Introduction
Renal cell carcinoma (RCC) is the third most common
urological cancer type after prostatic and bladder cancer,
accounting for ~3% of all malignant tumors in adults and almost 90%
of all renal tumors (1,2). The incidence of RCC shows a great
variation among different counties and the male-to-female ratio is
>2:1 (3). Cancer statistics
estimated for 2013 in the USA showed that due to its incidence of
>65,150 new cases and >13,680 mortalities, RCC is among the
10 leading cancer types (4).
Annual estimates of newly diagnosed cases of RCC have been
gradually increasing over recent years and the most prevalent
histological sub-type of RCC is clear-cell RCC with a prevalence of
85% (5). As RCC patients tend to
show no symptoms at the early stage, distant metastasis is present
in >30% of cases at the time of diagnosis (6,7), for
which only few and ineffective treatment options are available
(8). Therefore, it is urgently
required to identify novel biomarkers to facilitate the diagnosis
of RCC, as well as novel treatment strategies.
MicroRNAs (miRNAs/miRs) are a class of single
non-coding RNAs of ~22 nucleotides in length (9). Previous studies have shown that
miRNAs regulate various cellular processes, including
differentiation, migration, proliferation, apoptosis and metabolism
(10,11). miRNAs are epigenetic regulators
which bind to the 3′-untranslated regions of their target mRNAs and
degrade them or repress their translation (9,12).
In various cancer types, certain miRNAs are aberrantly expressed
and act as oncogenes or tumor suppressors (13–15).
Due to the imperfect complementarity between miRNAs and their
target mRNAs, each mRNA may be regulated by various miRNAs and each
miRNA may target various mRNAs (16,17).
However, the specific roles of certain miRNAs in cancer have
remained elusive. miRNAs have the potential to be used as
diagnostic and prognostic biomarkers, for clinical monitoring
purposes and as treatment targets for cancer.
Among these miRNAs, miR-196a has been reported to be
aberrantly expressed in various tumor types, including
osteosarcomas (18), pancreatic
cancer (19) and gastric cancer
(20), while its function has
remained elusive in RCC. However, certain miRNA profiling studies
have indicated that miR-196a is downregulated in RCC (21,22).
The purpose of the present study was therefore to assess the
expression of miR-196a in RCC and normal tissues and to explore the
effects of miR-196a on RCC cell proliferation, migration and
apoptosis.
Materials and methods
Sample collection
A total of 48 paired RCC tissues and adjacent normal
kidney tissues were collected from hospitals in Guangdong and Anhui
province (China). Adjacent normal tissues were extracted at a 2-cm
distance from visible RCC lesions. Written informed consent was
obtained from all patients from Peking University Shenzhen Hospital
(Shenzhen, China) between January 2012 and December 2014. Protocols
for the collection and use of the samples were reviewed and
approved by the ethics committee of Peking University Shenzhen
Hospital (Shenzhen, China). The tissues were dissected while being
immersed in RNAlater (Qiagen, Hilden, Germany) over 30 min and 1 g
of tissue was stored at −80°C for further use. The tissues
collected were reviewed and classified following hematoxylin and
eosin staining by three independent examiners using a previously
described method (23). The
clinical and pathological characteristics of the tissue donors are
presented in Table I.
 | Table IClinicopathological features of renal
cell carcinoma patients (mean age, 52 years; age range, 27–72
years). |
Table I
Clinicopathological features of renal
cell carcinoma patients (mean age, 52 years; age range, 27–72
years).
| Characteristic | Number of
patients |
|---|
| Males | 30 |
| Females | 18 |
| Histological
type |
| Clear cell | 39 |
| Papillary | 9 |
| pT-stage |
| T1 | 27 |
| T2 | 19 |
| T3 + T4 | 2 |
| Fuhrmann grade |
| I | 15 |
| II | 22 |
| III | 8 |
| IV | 3 |
| AJCC clinical
stage |
| I | 27 |
| II | 18 |
| III + IV | 3 |
RNA extraction and reverse-transcription
quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from RCC tissues and normal
adjacent tissues using TRIzol Reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and purified with the RNeasy
Maxi kit (Qiagen) following the manufacturer's instructions. The
RNA concentration was measured using a NanoDrop 2000/2000c (Thermo
Fisher Scientific, Inc.) and the RNA samples with optical density
(OD) ratios at 260/280 nm of 1.8–2.0 were used for further
experiments. For cDNA synthesis, 1 µg total RNA of each
sample subjected to reverse transcription with the miScript Reverse
Transcription kit (Qiagen) following the manufacturer's
instructions. The temperature protocol for the RT reaction was 37°C
for 60 min, 95°C for 5 min and storage at 4°C. Amplification of
cDNA for the quantification of miR-196a was performed using the
miScript SYBR®Green PCR kit (Qiagen) on a Roche
Lightcycler 480 Real-Time PCR System according to the
manufacturer's instructions. U6 was used as an internal control.
PCR thermocycling conditions were set as follows: 95°C for 1 min,
then 40 cycles of 95°C for 15 sec, 55°C for 30 sec and 72°C for 30
sec. The following primers (Invitrogen; Thermo Fisher Scientific,
Inc.) were used: miR-196a forward, 5′-TAGGTAGTTTCATGTTGTTGGG-3′ and
reverse as provided by the miScript SYBR® Green PCR kit;
U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-ACGCTTCACGAATTTGCGT-3′. The expression of miR-196a was analyzed
using the 2−ΔΔCq method (24).
Cell culture and transfection
The 786-O and ACHN human renal carcinoma cell lines
(American Type Culture Collection, Manassas, VA, USA) and 293T
human embryo kidney cell line (Type Culture Collection of the
Chinese Academy of Medical Sciences, Beijing, China) were cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum, 1% antibiotics (100
µl/ml penicillin and 100 mg/ml streptomycin sulfates) and 1%
glutamine (all from Gibco; Thermo Fisher Scientific, Inc.) at 37°C
in a humidified incubator containing 5% CO2. Cells were
transfected with miR-196a mimics, synthesized by Shanghai
GenePharma Co., Ltd. (Shanghai, China)
(5′-UAGGUAGUUUCAUGUUGUUGGGCAACAACAUGAAACUACCUAUU-3′) or negative
control (NC) miRNA (5′-CAGUACUUUUGUGUAGUACAA-3′) using
Lipofectamine 2000 (Invitrogen) in Opti-MEM® I Reduced
Serum Medium (Gibco; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. To determine the transfection
efficiency and miR-196a expression, fluorescence microscopy and
RT-qPCR were performed.
Wound healing assay for cell
migration
The migratory capacity of 786-O and ACHN cells was
assessed in vitro by performing a wound healing assay. At 24
h after seeding 3×105 cells into each well of a 12-well
plate, the cells were transfected with 100 pmol of miR-196a mimics
or negative control using Lipofectamine®2000. Following
6 h of transfection, the cell monolayer was scratched with a
sterile 200-µl pipette tip to generate a line-shaped wound.
Floating cells were removed by rinsing with phosphate-buffered
saline (PBS) and the cells were further cultured. A digital camera
system (Olympus Corporation, Tokyo, Japan) on a Leica DMIRB
inverted fluorescence microscope (Leica Microsystems GmbH, Wetzlar,
Germany) was used to acquire the images of the scratches at 0, 12
and 24 h. The experiments were performed in triplicate and repeated
at least three times.
Cell proliferation assay
The proliferation of 786-O and ACHN cells was
assessed in vitro by performing a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. Cells were seeded into a 96-well plate at 5,000 cells/well
and transfected with 5 pmol miR-196a mimics or negative control. At
0, 24, 48 and 72 h post-transfection, 20 µl MTT (5 mg/ml;
Sigma-Aldrich, St Louis, MO, USA) was added to the wells. followed
by incubation for 4 h. The medium was replaced with 150 µl
dimethylsulfoxide (Sigma-Aldrich), followed by agitation of the
plates for 30 min at room temperature. An ELISA microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) was then used to
measure the OD of each well at a wavelength of 490 nm.
Flow cytometric assay for apoptosis
The apoptotic rates of 786-O and ACHN cells were
measured in vitro by flow cytometry. In each well of a
six-well plate, 3×105 786-O or ACHN cells were seeded
and subsequently transfected with 200 pmol miR-196a mimics or
negative control. At 48 h post-transfection, all cells were
harvested, washed with cold PBS twice and re-suspended in 100
µl 1X binding buffer. To each cell suspension, 5 µl
Annexin V-fluorescein isothiocyanate (Invitrogen) and 5 µl
propidium iodide (Invitrogen) were added, followed by incubation at
room temperature in the dark for 15 min. Following addition of 400
µl binding buffer to each tube, flow cytometry (EPICS Xl-4,
Beckman Coulter, Brea, CA, USA) was used to assess the apoptotic
rate.
Statistical analysis
Paired Student's t-test was used to compare the
expression levels of miR-196a in matched tumor/normal tissues.
Student's t-test was used to analyze assays for characterizing
phenotypes of cells. The χ2 test was used to explore the
correlations between the pathological characteristics and the
expression levels of miR-196a in tumor tissues. All data are
expressed as the mean ± standard error. All statistical analyses
were preformed using the SPSS 19.0 statistical software package
(IBM SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-196a is downregulated in RCC tissues
compared with normal adjacent tissues
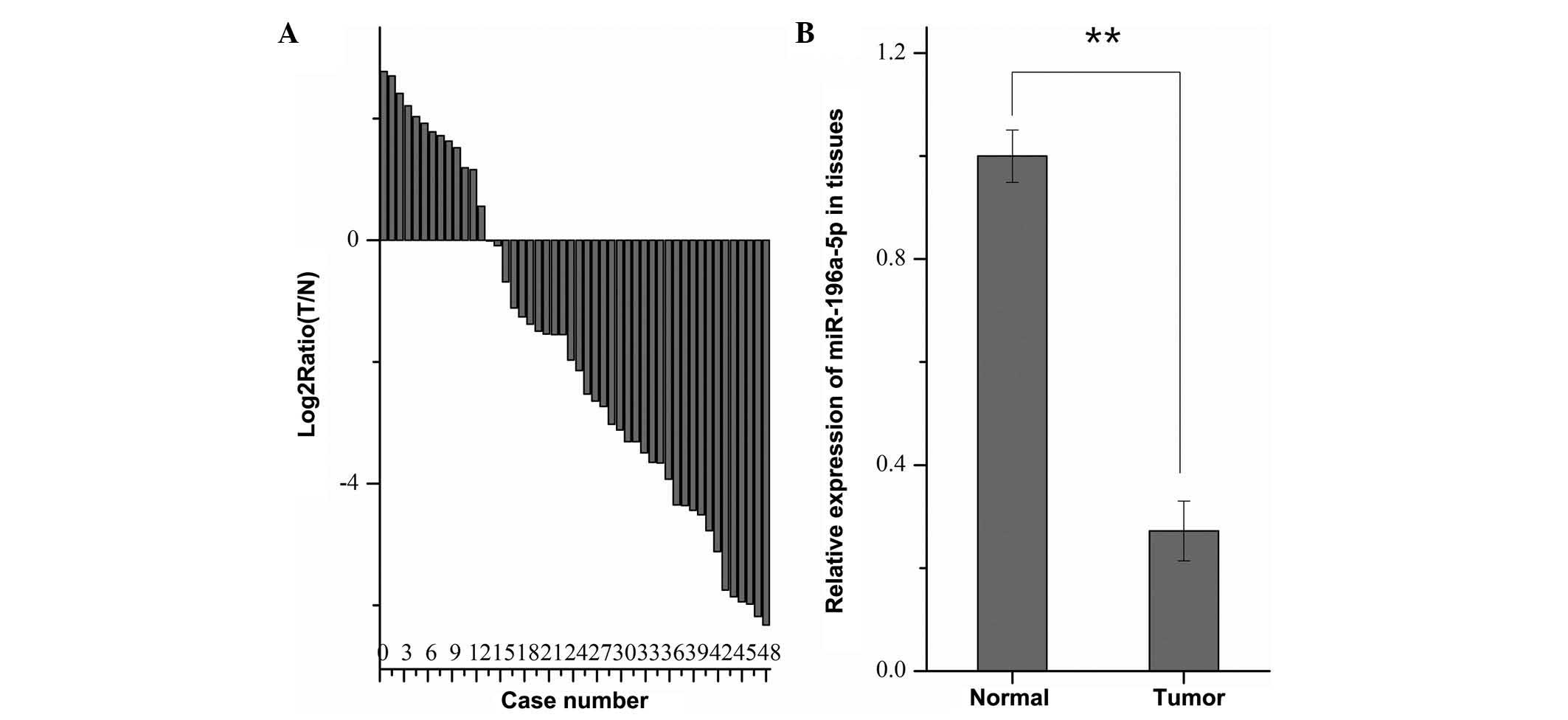
To explore the expression of miR-196a in 48 paired
RCC and normal adjacent tissues, RT-qPCR was performed. As shown in
Fig. 1A, miR-196a was
downregulated in the RCC tissues of 35 out of 48 patients.
Furthermore, the mean expression levels of miR-196a in RCC tissues
were significantly lower than those in the paired normal tissues
(P<0.01) (Fig. 1B). However,
χ2 analysis revealed that no correlation was present
between the pathological characteristics of the patients and the
expression levels of miR-196a in tumor tissues (results not
shown).
Validation of cell transfection
efficiency
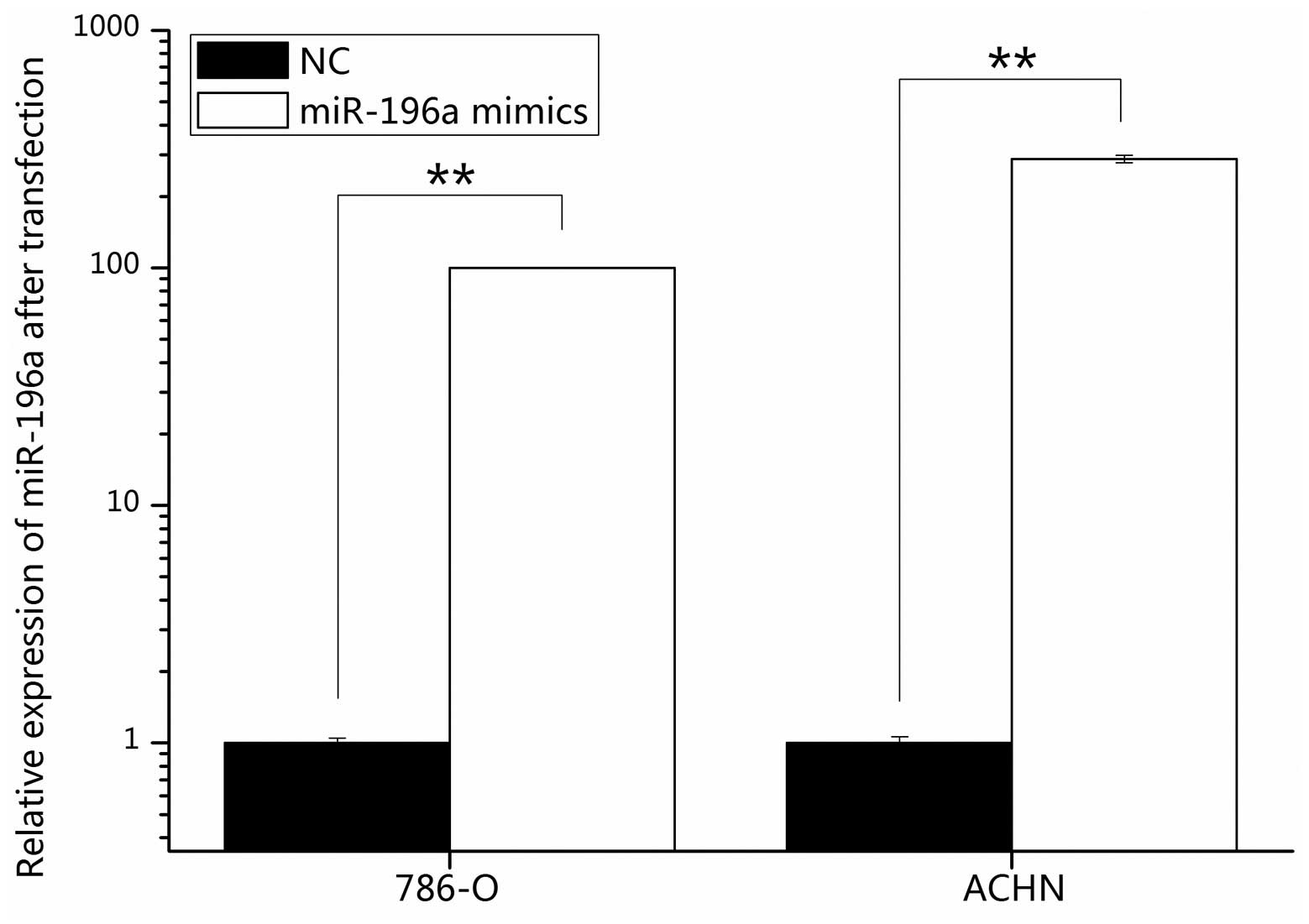
The transfection efficiency of miR-196a mimics was
determined by RT-qPCR, revealing that following transfection with
miR-196a mimics, miR-196a levels were increased by 100.08% in 786-O
and 287.02% in ACHN cells compared with those in NC-transfected
cells (Fig. 2).
miR-196a inhibits RCC cell migration
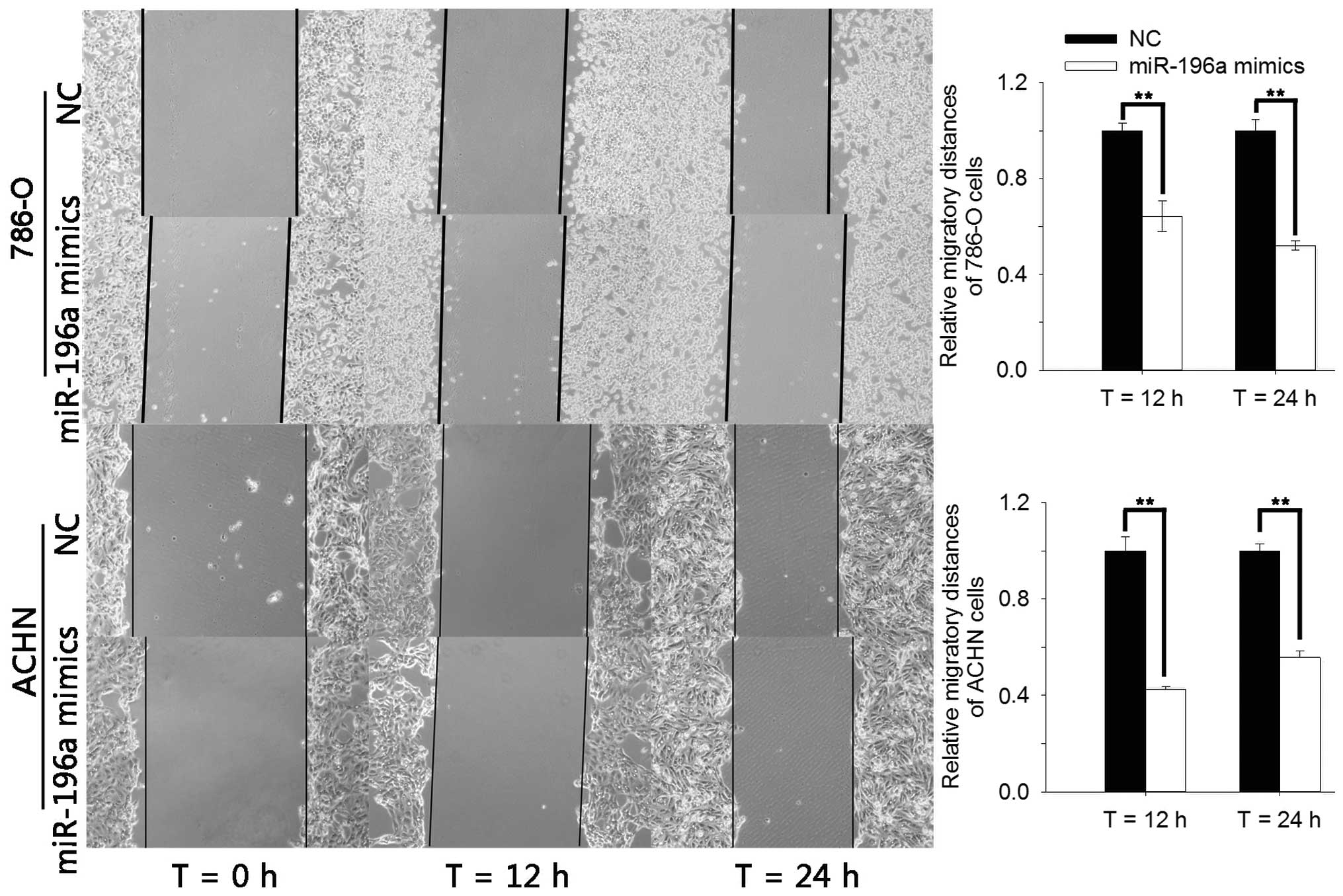
The effects of miR-196a on RCC cell migration were
explored using a wound healing assay. The results showed that the
migration of cells into the wounded area was reduced in the group
transfected with miR-196a mimics compared to that in the NC group
(Fig. 3). Compared with the NC
group, the width of the wound in the miR-196a mimics group was
decreased by 35.70% (P=0.01) and 47.97% (P<0.01) for 786-O cells
and by 57.42% (P<0.01) and 44.27% (P<0.01) for ACHN cells at
12 and 24 h of incubation, respectively. These results indicated
that miR-196a mimics had an inhibitory effect on RCC cell
migration.
miR-196a mimics suppress RCC cell
proliferation
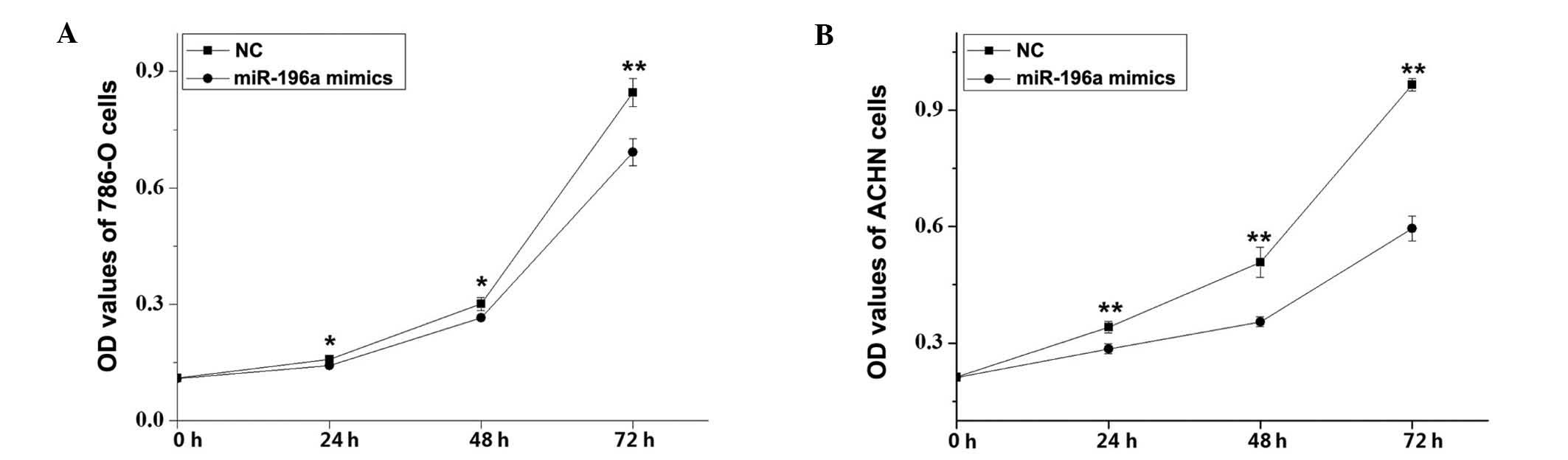
The effects of miR-196a on RCC cell proliferation
were assessed using an MTT assay. Compared with the NC control
group, the cell proliferation of 786-O cells was decreased by 9.91%
(P<0.05), 12.08% (P<0.05) and 18.08% (P<0.01) and that of
ACHN cells was decreased by 16.41, 28.93 and 38.37% (P<0.01 for
all) at 24, 48 and 72 h after transfection with miR-196a mimics,
respectively (Fig. 4). These
results revealed that miR-196a mimics inhibited RCC cell
proliferation.
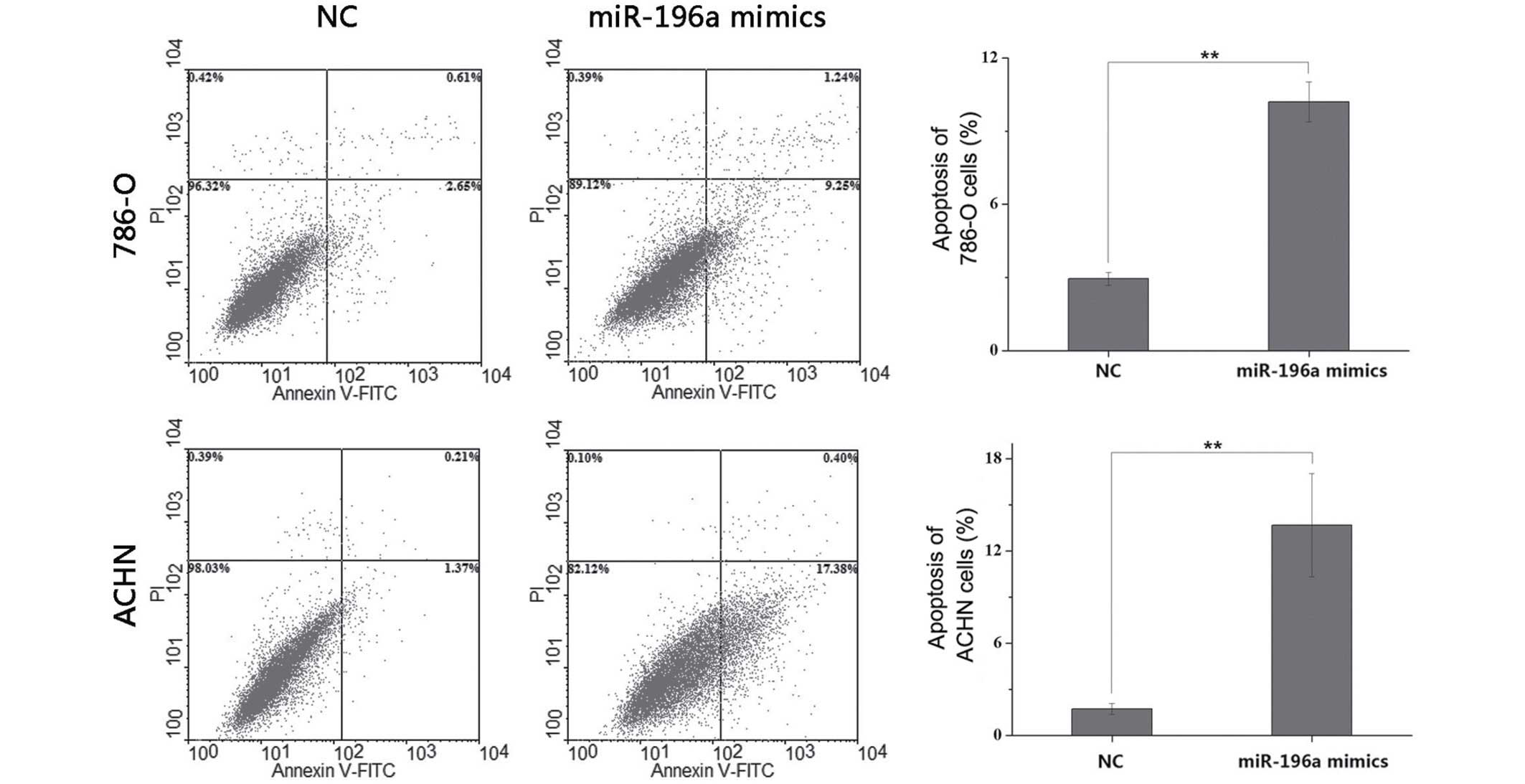
miR-196a mimics induce apoptosis in RCC
cells
Flow cytometric analysis was used to determine the
effects of miR-196a on the apoptotic rate of RCC cells at 48 h
post-transfection. The results indicated that the average early
apoptotic rate of 786-O cells was 2.95% in the NC group, which was
increased to 10.19% in the miR-196a mimics group (P<0.01).
Furthermore, the average early apoptotic rate of ACHN cells was
1.74% in the NC group, while it was increased to 13.69% in the
miR-196a mimics group (P<0.01) (Fig. 5). These results indicated that
miR-196a mimics induced apoptosis in RCC cells. The late apoptotic
rate of 786-O and ACHN cells transfected with miR-196a mimics was
significantly higher than cells transfected with NC (P<0.01),
while the late apoptotic rate of 786-O and ACHN was low in
comparison; therefore, mechanical error should not be excluded.
Nevertheless, the main effect of miR-196a on RCC cells was
manifested in early apoptosis.
Discussion
Tumorigenesis is associated with the activation of
cancer-promoting genes and the inactivation of a number of tumor
suppressor genes. >50% of genes encoding for miRNAs are located
at fragile genomic sites and genomic regions associated with
multiple cancer types, which indicates the relevance as well as
complex roles of miRNAs in cancer. An increasing number of studies
have shown that miRNAs have dual roles as oncogenes or tumor
suppressor genes in different types and stages of tumor (10,25,26).
For instance, miR-31 acts as an oncogene, as it has been implicated
in the development and drug resistance of tumors, and is a
biomarker associated with poor prognosis (27).
miR-196a was recently reported as an oncogene in
various tumor types (28–30). However, previous miRNA profiling
studies have indicated that miR-196a is downregulated in RCC
(21,22), therefore indicating its tumor
suppressor role in this type of cancer. The present study therefore
aimed to clarify the roles of miR-196a in RCC. RT-qPCR was
performed to quantify the relative expression of miR-196a in 48
paired RCC and adjacent normal tissues. Furthermore, the effects of
miR-196a mimics on RCC cell lines were assessed in vitro.
Wound healing, MTT and flow cytometric assays were performed to
assess the effects of miR-196a on cellular migration, proliferation
and apoptosis. The results confirmed that miR-196a was
downregulated in RCC tissues compared with that in paired normal
tissues. Furthermore, transfection with miR-196a mimics suppressed
cellular migration and proliferation and increased apoptosis in
786-O and ACHN cells, further confirming the tumor suppressor role
of miR-196a in RCC. Possibly due to the limited number of RCC
samples used in the present study, no correlation was found between
miR-196a expression and clinicopathological variables. Therefore,
miR-196a expression should be detected in a larger cohort of RCC
patients. Further studies will also be performed to elucidate the
underlying molecular mechanisms and target genes of miR-196a in
RCC.
Previous studies have reported on the roles of
miR-196a in cancer types other than RCC. In head and neck squamous
cell carcinoma, overexpression of miR-196a was found to produce an
oncogenic effect, while its knockdown resulted in decreased cell
proliferation, migration and invasion via suppression of annexin A1
(28). Another study demonstrated
that miR-196a was highly expressed in non-small cell lung cancer
and in which it regulates cell proliferation, migration and
invasion, partially via down-regulation of homeobox (HOX)A5
(31). Furthermore, Sun et
al (32) reported that
miR-196a was upregulated in gastric cancer tissues and promoted
cell proliferation by downregulating p27 (kip1).
Previous studies have also described miRNAs as novel
biomarkers. In pancreatic ductal adenocarcinoma, overexpression of
miR-196a was observed to be associated with disease progression and
patient prognosis (33), and
another study reported its potential use as a biomarker for the
early detection of familial pancreatic cancer (19). Aso et al (34) reported that miR-196a detected in
the pancreatic juice is a diagnostic biomarker for intestinal-type
intraductal papillary mucinous neoplasm. Furthermore, miR-196a and
miR-196b have been indicated to be correlated with aggressive
progression and unfavorable clinical outcome in colorectal cancer
patients (35).
miR-196a was also found to have a role in certain
other diseases or cellular processes. Zhang et al (36) revealed that the urine levels of
miR-196a are associated with focal segmental glomerulosclerosis. In
addition, miR-196a was found to regulate the differentiation and
proliferation of human adipose tissue-derived mesenchymal stem
cells by modulating the levels of the transcription factor HOXC8
(37).
In conclusion, the present study revealed that
miR-196a was downregulated in RCC tissues compared with that in
normal adjacent tissues. Transfection with miR-196a mimics
inhibited the proliferation and migration of the 786-O and ACHN RCC
cell lines, while inducing apoptosis. These results suggested that
miR-196a may function as a tumor suppressor in RCC. The results in
our study supported that miR-196a may not only be a promising
diagnostic biomarker but also a potential therapeutic target in
RCC. Further studies will identify target genes of miR-196a in
RCC.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (no. 81101922), the Medical
Scientific Research Foundation of Guangdong Province of China (nos.
A2012584 and A2013606), the Science and Technology Development Fund
Project of Shenzhen (no. JCYJ20130402114702124) and the fund of
Guangdong Key medical subject.
References
|
1
|
Yan Y, Yang FQ, Zhang HM, Che J and Zheng
JH: Up-regulation of flotillin-2 is associated with renal cell
carcinoma progression. Tumour Biol. 35:10479–10486. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patel C, Ahmed A and Ellsworth P: Renal
cell carcinoma: A reappraisal. Urol Nurs. 32:182–190; quiz 191.
2012.PubMed/NCBI
|
|
3
|
Du M, Lu D, Wang Q, Chu H, Tong N, Pan X,
Qin C, Yin C, Wang M and Zhang Z: Genetic variations in microRNAs
and the risk and survival of renal cell cancer. Carcinogenesis.
35:1629–1635. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yin B, Zeng Y, Wang X, Liu G, Zhang M and
Song Y: Expression and clinical significance of cancer-testis genes
in clear cell renal cell carcinoma. Int J Clin Exp Pathol.
7:4112–4119, eCollection 2014. 2014.PubMed/NCBI
|
|
6
|
Rasmussen F: Metastatic renal cell cancer.
Cancer Imaging. 13:374–380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tostain J, Li G, Gentil-Perret A and
Gigante M: Carbonic anhydrase 9 in clear cell renal cell carcinoma:
A marker for diagnosis, prognosis and treatment. Eur J Cancer.
46:3141–3148. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh P, Agarwal N and Pal SK: Sequencing
systemic therapies for metastatic kidney cancer. Curr Treat Options
Oncol. 16:3162015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu M, Tang Q, Qiu M, Lang N, Li M, Zheng
Y and Bi F: miR-21 targets the tumor suppressor RhoB and regulates
proliferation, invasion and apoptosis in colorectal cancer cells.
FEBS Lett. 585:2998–3005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang W, Li J, Zhu W, Gao C, Jiang R, Li W,
Hu Q and Zhang B: MicroRNA-21 and the clinical outcomes of various
carcinomas: A systematic review and meta-analysis. BMC Cancer.
14:8192014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou X, Wang X, Huang Z, Wang J, Zhu W,
Shu Y and Liu P: Prognostic value of miR-21 in various cancers: An
updating meta-analysis. PloS One. 9:e1024132014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pernaute B, Spruce T, Smith KM,
Sánchez-Nieto JM, Manzanares M, Cobb B and Rodríguez TA: MicroRNAs
control the apoptotic threshold in primed pluripotent stem cells
through regulation of BIM. Genes Dev. 28:1873–1878. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou J, Jiang Z, Wang Z, et al:
MicroRNA-142-3p is frequently upregulated in colorectal cancer and
may be involved in the regulation of cell proliferation. Chinese
Science Bulletin. 58:2836–2845. 2013. View Article : Google Scholar
|
|
14
|
Xu Y, Brenn T, Brown ER, Doherty V and
Melton DW: Differential expression of microRNAs during melanoma
progression: MiR-200c, miR-205 and miR-211 are downregulated in
melanoma and act as tumour suppressors. Br J Cancer. 106:553–561.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shibuya H, Iinuma H, Shimada R, Horiuchi A
and Watanabe T: Clinicopathological and prognostic value of
microRNA-21 and microRNA-155 in colorectal cancer. Oncology.
79:313–320. 2010. View Article : Google Scholar
|
|
16
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhai Z, Wu F, Dong F, Chuang AY, Messer
JS, Boone DL and Kwon JH: Human autophagy gene ATG16L1 is
post-transcriptionally regulated byMIR142-3p. Autophagy.
10:468–479. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Yao C, Li H, Wang G and He X:
Combined elevation of microRNA-196a and microRNA-196b in sera
predicts unfavorable prognosis in patients with osteosarcomas. Int
J Mol Sci. 15:6544–6555. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Slater EP, Strauch K, Rospleszcz S,
Ramaswamy A, Esposito I, Klöppel G, Matthäi E, Heeger K, Fendrich
V, Langer P and Bartsch DK: MicroRNA-196a and -196b as potential
biomarkers for the early detection of familial pancreatic cancer.
Transl Oncol. 7:464–471. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tsai MM, Wang CS, Tsai CY, Chen CY, Chi
HC, Tseng YH, Chung PJ, Lin YH, Chung IH, Chen CY and Lin KH:
MicroRNA-196a/-196b promote cell metastasis via negative regulation
of radixin in human gastric cancer. Cancer Lett. 351:222–231. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yi Z, Fu Y, Zhao S, Zhang X and Ma C:
Differential expression of miRNA patterns in renal cell carcinoma
and nontumorous tissues. J Cancer Res Clin Oncol. 136:855–862.
2010. View Article : Google Scholar
|
|
22
|
White NM, Khella HW, Grigull J, Adzovic S,
Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA,
et al: miRNA profiling in metastatic renal cell carcinoma reveals a
tumour-suppressor effect for miR-215. Br J Cancer. 105:1741–1749.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Edge S, Byrd DR, Compton C, Fritz A,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
Springer; New York, NY: 2010
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Luo J, Cai Q, Wang W, Huang H, Zeng H, He
W, Deng W, Yu H, Chan E, Ng CF, et al: A microRNA-7 binding site
polymorphism in HOXB5 leads to differential gene expression in
bladder cancer. PloS One. 7:e401272012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Juan D, Alexe G, Antes T, Liu H,
Madabhushi A, Delisi C, Ganesan S, Bhanot G and Liou LS:
Identification of a microRNA panel for clear-cell kidney cancer.
Urology. 75:835–841. 2010. View Article : Google Scholar
|
|
27
|
Wang S, Hu J, Zhang D, Li J, Fei Q and Sun
Y: Prognostic role of microRNA-31 in various cancers: A
meta-analysis. Tumor Biol. 35:11639–11645. 2014. View Article : Google Scholar
|
|
28
|
Suh YE, Raulf N, Güken J, Lawler K, Urbano
TG, Bullenkamp J, Gobeil S, Huot J, Odell E and Tavassoli M:
MicroRNA-196a promotes an oncogenic effect in head and neck cancer
cells by suppressing annexin A1 and enhancing radioresistance. Int
J Cancer. 137:1021–1034. 2015. View Article : Google Scholar
|
|
29
|
Pazzaglia L, Leonardi L, Conti A, Novello
C, Quattrini I, Montanini L, Roperto F, Del Piero F, Di Guardo G,
Piro F, et al: miR-196a expression in human and canine
osteosarcomas: A comparative study. Res Vet Sci. 99:112–119. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hong TH and Park IY: MicroRNA expression
profiling of diagnostic needle aspirates from surgical pancreatic
cancer specimens. Ann Surg Treat Res. 87:290–297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu XH, Lu KH, Wang KM, Sun N, Zhang EB,
Yang JS, Yin DD, Liu ZL, Zhou J, Liu ZJ, et al: MicroRNA-196a
promotes non-small cell lung cancer cell proliferation and invasion
through targeting HOXA5. BMC Cancer. 12:3482012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun M, Liu XH, Li JH, Yang JS, Zhang EB,
Yin DD, Liu ZL, Zhou J, Ding Y, Li SQ, et al: MiR-196a is
upregulated in gastric cancer and promotes cell proliferation by
downregulating p27 (kip1). Mol Cancer Ther. 11:842–852. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang J, Chen J, Chang P, LeBlanc A, Li D,
Abbruzzesse JL, Frazier ML, Killary AM and Sen S: MicroRNAs in
plasma of pancreatic ductal adenocarcinoma patients as novel
blood-based biomarkers of disease. Cancer Prev Res (Phila).
2:807–813. 2009. View Article : Google Scholar
|
|
34
|
Aso T, Ohtsuka T, Tamura K, Ideno N, Kono
H, Nagayoshi Y, Ohuchida K, Ueda J, Takahata S, Shindo K, et al:
Elevated expression level of microRNA-196a is predictive of
intestinal-type intraductal papillary mucinous neoplasm of the
pancreas. Pancreas. 43:361–366. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ge J, Chen Z, Li R, Lu T and Xiao G:
Upregulation of microRNA-196a and microRNA-196b cooperatively
correlate with aggressive progression and unfavorable prognosis in
patients with colorectal cancer. Cancer Cell Int. 14:1282014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang W, Zhang C, Chen H, Li L, Tu Y, Liu
C, Shi S, Zen K and Liu Z: Evaluation of microRNAs miR-196a,
miR-30a-5P and miR-490 as biomarkers of disease activity among
patients with FSGS. Clin J Am Soc Nephrol. 9:1545–1552. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim YJ, Bae SW, Yu SS, Bae YC and Jung JS:
miR-196a regulates proliferation and osteogenic differentiation in
mesenchymal stem cells derived from human adipose tissue. J Bone
Miner Res. 24:816–825. 2009. View Article : Google Scholar
|