Introduction
Malaria is a tropical disease that results in
millions of fatalities every year. The World Health Organization
estimated that there were 207 million malaria cases and 627,000
estimated malaria-related fatalities in 2012 in their Malaria
Report 2013 (1). Although the
incidence and associated mortality of malaria are decreasing, the
absolute number of malaria cases remains significantly large due to
the absence of an effective vaccine and the increase in
drug-resistant parasite strains (2).
Erythrocyte invasion is the first step in the
intraerythrocytic developmental cycle of Plasmodium
falciparum. This parasite invades red blood cells (RBCs)
through micronemes and rhoptry protein secretion, tight junction
formation, and actomyosin assembly (3). A previous study established that
disrupting the interactions between parasite and erythrocyte
proteins prevents malaria parasite infection (4). Therefore, the identification of novel
parasite proteins that are involved in RBC invasion has become a
hotspot in malaria research. The inhibitions of several parasite
proteins by gene knockout or antibodies has been demonstrated to
provide protection against malaria infection. For instance, rhoptry
neck protein 2 (PfRON2) can be secreted into RBCs and form
complexes under the RBC membrane. However, several regions of
PfRON2 can be left exposed to the extracellular environment. The
microneme protein PfAMA1 bound to the exposed region of PfRON2 can
form tight junctions between malaria parasite and RBCs. PfAMA1
knockout inhibits tight junction formation, thereby impairing
erythrocyte invasion (5). In
addition, the erythrocyte binding antigen PfEBA-175 is secreted
onto the apical merozoite surface of malaria parasite. The binding
of PfEBA175 to its RBC receptor gly-A triggers the secretion of
rhoptry proteins. Gly-A cannot trigger rhoptry release when EBA-175
knockout mice are used (6). Other
parasite proteins, such as PfRh5 and PfMtrap, also participate in
the invasion process (7–9). The identification of novel proteins
involved in the parasite invasion of RBCs is underway. Parasite
proteins that are highly expressed at the schizont stage or early
ring stage, or those that contain domains involved in cell-cell
adhesion, are usually considered to be candidate invasion-related
proteins.
The malaria parasite protein PfTip (Gene ID,
PF3D7_0529000) is a homolog of the human ITFG1, which is a T-cell
immunomodulatory protein (10).
The protein contains four successive VCBS domains, which exist in
multiple copies in long proteins from Vibrio,
Colwellia, Bradyrhizobium, and Shewanella
(hence the name VCBS), and fewer copies in proteins from bacteria.
This domain is involved in cell-cell adhesion (11,12).
Proteins possessing this domain are usually involved in cell-cell
contact. Whether or not PfTip can mediate parasite-erythrocyte
interaction has yet to be determined. Determining the role of PfTip
in RBC invasion by the malaria parasite may aid in elucidating the
mechanism of invasion and provide a novel intervention strategy for
malaria control.
The present study aimed to characterize the PfTip
protein. The sequence of PfTip was analyzed, its 3D structure was
modeled, its expression feature was identified, and its interactant
was predicted. The protective effect of PfTip inhibition was also
evaluated through in vitro and in vivo
experiments.
Materials and methods
Data sources and analysis tools
The protein sequences of PfTip and its homologs were
obtained from PlasmoDB (http://plasmodb.org/plasmo/). Domain identification
was performed using the PFAM database (http://pfam.xfam.org/) (13). Protein-protein interactions were
obtained from the BioGrid database (http://thebiogrid.org/) (14). Signal peptide and transmembrane
domain were predicted using SignaIP and TMHMM servers (http://www.cbs.dtu.dk/services/), respectively.
The secondary structure of PfTip was predicted using PSIPRED
(http://bioinf.cs.ucl.ac.uk/psipred/)
(15).
Structure modeling of PfTip
Building the structure of PfTip involves template
identification, alignment of the template with the target, building
of the model, and evaluation of the obtained structure (16). The PDB database was searched
through HHsearch (HHsuite 3.0; Max Planck Gesellschaft, Berlin,
Germany). Three templates with the highest sequence similarities
were selected as templates to build PfTip models. Building the
PfTip model included four steps: Template selection, sequence
alignment, model building and evaluation of the obtained 3D
structure. Initially, the PDB database was searched using
HHsearch/HHpred to identify the templates. Three templates with the
highest sequence similarities to the PfTip extracellular region
were selected, including 2c4d.1.A, 4tqj.1.A and 4igl.1.A. Secondly,
models were built based on selected templates using Swiss-model
workspace. Finally, the local quality of built models was accessed
by QMEAN6. The model built using 2c4d.1.A had the highest score and
was considered to be the best structure. The alignment was then
manually adjusted to improve the model quality further. The global
qualities of the built models were verified using QMEAN6 (17). The model with the highest QMEAN6
Z-score was considered as the best 3D structure of PfTip. The local
quality and structure of the best model were accessed through ProQ2
and PROCHECK (18,19).
Fast fourier transform (FFT)
analysis
FFT analysis was performed to detect genes
specifically involved in the cell cycle (20). The procedure maps an expression
signal in the time domain toward the frequency domain, thereby
showing the amplitude of each frequency present in the expression
signal. Through this method, it is possible to filter the
expression signal that is inherently noisy or lacks differential
expression and identify the major frequency of a particular gene in
the life cycle of the malaria parasite. The formula of a simple FFT
is expressed as follows:
Where N is the length of expression signal and
k represents the frequency. The expression signal of PfTip
was obtained from the published microarray data on the
intraerythrocytic developmental cycle of P. falciparum
(21). The major frequency of the
PfTip expression signal can be determined through FFT analysis
(22). The peak expression time
point can be estimated using the following formula:
Where P represents the phase of the major
frequency. FFT analysis was implemented in MATLAB 2012b (MathWorks,
Natick, MA, USA).
Expression and purification of the
PfTIP-GST protein
The genomic DNA was isolated using the Genomic DNA
Prep kit (Tiangen Biotech Co., Ltd., Beijing, China) and was set as
the template for amplification. The DNA sequence encoding the
ectodomain (48 to 674 aa) of PfTip was amplified through polymerase
chain reaction (PCR) using the forward primer 5′-GCGGATCCAGGATA
AAATCATTTTATGTTGAAGCA-3′ and the reverse primer 5′-GCCTCGAGTTA
TTTGGATGGATTAACTGAGAGTTG-3′. The components of the reaction system
were: PrimerSTAR Max Premix (25 µl), forward primer/reverse
primer (1 µl), template (1 µl), water (<50
µl). The thermal cycling conditions were as follows: 95°C
for 5 min, followed by 35 cycles of 94°C for 30 sec, 52°C for 30
sec, 72°C for 2 min, and a final extension for 5 min at 72°C.
Following being digested with BamHI and XhoI (Takara
Bio, Inc., Shiga, Japan), the fragment was inserted into pGEX-4T-1
(GE Healthcare, Fairfield, CT, USA) to express a GST fusion
protein. Firstly, the vector pGEX-4T-1 and PfTip DNA fragment were
digested by BamHI and XhoI for 2 h at 37°C, followed
by agarose gel electrophoresis. The target bands were cut and DNA
fragments were harvested using a gel extraction kit (Omega Bio-Tek
Inc., Norcross, GA, USA) and then the vector pGEX-4T-1 and PfTip
DNA fragment were ligated overnight at 16°C using T4 ligase (New
England Biolabs, Ipswich, MA, USA). The recombinant plasmids were
transformed into XL-10 competent cells and were incubated on agar
LB containing ampicillin for 12 h. The transformed clones were
picked and transferred into 10 ml of LB. Following being incubated
overnight at 37°C, bacteria cells were harvested and plasmids were
extracted. The correct open reading frame was confirmed by DNA
sequencing. Following checking the correct sequence, the plasmids
were transformed into Escherichia coli BL21 cells (Takara
Bio., Inc.). First, the recombinant plasmids and competent cells
were placed in tubes, mixed and then stored on ice for 30 min. The
tubes were then transferred into a preheated 42°C water bath for 90
sec. The tubes were immediately transferred to an ice bath and the
cells were allowed to chill for at least 2 min. Subsequently, 800
µl of LB was added to the tube and the cells were agitated
gently for 50 min in a rotary shaker. Finally, the transformed
competent cells were transferred onto agar LB containing ampicillin
and were incubated at 37°C for 12 h. To express the recombinant
protein PfTip-GST, IPTG (final concentration of 0.1 mM; Merck
Millipore, Darmstadt, Germany) was added to the medium. Following
further cultivation at 20°C for 10 h, the bacterial samples were
harvested. The parasite pellet was kept on ice and was sonicated
for three cycles with 20 sec pulses using an ultrasonicator (Ningbo
Scientz Biotechnology Co., Ltd., Ningbo, China) with a 2 mm tip (60
watt model, 30% of maximum power). Sodium dodecyl
sulfate-polyacrylamide gel electrophoresis was employed to
determine PfTip expression. To purify the PfTIP-GST protein, the
target band (~97 kD) was cut, destained, ground with liquid
nitrogen, and then dissolved in phosphate-buffered saline (PBS)
containing 8 M urea (Absin Bioscience, Shanghai, China). Following
centrifugation at 12,000 × g for 10 min, the supernatant was
collected and the protein concentration was determined through a
bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Generation of anti-PfTip sera
Two New Zealand white rabbits of the same breed were
purchased from the Animal Care Facility at the Fourth Military
Medical University (Xi'an, China). The present study was evaluated
and approved by The Fourth Military Medical University Animal Care
Committee. The rabbits were raised in standardized pathogen-free
conditions at 20±2°C and 60±5% humidity, under a 12/12 h dark/light
cycle. They were kept individually in steel cages with free access
to food and tap water. To harvest the antisera, the rabbits were
anesthetized by intramuscular injection of ketamine/xylazine (40
mg/kg; Sigma-Aldrich) and then blood was obtained through the
carotid artery. To generate anti-PfTip sera, the rabbits were
immunized by subcutaneously injecting 100 µg of purified
PfTip-GST protein mixed with complete Freund's adjuvant
(Sigma-Aldrich, St. Louis, MO, USA). Animals injected with the
adjuvant alone served as controls. Immunization was performed three
times every 2 weeks. Sera were extracted 2 weeks after each
immunization. The PfTip antibody titer of the sera was determined
through enzyme-linked immunosorbent assay (ELISA). When the
antibody titer was ideal, the rabbits were sacrificed and sera were
extracted.
ELISA
ELISA was performed to determine the PfTip antibody
titer of the sera. Each well was coated with 2 µg purified
PfTIP-GST protein at 37°C for 1 h and then at 4°C overnight. The
following day, the wells were washed three times with PBS with
Tween-20 (PBST) and then blocked at 37°C for 1 h with 5% non-fat
milk. Sera samples diluted with blocking buffer (1:50; Beyotime
Institute of Biotechnology, Shanghai, China) were added to each
well and then incubated at 37°C for 1 h. After washing with PBST,
100 µl diluted horseradish peroxidase (HRP)-conjugated
polyclonal sheep anti-rabbit IgG (cat. no. A0208; Beyotime
Institute of Biotechnology; 1:5,000) or polyclonal anti-mouse IgG
(cat. no. A0216; Beyotime Institute of Biotechnology; 1:5,000) was
added into each well and then incubated at 37°C for 1 h. Excess
IgG-HRP was removed by inverting the plate. TMB reagent (Beyotime
Institute of Biotechnology) was added to each well to produce
color. The absorbance of each well at 450 nm was measured and
analyzed using a BIOBASE-EL10A ELISA microplate reader (Biobase
Biodustry, Co., Ltd., Shandong, China).
Western blot analysis
Infected RBCs were lysed with 0.01% saponin (Sangon
Biotech Co., Ltd., Shanghai, China) and then centrifuged at 12,000
× g for 10 min. The parasite pellet was suspended in PBS, lysed
through four freeze/thaw cycles, and then sonicated for 3X 20 sec
on ice. The cell lysate was centrifuged at 12,000 × g for 20 min.
The supernatant was collected, and the protein concentration was
determined through a BCA assay. The cell lysate (50 µg) was
loaded per lane for separation through electrophoresis. The
proteins were transferred to polyvinylidene difluoride (PVDF)
membranes (EMD Millipore, Billerica, MA. USA), which were then
blocked with 5% non-fat milk in Tris-buffered saline for 1 h at
room temperature. The anti-PfTip sera were added to the membranes
and then incubated at 4°C overnight. After washing three times with
PBST, the membranes were hybridized with polyclonal rabbit
anti-human (1:1000; cat. no. T528; Signalway Antibody LLC, College
Park, MD, USA) GST-Tag secondary antibody conjugated with IRDye 800
(LI-COR Biosciences, Lincoln, NE, USA) and then incubated for 1 h.
Blots were imaged using an infrared imaging system
(Odyssey®; LI-COR Biosciences).
Reverse transcription-quantitative
PCR. Total RNA was extracted from the malaria
parasite using TRIzol (Takara Bio, Inc.) and then reverse
transcribed according to the manufacturer's instructions (Takara
Bio Inc., Shiga, Japan). The obtained cDNA was used as the template
for quantitative PCR. A PrimeScript™ RT reagent kit (Takara Bio,
Inc.) was used for cDNA synthesis. The reaction system consisted of
the following components: 5X PrimeScript Buffer (2 µl), 1X
PrimeScript RT Enzyme mix I (0.5 µl), 50 µM Oligo dT
Primer (0.5 µl), 100 µM random 6 mers (0.5
µl), total RNA (0.5 µg) and RNase Free
dH2O (up to 10 µl). The primers (Sangon Biotech
Co., Ltd.) used for the amplification of PfTip and the internal
control gene (PF07_0073, Seryl-tRNA synthetase) were as follows:
PfTip, 5′-GGGGTAATGCTCATGGACCT-3′ (forward) and 5′-GGGAAAT
GTGCTGACTGGCT-3′ (reverse); and PF07_0073, 5′-CAAGTAGCA
GGTCATCGTGGT-3′ (forward) and 5′-CAAGTTCGGCACATTCTTCCA-3′
(reverse). The PCR reaction system consisted of 12.5 µl of
SYBR Green I Master mix (Takara Bio, Inc.), 500 nM each primer, and
2 µl of the template in a total volume of 25 µl. An
Agilent Mx3000P QPCR System was used (Agilent Technologies, Inc.,
Santa Clara, CA, USA) for PCR. The thermal cycling conditions were
as follows: 95°C for 5 min, followed by 40 cycles of 94°C for 10
sec, and 60°C for 15 sec. The relative mRNA level of PfTip was
quantified using theΔΔCq method (23).
In vitro invasion assay
The invasion assay was conducted according a
previous method (24). P.
falciparum was cultured in RPMI-1640 medium supplemented with
10% human serum and erythrocytes (2% hematocrit) at 37°C and then
flushed with a gas mixture containing 5% O2, 5%
CO2 and 90% N2. The medium was changed every
48 h. For the invasion assay, P. falciparum was synchronized
as previously described (25).
When the parasite reached the trophozoite or schizont stage, MACS
(Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) magnetic bead
separation column was used for separation of parasitized RBCs. Then
the parasitemia was adjusted to 5% and was cultured in 96-well
plates with 180 µl of medium. Each well contained 20
µl of PfTip-immunized sera or control sera. After 20 h of
cultivation, Giemsa (Beijing Dingguo Biotechnology Co., Ltd.,
Beijing, China) staining smears were utilized to determine the ring
stage parasitemia. An Olympus BX51 microscope (Olympus Corporation
Tokyo, Japan) was used to observe Giemsa staining. The percent
inhibition was calculated using the following formula:
Where Ppf and
Pcon represent the ring stage parasitemia after
the addition of PfTip-GST sera and control sera, respectively.
In vivo protection test
A total of 14 female BALB/c mice aged 6–8 weeks were
purchased from the Animal Care Facility at the Fourth Military
Medical University and were divided into two groups, the treated
group (n=8) and the control group (n=6). The animal experiments
were approved by the Fourth Military Medical University Animal Care
Committee. The mice were raised in pathogen-free conditions at
20±4°C and 60±5% humidity, under a 12/12 h dark/light cycle. The
survival time of these mice were monitored for 25 days after
intraperitoneal injection with Plasmodium berghei. All mice
died within this period of time. The mice were subcutaneously
injected with 10 µg PfTIP-GST protein mixed with the
adjuvant three times every 2 weeks, and mice injected with the
adjuvant alone served as controls. Sera were extracted 2 weeks
after each injection. ELISA was performed to determine the antibody
titer of the obtained sera. Mice with high levels of antibody titer
were selected for further protection tests. The mice were
administered with 1.0×106 Plasmodium
berghei-infected RBCs through intraperitoneal injection. The
survival time in days of the experimental mice was recorded. The
animal experiments were permitted by the Fourth Military Medical
University Animal Care Committee, and all animal studies were
performed in compliance with the university institutional
guidelines.
Statistical analysis
Statistical analysis was performed using statistic
toolbox of MATLAB 2012b. Data are presented as the mean ± standard
deviation. One-way analysis of variance was used to compare two or
more groups. The survival ratio was compared using Log-Rank test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Sequential analysis and structural
modeling of PfTip protein
PfTip, consisting of 719 aa, was encoded by a single
exon in the P. falciparum genome. The protein sequence scans
of PfTip by TMHMM and SignaIP indicated a signal peptide in its
N-terminus (1 to 28 aa) and a transmembrane domain in its
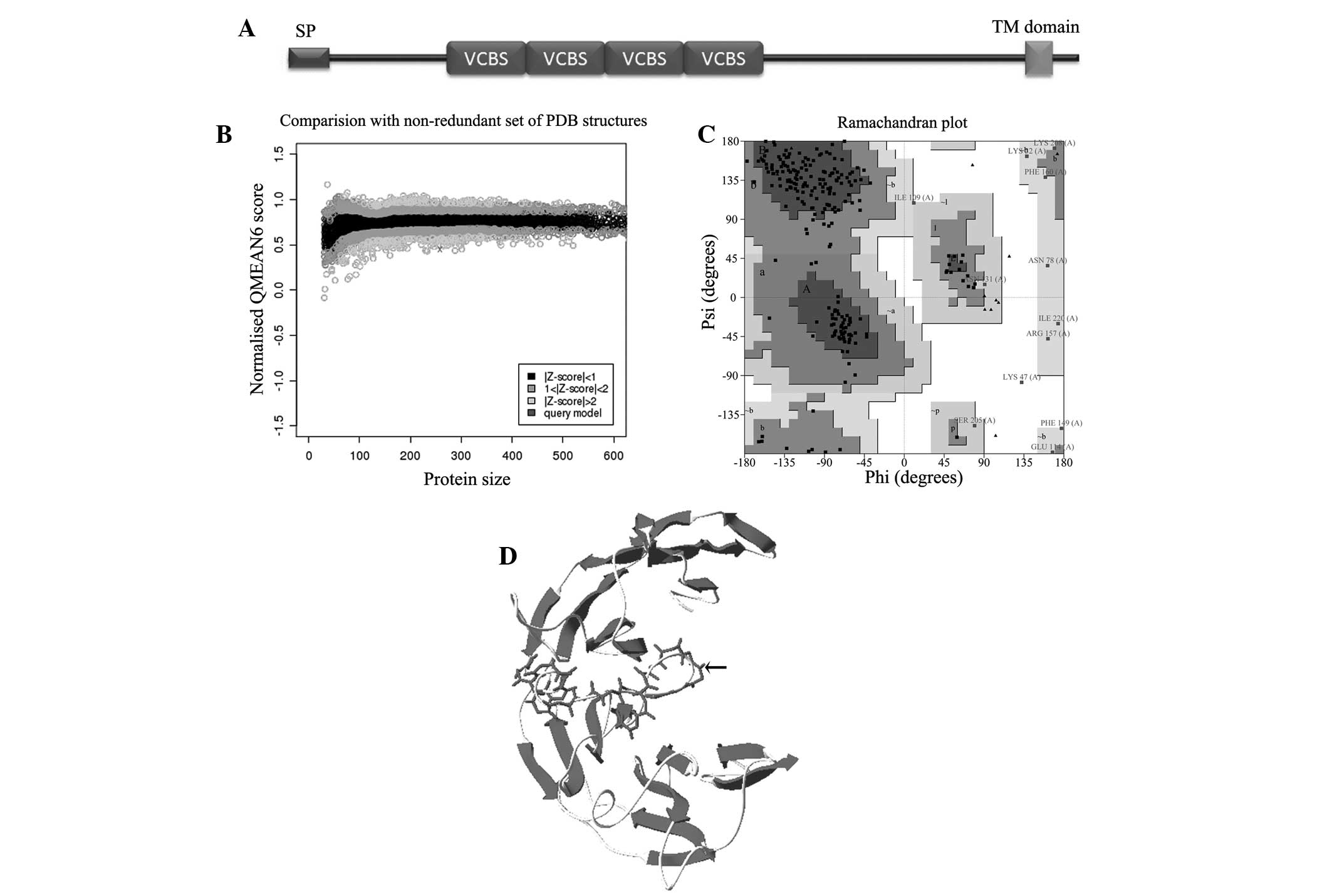
C-terminus (678 to 697 aa) (Fig.
1A). Thus, PfTip was considered as a membrane protein and was
expected to be expressed on the surface of the malaria parasite.
However, this protein may not function as a receptor signal
transducer from the outer to the inner parts of the cell as only 22
amino acids were predicted to be located in the cytoplasm.
Searching the Pfam database for the ectodomain of this protein
sequence, four successive VCBS domains (95 to 351 aa) were
identified in this protein (Fig.
1A). The results of homolog identification through sequence
alignment indicated that proteins containing the VCBS domain can be
found in viruses, bacteria, protista, fungi, and animals but not in
certain plants, including Oryza sativa and Arabidopsis
thaliana. The results of secondary structure prediction by
PSIPRED suggested the presence of 40 β-strands in the PfTip
ectodomain. However, no α-helix was predicted in this region,
implying that the structure of PfTip ectodomain is a member of the
β-fold superfamily.
To investigate the potential 3D structure of this
protein, the PfTip extracellular region (95 to 356 aa) consisting
of four successive VCBS domains was modeled using the Swiss-Model
Workspace. The model built using the template Psathyrella
velutina Lectin (PDB: 2c4d) had the highest QMEAN6 score (0.45,
normalized Z-score) and was selected as the 3D structure of PfTip
(Fig. 1B, upper panel). The
Ramachandran plot showed that 99.6% residues were placed in allowed
regions, except for one residue (LYS47, Table I, and Fig. 1C). The built 3D structure (Fig. 1D) shows the presence of β-sheets,
coils, loops, and turns but not α-helixes. This finding is
concurrent with the secondary structure prediction results.
Furthermore, the β-strands in PfTip formed a β-propeller fold,
which is also found in neuraminidase from the influenza virus
(26). Proteins consisting of this
fold usually participate in cell-cell contact and in pathogenic
organism invasion of host cells (27), suggesting that PfTip may be an
invasion-related protein. PfTip ectodomain has a low sequence
similarity to the template (only ~17.43% identity); thus,
accurately modeling the native structure of PfTip is difficult. The
assessment of the local quality of the obtained model revealed that
the loop (99–122 aa) connecting two β-meanders was not well built
(Fig. 1D, indicated by small
arrow). However, our model provided a clue for unraveling the
function of PfTip.
 | Table IRamachandran plot statistics of the
built 3D model of PfTip. |
Table I
Ramachandran plot statistics of the
built 3D model of PfTip.
| Ramachandran plot
statistics | No. of
residues | (%) |
|---|
| Residues in most
favoured regions [A,B,L] | 178 | 75.4 |
| Residues in
additional allowed regions | 46 | 19.5 |
| Residues in
generously allowed regions | 11 | 4.7 |
| Residues in
disallowed regions | 1 | 0.4 |
| Number of
non-glycine and non-proline residues | 236 | 100 |
| Number of
end-residues (excl. Gly and Pro) | 2 | |
| Number of glycine
residues (shown as triangles) | 14 | |
| Number of proline
residues | 7 | |
| Total number of
residues | 259 | |
Expression profile and interactant
prediction of PfTip
Investigating the expression profile of PfTip may
aid in elucidating the function of this protein. Proteins
associated with invasion should be highly expressed on the surface
of merozoites or in the schizont stage. Although gene expression is
highly dynamic in the intraerythrocytic developmental cycle of
P. falciparum, a striking nonstochastic periodicity can be
observed for the majority of expression profiles. Considering the
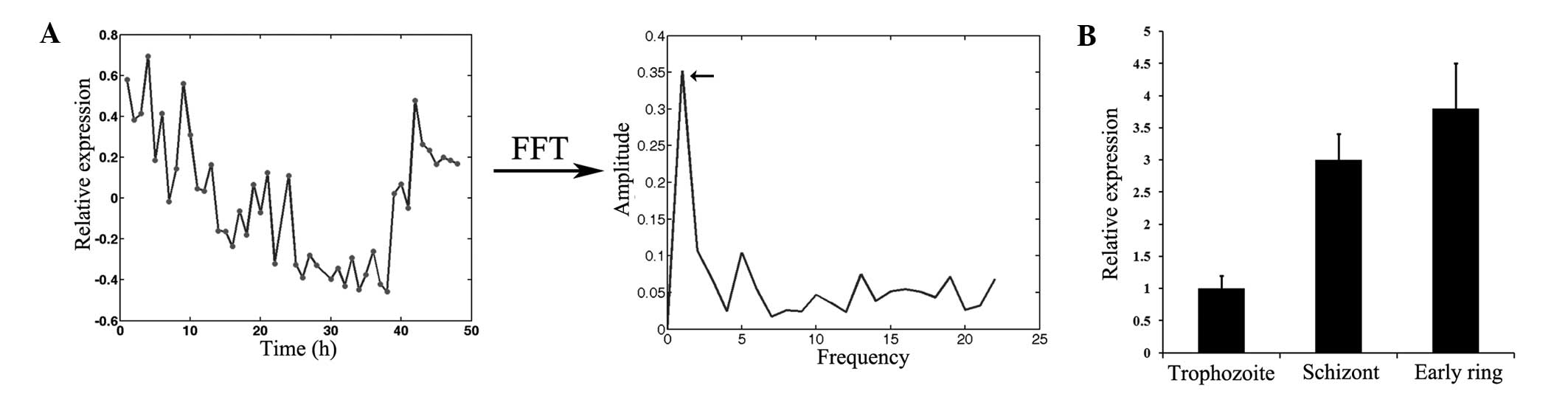
cyclical nature of gene expression, FFT analysis was performed to
evaluate PfTip expression and its peak expression time point. After
the parasite invasion of RBCs, PfTip expression decreased and had
the lowest value in the trophozoite stage (Fig. 2A, left panel). However, PfTip
expression began to rebound when the parasite reached the schizont
stage. Compared with several parasite genes with more than one
frequency in the asexual intraerythrocytic developmental cycle of
P. falciparum, PfTip has only one frequency (Fig. 2A, right panel). Its peak expression
time point was estimated to be 3.1 h (Tpeak
calculation in methods), suggesting that PfTip was highly expressed
at the early ring stage. Further experimental validation of PfTip
expression by RT-qPCR indicated that this protein was highly
expressed at the schizont and early ring stages. The expression
level of PfTip at the early ring stage was almost three times
higher than that at the trophozoite stage. Thus, based on its
expression pattern, PfTip is hypothesized to be an invasion-related
protein.
The prediction of proteins interacting with PfTip
may aid in elucidating the mechanism by which PfTip mediates RBC
invasion by the malaria parasite. Considering that the protein
domain is evolutionarily conserved and that protein-protein
interactions are predominantly achieved through domain-domain
interactions, a homolog method was employed to predict the
interactant of PfTip. In this method, the homologs of a protein are
considered to have the propensity to bind with the interactants of
the protein. Basing on this concept, the BioGrid database was
searched and 10 proteins that reportedly physically interact with
ITFG1, the homolog of PfTip in human were identified. However, no
protein was found to interact with the PfTip homologs from other
species, including Caenorhabditis elegans, Drosophila
melanogaster, and Mus musculus. The interaction partner
of PfTip, as a membrane protein, should be expressed on the surface
of erythrocytes. Combining the prediction results of SignaIP and
TMHMM, nine proteins were removed that were not expressed on the
surface of RBCs, leaving TNFRSF14 as the candidate partner of PfTip
(Table II). TNFRSF14 has
previously been demonstrated to be a receptor of BTLA (28). A previous study showed that
TNFRSF14 functions as a co-stimulator of T cells (29). Whether or not TNFRSF14 physically
interacts with PfTip requires further experimental validation.
 | Table IIPrediction of PfTip interactant. |
Table II
Prediction of PfTip interactant.
|
Interactantsa | Signal
peptideb | TM domainc |
|---|
| CDC73 | N | Y |
| FAM118B | N | N |
| FBXO6 | N | N |
| HMOX2 | N | Y |
| MRPL44 | N | N |
| NUDT3 | N | N |
| SERBP1 | N | N |
| TAF1D | N | N |
| TNFRSF14 | Y | Y |
| UBC | N | N |
Experimental validation of PfTip function
in invasion
Considering that PfTip contains the VCBS domain in
its structure and is highly expressed in the early ring and
schizont stages, it was hypothesized that PfTip is an
invasion-related protein. Thus, PfTip blockage may inhibit malaria
parasite invasion and confer protection. To experimentally validate
the function of PfTip in erythrocyte invasion, a plasmid expressing
the fusion protein PfTip-GST (Fig.
3A, left panel) was constructed. Then, rabbits were immunized
three times using the purified PfTip-GST protein to generate an
antibody recognizing the PfTip protein. The results of western blot
analysis indicated that the obtained rabbit serum can specifically
recognize PfTip (Fig. 3A, right
panel).
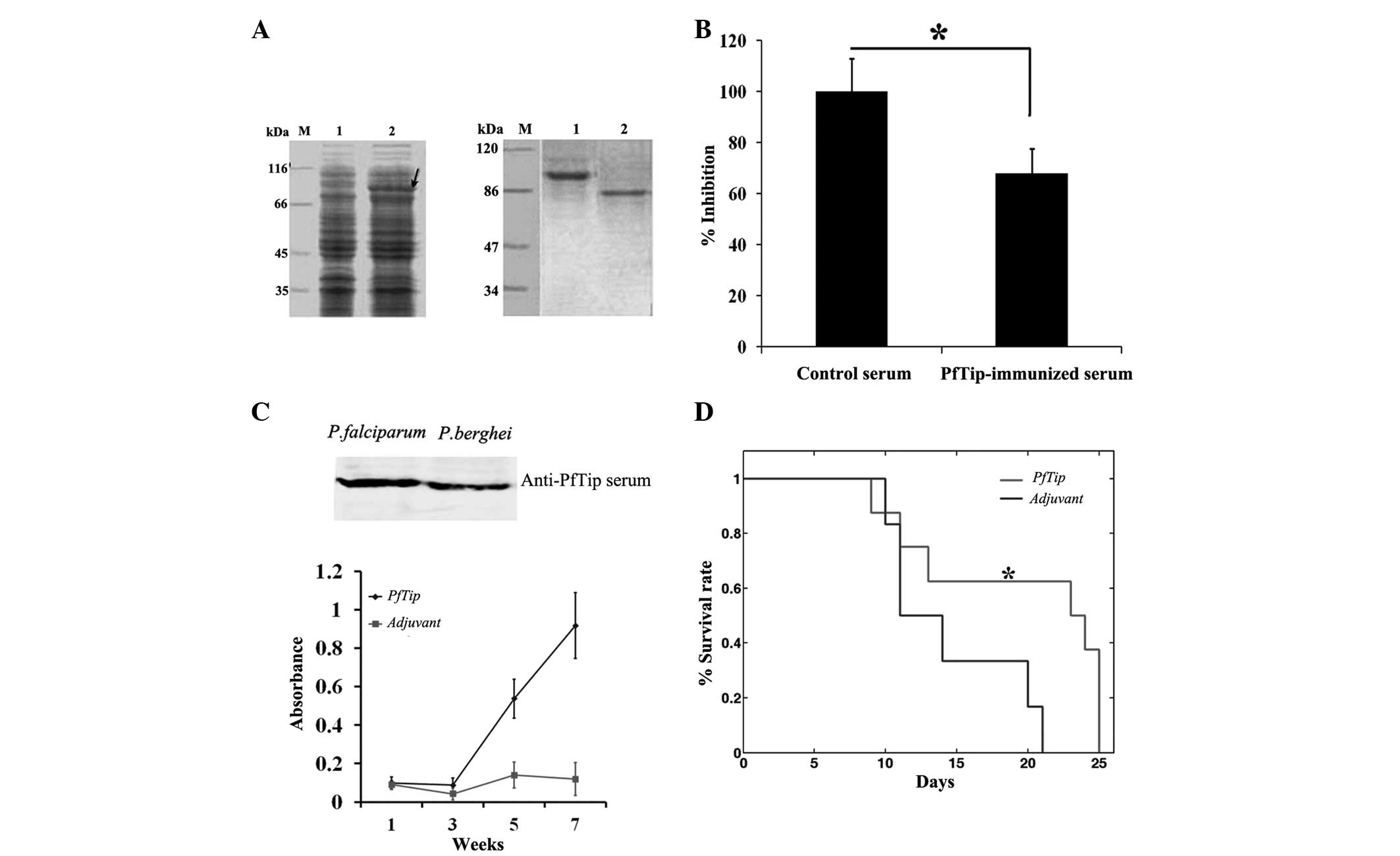 | Figure 3Blockage of PfTip protein inhibits
parasite invasion of host cells and provides protection activity.
(A) Preparation of PfTip-immunized serum. Left panel, sodium
dodecyl sulfate-polyacrylamide gel electrophoresis of PfTip
expression in E. coli BL21 cells. Arrow indicates PfTip
expression. M, marker; Lane 1, without IPTG; Lane 2, 0.1 mM IPTG.
Right panel, sera specificity examination using western blot
analysis. Lane 1, lysate of E. coli BL21 cells
overexpressing PfTip; lane 2, P. falciparum lysate. (B)
Suppression of parasite invasion ability using PfTip-immunized
serum. One-way analysis of variance was used,
*P<0.05. (C) Determination of antibody specificity
and titer using western blot analysis and ELISA. Upper panel,
PfTip-immunized serum recognize PfTip and PbTip antigen; Lower
panel, determination of antibody titer using ELISA. (D) Comparison
of survival rates between two groups of experimental mice (PbTip
group, n=8; adjuvant group, n=6). Log-rank test was used,
P<0.05. ELISA, enzyme-linked immunosorbent assay. |
The interaction between PfTip and its receptor was
blocked using the obtained immune serum to evaluate whether or not
PfTip blockage inhibits parasite invasion. The ring stage
parasitemia was examined and used for percent inhibition
computation. As shown in Fig. 3B,
the ring stage parasitemia observed in the PfTip-immunized group
was significantly lower than that in the control group (P<0.05).
The parasitemia decreased by almost 35%. This finding demonstrated
that PfTip blockage by the immune serum inhibited RBC invasion by
the malaria parasite. However, the underlying molecular mechanism
requires further investigation as the surface protein of RBCs that
interacts with PfTip has not yet been identified.
To test the protection activity of PfTip, an in
vivo experiment was performed. Given that the protective effect
of PfTip cannot be validated in humans, a mouse model was used
instead. Mice were immunized with the PfTip-GST protein three times
to produce a relatively high level of antibodies in mouse sera
(Fig. 3C, upper panel). The
PfTip-immunized serum can also recognize PbTip due to the high
sequence similarity (~85%) between PbTip and PfTip (Fig. 3C, lower panel). This result
suggests that the antibody induced by PfTip can effectively block
the interactions between PbTip and its partner. To confirm the
protective effect of PbTip blockage, the mice intraperitoneally
injected with P. berghei were monitored for up to 25 days.
Despite the lack of difference in survival rate between the two
groups, the survival curve was delayed when PfTip was injected
(Fig. 3D, lower panel). The mean
survival time of these two groups were 12.5 and 23.5 days,
respectively. Statistical analysis indicates that the difference
was significant (P=0.0496). This result demonstrates that PfTip
injection into mice confer a slight resistance to the lethal
malaria parasite infection.
Discussion
Erythrocyte invasion by malaria parasite is a
complex biological process. Various parasite proteins, including
PfAMA1 and PfRh5, participate in this invasion process (30,31).
The interactions between parasite and erythrocyte proteins
facilitate the adhesion of the parasite to RBCs, subsequently
initiating the invasion process. The identification of proteins
associated with this process may aid in elucidating the molecular
mechanism related to the parasite invasion of RBCs and potentially
provide a novel strategy for malaria control. In the present study,
PfTip was characterized and demonstrated that PfTip consists of
four successive VCBS domains and PfTip expression at the schizont
and early ring stage was relatively higher compared with that
observed in the trophozoite stage. In addition, PfTip blockage by
sera inhibits parasite invasion and provides protection. However,
further studies regarding the protection mechanism employed by
PfTip are required.
Regarding the structure of PfTip, the model of PfTip
ectodomain was generated through homology modeling. Although the
local quality of several regions of this model was not ideal due to
the low sequence similarity with the template, the score of the
global quality revealed that the developed model was relatively
reliable. The structure of the ectodomain composed of four
successive VCBS domains shows a part of the β-propeller fold. This
fold is a type of a β-strand architecture characterized by several
β-sheets toroidally arranged around a central axis (12). The assembly of multiple β-sheets
repeatedly forms the structure domain responsible for
protein-protein interaction. This fold can be found in several
enzymes and membrane proteins (26,32).
For instance, the influenza virus protein neuraminidase possesses a
β-propeller fold. This protein is present in the virus envelope and
catalyzes the cleavage of sialic acid moieties from cell membrane
proteins to aid in the invasion of host cells. Thus, the built
model suggested that PfTip may be an invasion-related protein. The
study on the expression pattern of PfTip also supported this
hypothesis as PfTip is highly expressed at the early ring and
schizont stages.
If PfTip is an invasion-related protein, then its
blockage should theoretically inhibit RBC invasion by the malaria
parasite. This speculation was supported by the results of in
vivo and in vitro experiments. In the in vitro
experiment, PfTip blockage using sera significantly suppressed the
parasite invasion of RBCs and subsequently reduced ring
parasitemia. In the in vivo experiment, the survival curve
of the PfTip-immunized mice was delayed, indicating the protective
effect of PfTip injection. The delay in the survival curve may be
attributed to two reasons. First, the antibody induced through
PfTip injection impaired the invasion ability of malaria parasite
and thus reduced the parasite burden required to initiate the
pathways of severe malaria. Second, PfTip probably enhanced host
immunity against parasite infection. This phenomenon can be
attributed to the fact that ITFG1, a homolog of PfTip in humans,
can stimulate the T cell secretion of γ-interferon (10). The interactions between the T-cell
receptor and the VCBS domain were evolutionarily conserved. Thus,
the injection of PfTip in mice may also lead to T cell activation,
subsequently enhancing the host immunity against parasite
infection. Although the co-stimulator TNFRSF14 was predicted to be
the receptor of PfTip protein, we did not experimentally validate
this interaction as the expression of this protein in mammalian
cells was not viable even under code optimization. The validation
of this predicted interaction will be a focus of future
studies.
In conclusion, the present study revealed that PfTip
was a novel invasion-related protein. As inhibition of PfTip
protein inhibited malaria parasite invasion and conferred
protection, this protein may be considered a candidate vaccine.
Identification of the PfTip-binding receptor on the surface of
erythrocytes is required in order to understand how PfTip protein
aids parasites invading RBCs.
Acknowledgments
This study was supported by the Nature Science
Foundation of China (grant no. 81572013) and the China Postdoctoral
Science Foundation (grant no. 2015M582796). The authors would like
to thank Professor Ya Zhao for critically revising the
manuscript.
References
|
1
|
World health organization: World malaria
report 2013. http://www.who.int/malaria/publications/world_malaria_report_2013/en/.
2013
|
|
2
|
Ashley EA, Dhorda M, Fairhurst RM,
Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, et
al: Spread of artemisinin resistance in Plasmodium falciparum
malaria. N Engl J Med. 371:411–423. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharma P and Chitnis CE: Key molecular
events during host cell invasion by Apicomplexan pathogens. Curr
Opin Microbiol. 16:432–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Patarroyo ME, Bermúdez A and Patarroyo MA:
Structural and immunological principles leading to chemically
synthesized, multiantigenic, multistage, minimal subunit-based
vaccine development. Chem Rev. 111:3459–3507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Giovannini D, Späth S, Lacroix C, Perazzi
A, Bargieri D, Lagal V, Lebugle C, Combe A, Thiberge S, Baldacci P,
et al: Independent roles of apical membrane antigen 1 and rhoptry
neck proteins during host cell invasion by apicomplexa. Cell Host
Microbe. 10:591–602. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singh S, Alam MM, Pal-Bhowmick I,
Brzostowski JA and Chitnis CE: Distinct external signals trigger
sequential release of apical organelles during erythrocyte invasion
by malaria parasites. PLoS Pathog. 6:e10007462010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baum J, Chen L, Healer J, Lopaticki S,
Boyle M, Triglia T, Ehlgen F, Ralph SA, Beeson JG and Cowman AF:
Reticulocyte-binding protein homologue 5-an essential adhesin
involved in invasion of human erythrocytes by plasmodium
falciparum. Int J Parasitol. 39:371–380. 2009. View Article : Google Scholar
|
|
8
|
Crosnier C, Bustamante LY, Bartholdson SJ,
Bei AK, Theron M, Uchikawa M, Mboup S, Ndir O, Kwiatkowski DP,
Duraisingh MT, et al: Basigin is a receptor essential for
erythrocyte invasion by plasmodium falciparum. Nature. 480:534–537.
2011.PubMed/NCBI
|
|
9
|
Bartholdson SJ, Bustamante LY, Crosnier C,
Johnson S, Lea S, Rayner JC and Wright GJ: Semaphorin-7A is an
erythrocyte receptor for P. falciparum merozoite-specific TRAP
homolog, MTRAP. PLoS Pathog. 8:e10030312012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fiscella M, Perry JW, Teng B, Bloom M,
Zhang C, Leung K, Pukac L, Florence K, Concepcion A, Liu B, et al:
TIP, a T-cell factor identified using high-throughput screening
increases survival in a graft-versus-host disease model. Nat
Biotechnol. 21:302–307. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cioci G, Mitchell EP, Chazalet V, Debray
H, Oscarson S, Lahmann M, Gautier C, Breton C, Perez S and Imberty
A: Beta-propeller crystal structure of psathyrella velutina lectin:
An integrin-like fungal protein interacting with monosaccharides
and calcium. J Mol Biol. 357:1575–1591. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kopec KO and Lupas AN: β-Propeller blades
as ancestral peptides in protein evolution. PloS One. 8:e770742013.
View Article : Google Scholar
|
|
13
|
Finn RD, Bateman A, Clements J, Coggill P,
Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J,
et al: Pfam: The protein families database. Nucleic Acids Res.
42(Database Issue): D222–D230. 2014. View Article : Google Scholar :
|
|
14
|
Chatr-Aryamontri A, Breitkreutz BJ,
Heinicke S, et al: The BioGRID interaction database: 2013 update.
Nucleic Acids Res. 41(Database Issue): D816–D823. 2013. View Article : Google Scholar :
|
|
15
|
Buchan DW, Minneci F, Nugent TC, Bryson K
and Jones DT: Scalable web services for the PSIPRED protein
analysis workbench. Nucleic Acids Res. 41(Web Server Issue):
W349–W357. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bordoli L, Kiefer F, Arnold K, Benkert P,
Battey J and Schwede T: Protein structure homology modeling using
SWISS-MODEL workspace. Nat Protoc. 4:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Benkert P, Biasini M and Schwede T: Toward
the estimation of the absolute quality of individual protein
structure models. Bioinformatics. 27:343–350. 2011. View Article : Google Scholar :
|
|
18
|
Laskowski RA, Moss DS and Thornton JM:
Main-chain bond lengths and bond angles in protein structures. J
Mol Biol. 231:1049–1067. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ray A, Lindahl E and Wallner B: Improved
model quality assessment using ProQ2. BMC Bioinformatics.
13:2242012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim BR, Zhang L, Berg A, Fan J and Wu R: A
computational approach to the functional clustering of periodic
gene-expression profiles. Genetics. 180:821–834. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bozdech Z, Llinás M, Pulliam BL, Wong ED,
Zhu J and DeRisi JL: The transcriptome of the intraerythrocytic
developmental cycle of plasmodium falciparum. PLoS Biol. 1:E52003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu X, Huang Y, Liang J, Zhang S, Li Y,
Wang J, Shen Y, Xu Z and Zhao Y: Computational prediction of
protein interactions related to the invasion of erythrocytes by
malarial parasites. BMC Bioinformatics. 15:3932014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Recuenco FC, Kobayashi K, Ishiwa A,
Enomoto-Rogers Y, Fundador NG, Sugi T, Takemae H, Iwanaga T,
Murakoshi F, Gong H, et al: Gellan sulfate inhibits plasmodium
falciparum growth and invasion of red blood cells in vitro. Sci
Rep. 4:47232014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iwanaga T, Sugi T, Kobayashi K, Takemae H,
Gong H, Ishiwa A, Murakoshi F, Recuenco FC, Horimoto T, Akashi H
and Kato K: Characterization of plasmodium falciparum cdc2-related
kinase and the effects of a CDK inhibitor on the parasites in
erythrocytic schizogony. Parasitol Int. 62:423–430. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smith BJ, Colman PM, Von Itzstein M,
Danylec B and Varghese JN: Analysis of inhibitor binding in
influenza virus neuraminidase. Protein Sci. 10:689–696. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chao KL, Tsai IW, Chen C and Herzberg O:
Crystal structure of the Sema-PSI extracellular domain of human RON
receptor tyrosine kinase. PloS One. 7:e419122012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shui JW, Steinberg MW and Kronenberg M:
Regulation of inflammation, autoimmunity and infection immunity by
HVEM-BTLA signaling. J Leukoc Biol. 89:517–523. 2011. View Article : Google Scholar :
|
|
29
|
Chen L and Flies DB: Molecular mechanisms
of T cell co-stimulation and co-inhibition. Nat Rev immunol.
13:227–242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yap A, Azevedo MF, Gilson PR, Weiss GE,
O'Neill MT, Wilson DW, Crabb BS and Cowman AF: Conditional
expression of apical membrane antigen 1 in plasmodium falciparum
shows it is required for erythrocyte invasion by merozoites. Cell
Microbiol. 16:642–656. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shen Y, Wang J, Liu X, Liang J, Huang Y,
Liu Z, Zhao YA and Li Y: Blockade of plasmodium falciparum
erythrocyte invasion: New assessment of anti-Plasmodium falciparum
reticulocyte-binding protein homolog 5 antibodies. Exp Ther Med.
9:1357–1362. 2015.PubMed/NCBI
|
|
32
|
Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X,
Zhang J, Yang P, Deng H, Wang J, et al: Structural basis of
ultraviolet-B perception by UVR8. Nature. 484:214–219. 2012.
View Article : Google Scholar : PubMed/NCBI
|