Introduction
Chromosome replication is a risky process for
maintaining genome integrity, as unrepaired DNA lesions at S phase
interfere with the progress of replication forks and thereby result
in excessive formation of single-strand DNA (ssDNA) that could be a
major cause of deleterious lesions, such as DNA double-strand
breaks. To preserve genome integrity during chromosome replication,
eukaryotic cells have acquired several adaptive responses to DNA
damage (1,2). One of the most studied pathways is
the S-phase checkpoint response, which is evoked by an exposed
ssDNA at stalled replication forks, attributed to a deficiency in
DNA synthesis. The checkpoint kinase ATM and Rad3-related (ATR) is
recruited on ssDNA where it is coated with ssDNA-binding protein,
replication protein A (RPA). It then causes the phosphorylation and
activation of downstream checkpoint kinase 1 (Chk1), which in turn
stabilizes replication forks for genome integrity (3–5).
Another is a damage tolerance mechanism termed translesion DNA
synthesis (TLS), the major process with which cells replicate past
the unrepaired DNA lesion during S phase (6). When the normal replication machinery
is blocked at ultraviolet (UV)-induced cyclobutane pyrimidine
dimers (CPDs), the Y-family DNA polymerase Polh replaces the
stalled replicative DNA polymerase. This is dependent upon
monoubiquitination of the ring-shaped clamp protein, proliferating
cell nuclear antigen (PCNA), by the E3 ubiquitin ligase RAD18.
Monoubiquitinated PCNA has an increased affinity for Polh, thus
aiding the recruitment of Polh to stalled replication forks and
allowing accurate replicative bypass of CPDs by incorporating
correct bases on the opposite strand (7,8).
Consequently, TLS overcomes UV-induced replication blocks, thereby
preventing sustained activation of the S-phase checkpoint in
response to excessive formation of ssDNA (9).
Accumulating evidence has shown that the S-phase
checkpoint and TLS are activated by conserved clamp loader complex
termed chromosome transmission fidelity protein 18 and replication
factor C (CTF18-RFC) (10–13). CTF18-RFC is one of four
'heteropentameric RFC complexes' each of which contains a common
small subunit comprising RFC2-4 together with a unique larger
subunit, including either RFC1, Elg1, RAD17 or CTF18. RFC1-RFC is
important in normal DNA replication as it loads the homotrimeric
PCNA clamp around the junction of primers with template DNA at
replication forks (14). Elg1-RFC
is involved in the maintenance of genome stability (15,16),
while RAD17-RFC contributes to the activation of the DNA damage
checkpoint by loading the heterotrimeric 9-1-1 checkpoint clamp at
sites of damaged DNA (17). In
addition, although CTF18-RFC was originally reported to be
important in establishing sister chromatid cohesion (18), recent studies with budding yeast
have shown that CTF18-RFC mediates activation of the S-phase
checkpoint depending on the association with DNA polymerase ε
(10). By contrast, a biochemical
study with an in vitro reconstitution system has
demonstrated that CTF18-RFC binds to and stimulates the DNA
synthetic activity of DNA polymerase η (11). However, the molecular mechanisms
underlying these alternative functions of CTF18-RFC remain largely
unknown.
In the present study, it was demonstrated that RPA
is a novel binding partner of CTF18 in mammalian cells. Among the
heterotrimeric subunits, RPA1 and RPA2 are detected as a complex
with CTF18. Notably, the DuoLink in situ proximity ligation
assay (PLA) demonstrated the nuclear interaction between CTF18 and
RPA in response to replication stresses induced by hydroxyurea (HU)
treatment and UV irradiation. Furthermore, the kinetics of
CTF18-RPA interaction were positively correlated with the sustained
activation of the ATR-Chk1 pathway after UV irradiation. The
present findings provide insight into the mechanism underlying the
functional role of CTF18 in replication stress responses.
Materials and methods
Cell culture
HEK293 cells (RIKEN BioResource Center, Tsukuba,
Japan) were kept at 37°C in a humidified 5% CO2
atmosphere and cultured in Dulbecco's modified Eagle's medium
(Nacalai Tesque, Inc., Kyoto, Japan) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and 100 units penicillin/streptomycin (Sigma-Aldrich, St.
Louis, MO, USA).
Antibodies
The following antibodies were used in the present
study: Rabbit polyclonal anti-CTF18 (cat no. A301-883A; 1:1,000 for
western blot analysis; 1:100 for PLA; Bethyl Laboratories, Inc.,
Montgomery, TX, USA); rabbit monoclonal anti-RPA1 (cat. no.
ab79398; 1:3,000 for western blot analysis) and mouse monoclonal
anti-RPA2 (cat. no. ab2175; 1:2,000 for western blot analysis;
1:300 for PLA) (Abcam, Cambridge, MA, USA); mouse monoclonal anti-α
tubulin (cat. no. B-5-1-2; 1:3,000 for western blot analysis;
Sigma-Aldrich); mouse monoclonal anti-Chk1 (cat. no. K0086-3;
1:2,000 for western blot analysis; Medical & Biological
Laboratories, Nagoya, Japan); and rabbit polyclonal
anti-phospho-Chk1 at Ser345 (cat. no. 2341 1:1,000 for western blot
analysis; Cell Signaling Technology, Danvers, MA, USA). Secondary
antibodies were polyclonal HRP-conjugated sheep anti-mouse (cat.
no. NA931V; 1:30,000) and donkey anti-rabbit (cat. no. NA934V;
1:30,000) obtained from GE Healthcare Life Sciences (Little
Chalfont, UK).
Cell synchronization
Cell cycle synchronization was performed by the
double thymidine block method as reported previously (19). Briefly, exponentially growing
HEK293 cells were treated with 2 mM thymidine (Nacalai Tesque,
Inc.) for 16 h, thymidine-free media for 10 h, and 2 mM thymidine
for 18 h to arrest the cell cycle at the G1/S boundary. Then, cells
were released by changing the medium and analyzed at various time
intervals. Cell synchronization was conducted prior to the in
situ proximity ligation assay and western blotting to detect
phosphorylation of Chk1, however, the co-immunoprecipitation assay
was performed with unsynchronized cells.
Co-immunoprecipitation assay and western
blotting
Co-immunoprecipitation was performed as previously
described (20). Briefly,
whole-cell lysates were obtained from HEK293 cells using a lysis
buffer [containing 20 mM Hepes (pH 7.9), 150 mM NaCl, 0.1% Triton
X-100 ((Nacalai Tesque, Inc.) and protease inhibitor cocktail
(Nacalai Tesque, Inc.)] were immunoprecipitated with normal IgG or
anti-CTF18 antibody, and then separated by 10% SDS-PAGE. The
proteins were transferred to a polyvinylidene difluoride membrane
(EMD Millipore, Billerica, MA, USA) and blocked for 1 h with 0.3%
skim milk at room temperature. The membrane was incubated overnight
at 4°C with antibodies against RPA1, RPA2 and CTF18. Following
washing with Tris-buffered saline with Tween 20 (TBST), the
membrane was probed with the secondary antibodies for 1 h at room
temperature and then washed again with TSBT. The membranes were
visualized using the enhanced chemiluminescence system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and exposed to X-ray film
(Fujifilm Corporation, Tokyo, Japan).
In situ proximity ligation assay
(PLA)
PLA was performed according to the manufacturer's
instructions (Sigma-Aldrich). Briefly, synchronized HEK293 cells at
S phase were treated with HU (0, 2 or 5 mM; Wako Pure Chemical
Industries, Ltd., Osaka, Japan) or irradiated with UV light (0, 20
or 100 J/m2), after 2 h, cells were extracted with CSK
buffer (10 mM PIPES-NaOH, pH 6.8; 300 mM sucrose and 100 mM NaCl)
containing 0.5% Triton X-100 for 5 min for detection of chromatin
bound proteins (21). After
washing with CSK buffer without Triton X-100, cells were fixed with
3.7% formalin (Wako Pure Chemical Industries, Ltd.) for 20 min,
followed by permeabilization with ice-cold methanol for 10 min.
After blocking with Duolink Blocking solution (Sigma-Aldrich),
cells were probed with mouse monoclonal anti-RPA2 and rabbit
polyclonal anti-CTF18 antibodies, and then mouse or rabbit PLA
probes were added. Hybridization of the oligonucleotide arms of the
PLA probes creates a template for rolling circle amplification
(RCA) only when the epitopes of the target proteins are in close
proximity (<40 nm). Following amplification of the RCA, an
oligonucleotide probe labeled with Texas Red fluorophore is added
and hybridizes with the RCA product. All fluorescence data were
obtained with a confocal microscope FV10i (Olympus Corporation,
Tokyo, Japan) and z-stacked images (collected in 1 μm steps)
were used for quantification of PLA signals with the Duolink Image
Tool (version 1.0; Sigma-Aldrich). The PLA signals were calculated
from three different fields and the scores were converted into fold
change compared with the control.
Statistical analyses
All experiments were repeated in triplicate.
Statistical significance for in situ PLA was determined by
two-tailed unpaired Student's t-test using Graphpad Prism 5
(Graphpad Software, Inc., La Jolla, CA, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
CTF18 interacts with the RPA complex in
HEK293 cells
To elucidate the mechanism underlying replication
stress responses by CTF18-RFC, the present study aimed to identify
a new binding partner for CTF18. It focused on a ssDNA-binding
protein RPA that stabilizes the ssDNA region during DNA replication
and repair, and also acts as a scaffold for DNA processing
proteins. Although RPA is a heterotrimeric complex composed of 70,
32 and 14 kDa subunits, referred to as RPA1, RPA2 and RPA3,
respectively (22), the present
study investigated the interaction between endogenous CTF18 with
two RPA subunits, RPA1 and RPA2, which are detectable with specific
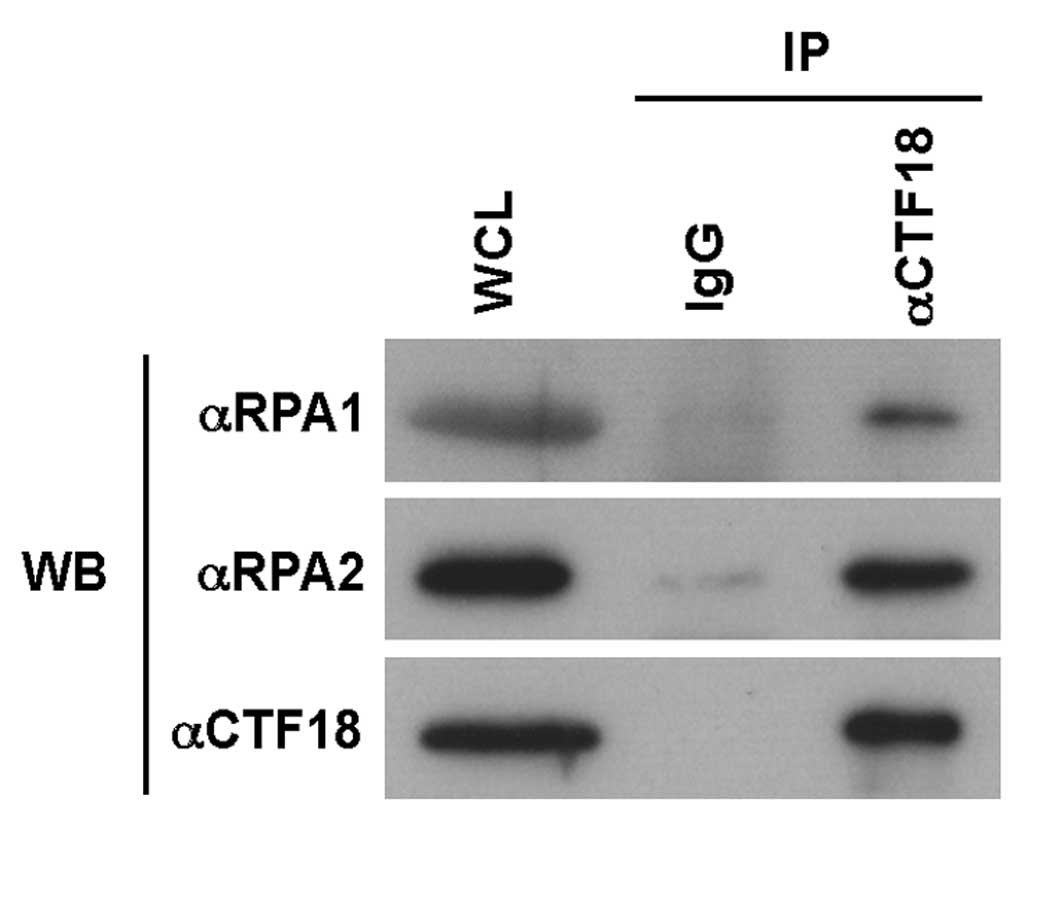
antibodies. As shown in Fig. 1, an
immune-complex of CTF18 from whole cell lysates of HEK293 cells
included RPA1 and RPA2, suggesting that CTF18 forms a complex with
RPA in vivo.
CTF18 is associated with RPA in response
to replication stress
If the interaction between CTF18 and RPA occurs in
the replication stress responses, it is expected that the CTF18-RPA
complexes could be observed in the nucleus when replication forks
are stalled during S phase. To test this hypothesis, the PLA assay
was conducted, where two endogenous proteins are immunostained with
secondary antibodies, originating from different species,
conjugated to complementary oligonucleotides. In this assay, when
two distinct antibodies locate in close proximity (<40 nm), the
conjugated DNA can be amplified and detected with a fluorescent
probe as foci that represent molecules of each of two interacting
proteins (23,24). To induce the replication checkpoint
response, double-thymidine arrested HEK293 cells were released into
S phase and subsequently treated with HU, which causes a reversible
inhibition of DNA synthesis and thus blocks the progression of
replication forks (25). The
DuoLink assay showed that there is no significant signal in the
absence of HU, whereas HU treatment resulted in the formation of
nuclear PLA foci in a dose-dependent manner (Fig. 2A). These results suggest that the
S-phase checkpoint response elicited by stalled replication forks
leads to the interaction between CTF18 and RPA in the nucleus.
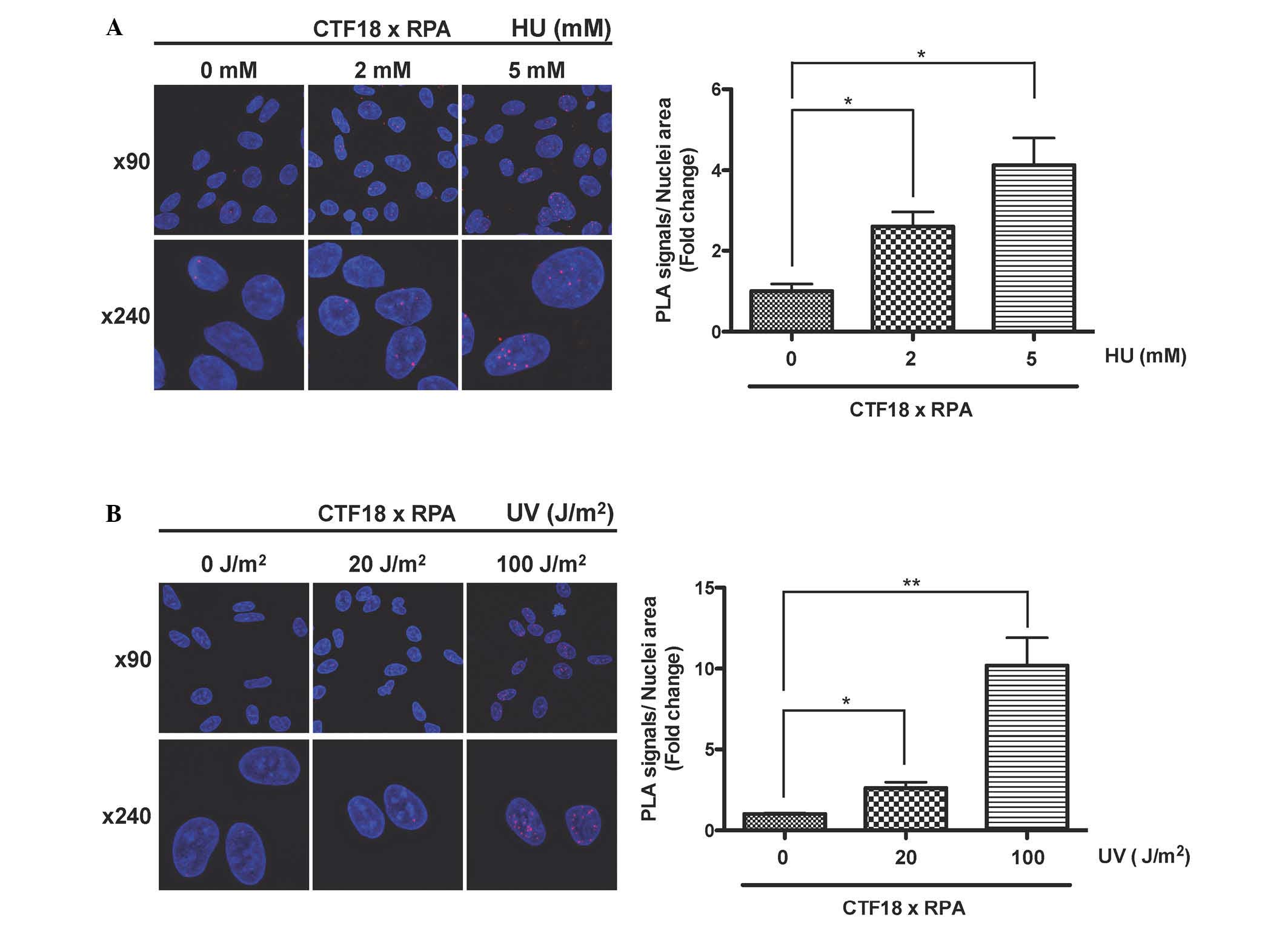 | Figure 2CTF18 binds to RPA in response to
replication stresses. (A) The interaction between CTF18 and RPA
occurs after treatment with HU. HEK293 cells were synchronized to S
phase by double thymidine block and then treated with the indicated
concentration of HU. After 2 h, cells were treated with
CSK/Triton-X buffer followed by fixation, and then an in
situ PLA was performed with anti-RPA2 and anti-CTF18
antibodies. The red fluorescent foci indicate the proximity of the
two proteins (magnification, ×90 or ×240). Duolink Image Tool was
used to quantify PLA signals (n=3, ×90 magnification). The vertical
axis shows the total nuclear PLA signals divided by nuclei area and
normalized to non-treatment group, and the horizontal axis
indicates the concentration of HU. Error bars indicate the standard
error of the mean of three different fields. (B) The CTF18-RPA
interaction occurs after exposure to UV irradiation. HEK293 cells
were synchronized to S phase by double thymidine block and then
irradiated with indicated dose of UV light. The PLA was performed
as shown in (A). Nuclei were stained with Hoechst 33342.
*P<0.05, **P<0.01. CTF18, chromosome
transmission fidelity protein 18; RPA, replication protein A; PLA,
proximity ligation assay; HU, hydroxyurea; UV, ultraviolet. |
Interaction between CTF18 and RPA occurs
after UV irradiation
In addition to the S-phase checkpoint pathway,
eukaryotic cells can tolerate replication stress by bypassing DNA
lesions via TLS (6). Since CTF18
has been shown to be implicated in TLD (11), it was investigated whether
UV-induced DNA damage triggered CTF18-RPA interaction during S
phase. Synchronized HEK293 cells at early S phase were exposed to
UV irradiation at 20 or 100 J/m2, and after 2 h, the
CTF18-RPA interaction was assessed by counting PLA foci. A few foci
were observed in the nucleus following exposure to 20
J/m2 UV, and the formation of nuclear foci was
significantly augmented when irradiated with a high dose (100
J/m2) of UV light compared with a low dose (20
J/m2; Fig. 2B). These
findings suggest that the CTF18-RPA interaction occurs in response
to the initiation of translesion DNA synthesis, although it is
impossible to exclude the possibility that the interaction may
result from the S-phase checkpoint response by UV irradiation.
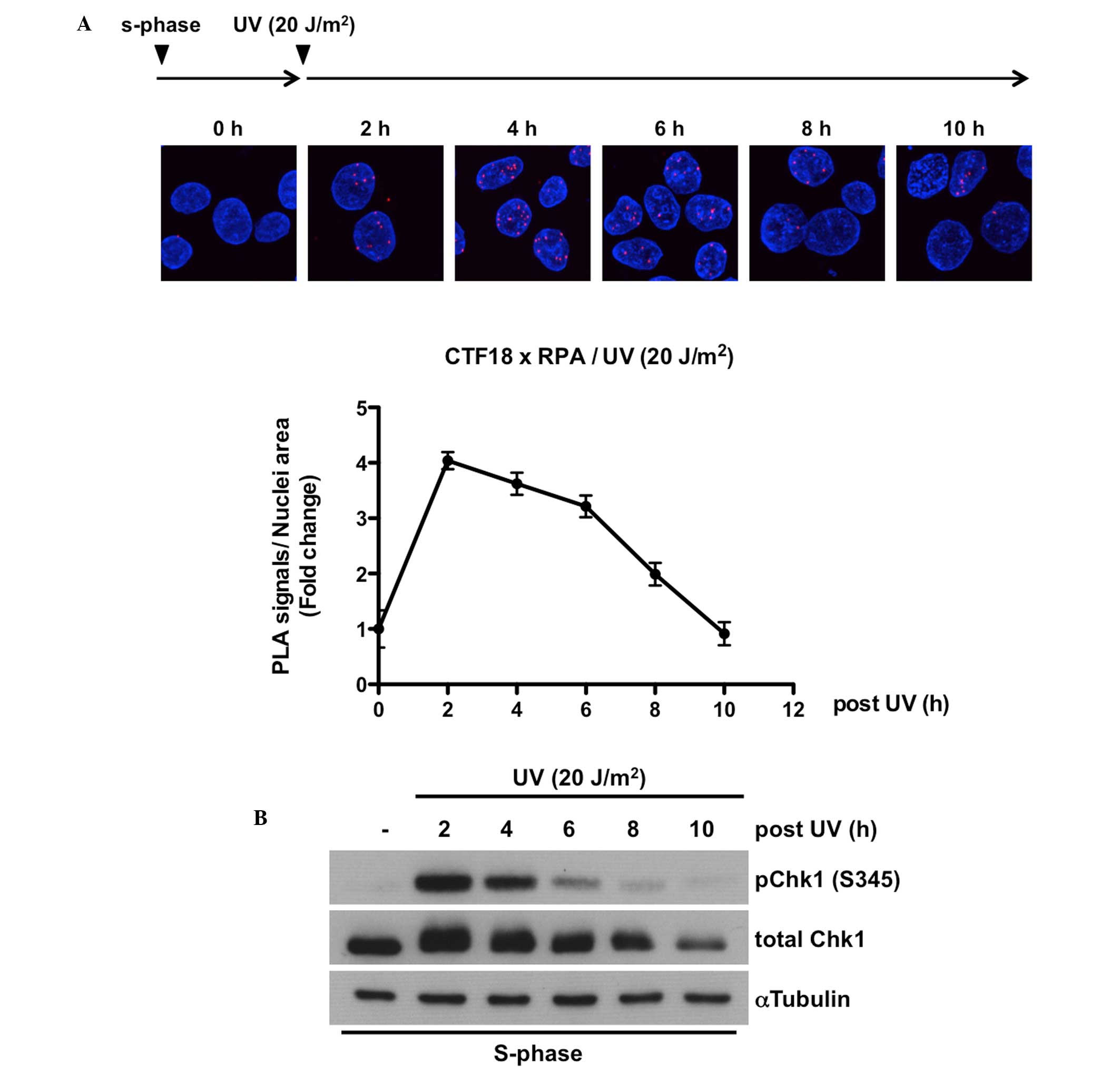
Kinetics of the CTF18-RPA interaction
correlate positively with that of Chk1 phosphorylation
Finally, it was hypothesized that if the CTF18-RPA
interaction is required for the replication stress responses, it
may be sustained until the attenuation of the S-phase checkpoint
pathway. Therefore the dissociation kinetics of the CTF18-RPA
complex were examined by tracking the time course of PLA signals
and comparing it with the activation status of the ATR-Chk1
signaling pathway following UV-induced replication stress. As shown
in Fig. 3A, it was demonstrated
that while the number of foci peaks at 2 h after UV irradiation, it
gradually decreases with time and almost disappears before 10 h. In
addition, this time-dependent decline in UV-induced binding of
CTF18 to RPA was similar to that of Chk1 phosphorylation at Ser345
(Fig. 3B). Collectively, these
data imply that the CTF18-RPA interaction is involved in the
replication stress response, including translesion DNA synthesis
and S-phase checkpoint pathway.
Discussion
The present data demonstrated that RPA is a novel
binding partner of CTF18 in mammalian cells. It was shown that this
interaction was triggered when replication stress occurred and then
gradually diminished in accordance with a decrease in the
phosphorylation levels of Chk1 at Ser345. Accumulating evidence has
shown that the CTF18-RFC complex is critical in activation of the S
phase checkpoint and translesion DNA synthesis by interacting with
DNA polymerase ε and η, respectively (10–13).
However, the mechanism whereby CTF18-RFC responds to replication
stress and targets the stalled replication forks remains to be
determined. In this study, it was hypothesized that RPA may serve
as a platform for the molecular assembly of CTF18-RFC together with
DNA polymerase ε and η, which in turn aids in producing an
efficient response to replication stress.
The in situ PLA demonstrated that replication
stress induced by HU treatment or UV irradiation triggers the
interaction between CTF18 and RPA in the nucleus. It remains to be
determined how CTF18 senses replication stress and binds
preferentially to RPA on ssDNA. A possible mechanism could be the
phosphorylation of the RPA2 subunit in response to replication
stress. Several studies have shown that stalled replication forks
cause hyperphosphorylation of RPA2 at the N-terminal region through
the DNA damage response pathways involving the ATR and the
DNA-dependent protein kinase (21,26–28).
Moreover, phosphorylation of RPA2 is known to prevent its
association with the replication machinery and thus be considered
as a trigger for redirecting RPA functions from DNA replication to
DNA damage responses (28,29). In agreement with this, RPA2
phosphorylation has been reported to enhance its interactions with
the ATR and the 9-1-1 checkpoint clamp (30,31).
Hence, although further studies are required to address the link
between RPA2 phosphorylation and CTF18-RPA interaction, the present
results have provided insight into the molecular basis of the
initiation of the replication stress response in mammalian
cells.
Among the four clamp loader complexes, the Elg1-RFC
is hypothesized to act principally as an unloader for PCNA from
nascent DNA after the passage of replication forks and thereby
regulate PCNA levels in chromatin (32–34).
In addition, Bylund and Burgers (35) demonstrated that CTF18-RFC also
unloads PCNA specifically when ssDNA is coated with RPA, and they
proposed a model in which this unloading activity of CTF18-RFC may
contribute to establishing sister chromatid cohesion. However,
considering the present result that CTF18 binds to RPA after
UV-irradiation during S phase, it is possible that CTF18-RFC may
remove monoubiquitinated PCNA after replicative bypass of
UV-induced CPD with Polh and subsequently reload unmodified PCNA to
restart normal DNA replication. Thus, the present results provide
insight into the mechanism how DNA polymerases switch during
TLS.
In conclusion, the present study demonstrated that
CTF18 forms a complex with RPA when replication stress is elicited
by hydroxyurea treatment or UV exposure during S phase. The
interaction kinetics between CTF18 and RPA is positively associated
with the phosphorylation status of Chk1. These results suggest that
RPA may be a scaffold for CTF18-RFC to be recruited to stalled
replication forks and respond to replication stress.
Acknowledgments
The authors would like to thank the members of
Fukamizu laboratory for their helpful discussion. This study was
supported by Grants-in-Aid for Scientific Research on Priority
Areas (grant no. 17054004 to Professor Akiyoshi Fukamizu),
Grants-in-Aid for Young Scientists (grant no. 20780237, 22688029 to
Dr Hiroaki Daitoku), and Grants-in-Aid for JSPS Fellows (grant no.
25.452 to Mr. Yuta Kaneko) from the Ministry of Education, Culture,
Sports, Science, and Technology, Japan.
References
|
1
|
Branzei D and Foiani M: Regulation of DNA
repair throughout the cell cycle. Nat Rev Mol Cell Biol. 9:297–308.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeman MK and Cimprich KA: Causes and
consequences of replication stress. Nat Cell Biol. 16:2–9. 2014.
View Article : Google Scholar
|
|
3
|
Zou L and Elledge SJ: Sensing DNA damage
through ATRIP recognition of RPA-ssDNA complexes. Science.
300:1542–1548. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Choi JH, Lindsey-Boltz LA, Kemp M, Mason
AC, Wold MS and Sancar A: Reconstitution of RPA-covered
single-stranded DNA-activated ATR-Chk1 signaling. Proc Natl Acad
Sci USA. 107:13660–13665. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smits VA and Gillespie DA: DNA damage
control: Regulation and functions of checkpoint kinase 1. FEBS J.
282:3681–3692. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang W: An overview of Y-Family DNA
polymerases and a case study of human DNA polymerase eta.
Biochemistry. 53:2793–2803. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davies AA, Huttner D, Daigaku Y, Chen S
and Ulrich HD: Activation of ubiquitin-dependent DNA damage bypass
is mediated by replication protein a. Mol Cell. 29:625–636. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Watanabe K, Tateishi S, Kawasuji M,
Tsurimoto T, Inoue H and Yamaizumi M: Rad18 guides poleta to
replication stalling sites through physical interaction and PCNA
monoubiquitination. EMBO J. 23:3886–3896. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Despras E, Daboussi F, Hyrien O,
Marheineke K and Kannouche PL: ATR/Chk1 pathway is essential for
resumption of DNA synthesis and cell survival in UV-irradiated XP
variant cells. Hum Mol Genet. 19:1690–1701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
García-Rodríguez LJ, De Piccoli G,
Marchesi V, Jones RC, Edmondson RD and Labib K: A conserved Polε
binding module in Ctf18-RFC is required for S-phase checkpoint
activation downstream of Mec1. Nucleic Acids Res. 43:8830–8838.
2015. View Article : Google Scholar
|
|
11
|
Shiomi Y, Masutani C, Hanaoka F, Kimura H
and Tsurimoto T: A second proliferating cell nuclear antigen loader
complex, Ctf18-replication factor C, stimulates DNA polymerase eta
activity. J Biol Chem. 282:20906–20914. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Crabbé L, Thomas A, Pantesco V, De Vos J,
Pasero P and Lengronne A: Analysis of replication profiles reveals
key role of RFC-Ctf18 in yeast replication stress response. Nat
Struct Mol Biol. 17:1391–1397. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kubota T, Hiraga S, Yamada K, Lamond AI
and Donaldson AD: Quantitative proteomic analysis of chromatin
reveals that Ctf18 acts in the DNA replication checkpoint. Mol Cell
Proteomics. 10:M1102011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mailand N, Gibbs-Seymour I and
Bekker-Jensen S: Regulation of PCNA-protein interactions for genome
stability. Nat Rev Mol Cell Biol. 14:269–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanellis P, Agyei R and Durocher D: Elg1
forms an alternative PCNA-interacting RFC complex required to
maintain genome stability. Curr Biol. 13:1583–1595. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bellaoui M, Chang M, Ou J, Xu H, Boone C
and Brown GW: Elg1 forms an alternative RFC complex important for
DNA replication and genome intergrity. EMBO J. 22:4304–4313. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Parrilla-Castellar ER, Arlander SJ and
Karnitz L: Dial 9-1-1 for DNA damage: The Rad9-Hus1-Rad1 (9-1-1)
clamp complex. DNA Repair (Amst). 3:1009–1014. 2004. View Article : Google Scholar
|
|
18
|
Hanna JS, Kroll ES, Lundblad V and Spencer
FA: Saccharomyces cerevisiae CTF18 and CTF4 are required for sister
chromatid cohesion. Mol Cell Biol. 21:3144–3158. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Taniguchi T, Garcia-Higuera I, Andreassen
PR, Gregory RC, Grompe M and D'Andrea AD: S-phase-specific
interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and
RAD51. Blood. 100:2414–2420. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamagata K, Daitoku H, Takahashi Y, Namiki
K, Hisatake K, Kako K, Mukai H, Kasuya Y and Fukamizu A: Arginine
methylation of FOXO transcription factors inhibits their
phosphorylation by Akt. Mol Cell. 32:221–231. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vassin VM, Anantha RW, Sokolova E, Kanner
S and Borowiec JA: Human RPA phosphorylation by ATR stimulates DNA
synthesis and prevents ssDNA accumulation during DNA-replication
stress. J Cell Sci. 122:4070–4080. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fan J and Pavletich NP: Structure and
conformational change of a replication protein A heterotrimer bound
to ssDNA. Genes Dev. 26:2337–2347. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Söderberg O, Gullberg M, Jarvius M,
Ridderstråle K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P,
Bahram F, Larsson LG and Landegren U: Direct observation of
individual endogenous protein complexes in situ by proximity
ligation. Nat Methods. 3:995–1000. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gullberg M and Andersson AC: Visualization
and quantification of protein-protein interactions in cells and
tissues. Nat Methods. 72010.
|
|
25
|
Mazouzi A, Velimezi G and Loizou JI: DNA
replication stress: Causes, resolution and disease. Exp Cell Res.
329:85–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liaw H, Lee D and Myung K:
DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair
and suppresses sister chromatid exchange. PLoS One. 6:e214242011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu S, Opiyo SO, Manthey K, Glanzer JG,
Ashley AK, Amerin C, Troksa K, Shrivastav M, Nicholoff JA and
Oakley GG: Distinct roles for DNA-PK, ATM and ATR in RPA
phosphorylation and checkpoint activation in response to
replication stress. Nucleic Acids Res. 40:10780–10794. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Olson E, Nievera CJ, Klimovich V, Fanning
E and Wu X: RPA2 is a direct downstream target for ATR to regulate
the S-phase checkpoint. J Biol Chem. 281:39517–39533. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maréchal A and Zou L: RPA-coated
single-stranded DNA as a platform for post-translational
modifications in the DNA damage response. Cell Res. 25:9–23. 2015.
View Article : Google Scholar :
|
|
30
|
Vassin VM, Wold MS and Borowiec JA:
Replication protein A (RPA) phosphorylation prevents RPA
association with replication centers. Mol Cell Biol. 24:1930–1943.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu X, Shell SM and Zou Y: Interaction and
colocalization of Rad9/Rad1/Hus1 checkpoint complex with
replication protein A in human cells. Oncogene. 24:4728–4735. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu X, Yang Z, Liu Y and Zou Y:
Preferential localization of hyperphosphorylated replication
protein A to double-strand break repair and checkpoint complexes
upon DNA damage. Biochem J. 391:473–480. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kubota T, Nishimura K, Kanemaki MT and
Donaldson AD: The Elg1 replication factor C-like complex functions
in PCNA unloading during DNA replication. Mol Cell. 50:273–280.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee KY, Fu H, Aladjem MI and Myung K:
ATAD5 regulates the lifespan of DNA replication factories by
modulating PCNA level on the chromatin. J Cell Biol. 200:31–44.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bylund GO and Burgers PM: Replication
protein A-directed unloading of PCNA by the Ctf18 cohesion
establishment complex. Mol Cell Biol. 25:5445–5455. 2005.
View Article : Google Scholar : PubMed/NCBI
|