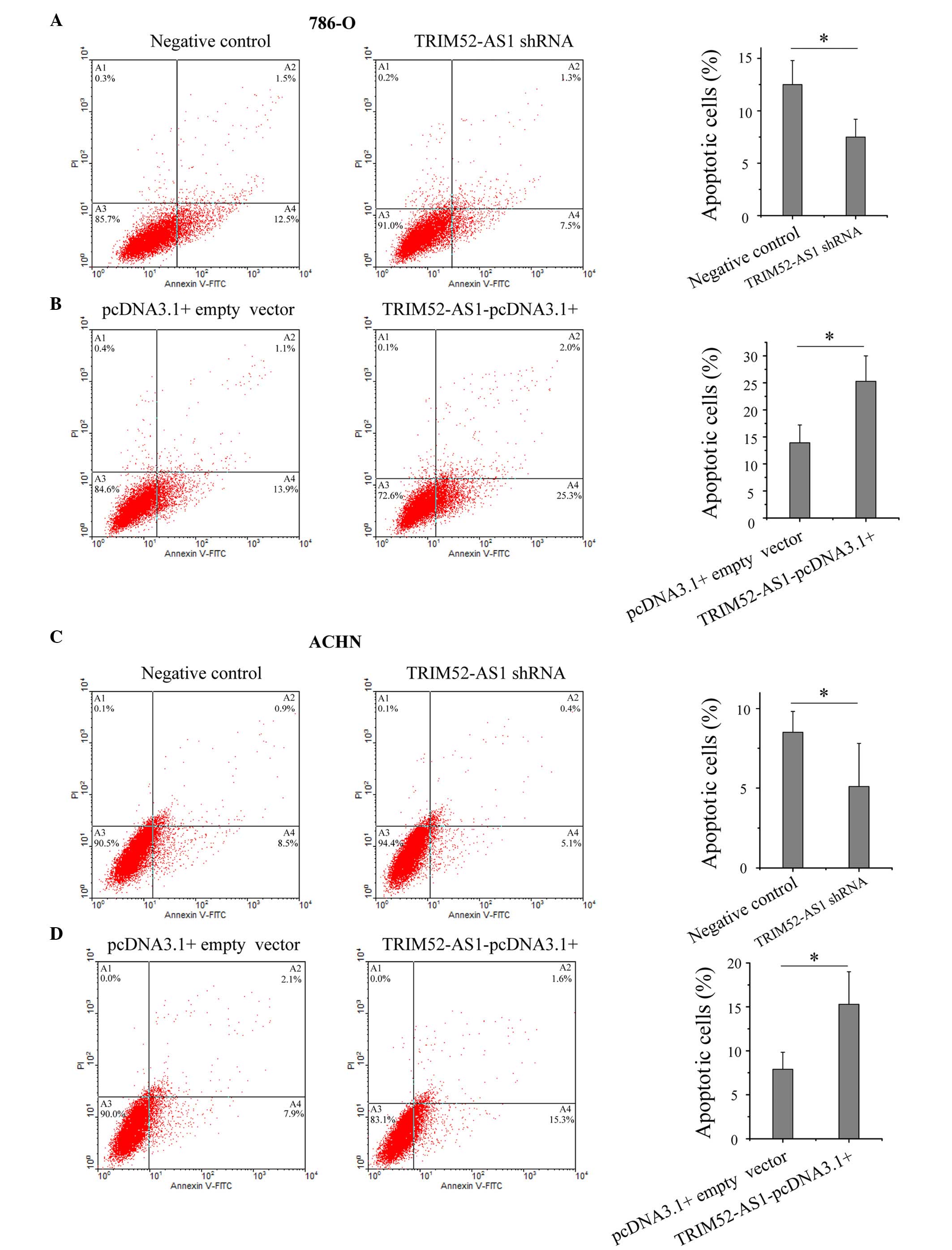
LncRNAs have been reported to be important in cell
apoptosis, particularly in the escape of cancer cells from
apoptosis (20–22). To determine the impact of
TRIM52-AS1 on RCC cell apoptosis, flow cytometry was performed to
detect the rate of apoptosis in the cells. As shown in Fig. 4, the apoptotic rates of the 786-O
cells transfected with the TRIM52-AS1 shRNA and
TRIM52-AS1-pcDNA3.1+ vector were 7.5 and 25.3%, respectively, and
the apoptotic rates of the ACHN cells were 5.1 and 15.3%,
respectively, which demonstrated that the downregulation of
TRIM52-AS1 promoted RCC cell apoptosis.
Discussion
RCC is the second leading contributor to mortality
rates among urological tumors. The reported incidence of RCC has
increased in the USA over the past two decades (23,24).
Despite substantial improvements in cancer therapy, major
limitations in the management RCC remain. RCC is characterized by
its resistance to current standard therapies (25), and the identification of
alternative treatment strategies remains a top priority.
LncRNAs are RNA transcripts of >200 nucleotides
with no protein encoding functions. Increasing studies have
indicated that the molecular mechanisms of carcinogenesis are not
only relevant to protein-coding genes, but are also relevant to
non-coding regulatory RNAs. Previous studies have shown that
numerous lncRNAs are deregulated in various types of solid tumor,
and that several lncRNAs can regulate cancer metastasis by directly
targeting chromatin modification complexes, indicating that the
abnormal expression of lncRNAs increases the chances of
tumorigenesis and cancer development (26–29).
For example, the overexpression of lncRNA HOTAIR is associated with
breast cancer (30), upregulated
MALAT1 contributes to bladder cancer (31), downexpression of lncRNA XIST is
associated with glioblastoma (32)
and upregulated PCAT-1 contributes to prostate cancer (33).
Previous studies have indicated that TRIM52-AS1 is
down-regulated in RCC tissues, as detected by lncRNA expression
profiling, As a common type of urological cancer, whether the
abnormal expression of TRIM52-AS1 is associated with RCC
carcinogenesis has not been reported previously. Therefore, the
present study is the first, to the best of our knowledge to certify
the expression of TRIM52-AS1 and examine its function in RCC. The
results were consistent with previous sequencing, demonstrating
that TRIM52-AS1 was significantly downregu-lated in RCC. In
addition, the overexpression of TRIM52-AS1 in the 786-O and ACHN
cell lines significantly inhibited cell proliferation and
migration, and induced cell apoptosis. By contrast, the knockdown
of its expression had the opposite effects. Together, these results
suggested that TRIM52-AS acted as a tumor suppressor gene in the
occurrence and development of RCC.
In conclusion, the present study revealed that
TRIM52-AS1 was downregulated in RCC and was significantly involved
in RCC by affecting cellular migration, proliferation and
apop-tosis. These results indicated TRIM52-AS1 as a promising
biomarker and/or a therapeutic target for RCC, Further
investigation is required to determine the molecular mechanisms
underlying the effect of TRIM52-AS1 in RCC.
Acknowledgments
This study was supported by the Overseas Issue
Foundation of Education Department In Heilongjiang Province (grant
no. 1254HQ014) and The First Affiliated Hospital Of Harbin Medical
University Foundation (grant no. 2014L01).
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patel C, Ahmed A and Ellsworth P: Renal
cell carcinoma: A reappraisal. Urol Nurs. 32:182–190; quiz 191.
2012.PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Milowsky MI and Nanus DM: Chemotherapeutic
strategies for renal cell carcinoma. Urol Clin North Am.
30:601–609. x2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rouviere O, Bouvier R, Négrier S, Badet L
and Lyonnet D: Nonmetastatic renal-cell carcinoma: is It really
possible to define rational guidelines for post-treatment
follow-up? Nat Clin Pract Oncol. 3:200–213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costa FF: Non-coding RNAs: Meet thy
masters. Bioessays. 32:599–608. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice
KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally
expressed gene 3, an imprinted noncoding RNA gene, is associated
with meningioma pathogenesis and progression. Cancer Res.
70:2350–2358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Deng K, Guo X, Wang H and Xia J: The
lncRNA-MYC regulatory network in cancer. Tumour Biol. 35:9497–9503.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taft RJ, Pang KC, Mercer TR, Dinger M and
Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar
|
|
11
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar
|
|
12
|
Zhang HM, Yang FQ, Yan Y, Che JP and Zheng
JH: High expression of long non-coding RNA SPRY4-IT1 predicts poor
prognosis of clear cell renal cell carcinoma. Int J Clin Exp
Pathol. 7:5801–5809. 2014.PubMed/NCBI
|
|
13
|
Zhang ZZ, Shen ZY, Shen YY, Zhao EH, Wang
M, Wang CJ, Cao H and Xu J: HOTAIR long noncoding RNA promotes
gastric cancer metastasis through suppression of Poly r(C) Binding
Protein (PCBP) 1. Mol Cancer Ther. 14:1162–1170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218–5p. Cell
Death Dis. 6:e15832015. View Article : Google Scholar
|
|
15
|
Li JY, Ma X and Zhang CB: Overexpression
of long non-coding RNA UCA1 predicts a poor prognosis in patients
with esophageal squamous cell carcinoma. Int J Clin Exp Pathol.
7:7938–7944. 2014.
|
|
16
|
Yu G, Yao W, Wang J, Ma X, Xiao W, Li H,
Xia D, Yang Y, Deng K, Xiao H, et al: LncRNAs expression signatures
of renal clear cell carcinoma revealed by microarray. PLoS One.
7:e423772012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fachel AA, Tahira AC, Vilella-Arias SA,
Maracaja-Coutinho V, Gimba ER, Vignal GM, Campos FS, Reis EM and
Verjovski-Almeida S: Expression analysis and in silico
characterization of intronic long noncoding RNAs in renal cell
carcinoma: Emerging functional associations. Mol Cancer.
12:1402013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martínez- Salamanca JI, Huang WC, Millán
I, Bertini R, Bianco FJ, Carballido JA, Ciancio G, Hernández C,
Herranz F and Haferkamp A: Prognostic impact of the 2009 UICC/AJCC
TNM staging system for renal cell carcinoma with venous extension.
Eur Urol. 59:120–127. 2011. View Article : Google Scholar
|
|
19
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia
R, Liu YW, Liu XH, Zhang EB, Lu KH and Shu YQ: Long noncoding RNA
ANRIL promotes non-small cell lung cancer cell proliferation and
inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer
Ther. 14:268–277. 2015. View Article : Google Scholar
|
|
21
|
Xu WH, Zhang JB, Dang Z, Li X, Zhou T, Liu
J, Wang DS, Song WJ and Dou KF: Long non-coding RNA URHC regulates
cell proliferation and apoptosis via ZAK through the ERK/MAPK
signaling pathway in hepatocellular carcinoma. Int J Biol Sci.
10:664–676. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han Y, Yang YN, Yuan HH, Zhang TT, Sui H,
Wei XL, Liu L, Huang P, Zhang WJ and Bai YX: UCA1, a long
non-coding RNA up-regulated in colorectal cancer influences cell
proliferation, apoptosis and cell cycle distribution. Pathology.
46:396–401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Taneja SS: Re: Cancers with increasing
incidence trends in the United States: 1999 through 2008. J Urol.
188:1120–1121. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Underwood JM, Richards TB, Henley SJ,
Momin B, Houston K, Rolle I, Holmes C and Stewart SL: Decreasing
trend in tobacco-related cancer incidence, United States 2005–2009.
J Community Health. 40:414–418. 2015. View Article : Google Scholar
|
|
25
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen LL and Zhao JC: Functional analysis
of long noncoding RNAs in development and disease. Adv Exp Med
Biol. 825:129–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang T, Alvarez A, Hu B and Cheng SY:
Noncoding RNAs in cancer and cancer stem cells. Chin J Cancer.
32:582–593. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Zhang P, Wang L, Piao HL and Ma
L: Long non-coding RNA HOTAIR in carcinogenesis and metastasis.
Acta Biochim Biophys Sin (Shanghai). 46:1–5. 2014. View Article : Google Scholar
|
|
31
|
Han Y, Liu Y, Zhang H, Wang T, Diao R,
Jiang Z, Gui Y and Cai Z: Hsa-miR-125b suppresses bladder cancer
development by down-regulating oncogene SIRT7 and oncogenic long
non-coding RNA MALAT1. FEBS Lett. 587:3875–3882. 2013. View Article : Google Scholar
|
|
32
|
Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J,
Chen L, Xi Z, Teng H, Wang Z, et al: Knockdown of long non-coding
RNA XIST exerts tumor-suppressive functions in human glioblastoma
stem cells by up-regulating miR-152. Cancer Lett. 359:75–86. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Prensner JR, Chen W, Han S, Iyer MK, Cao
Q, Kothari V, Evans JR, Knudsen KE, Paulsen MT, Ljungman M, et al:
The long non-coding RNA PCAT-1 promotes prostate cancer cell
proliferation through cMyc. Neoplasia. 16:900–908. 2014. View Article : Google Scholar : PubMed/NCBI
|