Introduction
Melanoma is the most serious type of skin cancer,
with an increasing incidence worldwide (1). The transition from the radial growth
phase to the vertical growth phase is responsible for the majority
of cancer-associated mortalities caused by melanoma (2). Sun exposure and acquired genetic
alterations are two well-established etiologies for melanoma, and
previous studies have revealed that congenital germline mutation of
certain critical genes, such as cyclin-dependent kinase inhibitor
2A, is another risk factor (3–5). The
diagnostic technology and health consciousness regarding melanoma
have greatly improved in recent years and, furthermore, the
surgical techniques and adjuvant treatment measures have progressed
(6,7). However, the survival rate of patients
with melanoma remains a cause for concern, and the majority of
patients will exhibit recurrence within 5 years (8). Thus, effective therapeutic measures
based on novel molecular targets that are involved in the progress
of melanoma require investigation.
Histone arginine methylation is a dynamic process
that transfers a methyl group from S-adenosylmethionine (AdoMet) to
the guanidine group of targeted arginine residues (9). Protein arginine methylation regulates
multiple biological processes, including gene transcription and
signaling transduction (10). This
modification process is mainly catalyzed by nine protein arginine
methyltransferases (PRMTs) (11).
Dysregulation of PRMTs has previously been identified in several
human types of tumor (11). PRMT5
was previously identified as a prognostic marker of lung cancer,
which regulates tumor growth and the cell cycle (12). Additionally, PRMT2 regulates the
activity of E2F transcription factor and the estrogen receptor α
status in breast cancer (13,14).
Regarding melanoma, overexpression of PRMT5 was correlated with
tumor progression and metastasis; furthermore, PRMT5 was
demonstrated to regulate the level of microphthalmia-associated
transcription factor and the cell cycle regulator, cyclin-dependent
kinase inhibitor 1B (p27) (15,16).
However, whether PRMT1 regulates the aggressive behavior of human
melanoma remains elusive. The present study aimed to clarify the
role of PRMT1 in melanoma and to explore the potential mechanisms
involved.
Materials and methods
Ethical statement
All specimens were obtained with the informed
consents of patients prior to surgery. All human and animal studies
had been approved by the Ethical Committee of Shandong Provincial
Hospital, Shandong University School of Medicine (Jinan, China),
and were therefore performed in accordance with the ethical
standards of the 1964 Declaration of Helsinki and its later
amendments.
Clinical specimens and cell culture
Melanoma specimens and adjacent normal tissues
(n=28) were obtained from Shandong Provincial Hospital (Jinan,
China). Human melanoma cell lines, A375, Hs294T and SK-MEL-28, and
human immortalized keratinocytes (HaCaTs) and immortalized human
melanocyte cell line, PIG1 (control cell lines), were obtained from
the American Type Culture Collection (Manassas, VA, USA). All cells
were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10%
fetal bovine serum (FBS) at 37°C in a 5% CO2
atmosphere.
Lentiviral transfection for stable
expression clones
Lenti-virus products expressing short hairpin RNA
targeting (shPRMT1) or negative control (shNC) were purchased from
GeneChem Co., Ltd. (Shanghai, China). The interference effect of
three shRNAs was verified and the effective interference shRNA was
used for silencing PRMT1. The sequence in the PRMT1 gene recognized
by shPRMT1 is 5′-GAC ATG ACA TCC AAA GACT-3′. Stable cell lines
were obtained according to the manufacturer's infection and
selection protocol.
Immunoprecipitation (IP), liquid
chromatography-mass spectrometry (LC-MS) and western blot
analysis
IP was conducted using protein G-agarose (EMD
Millipore, Billerica, MA, USA). LC-MS was used to detect proteins
that interact with PRMT1. For western blot analysis, cells were
lysed with radioimmunoprecipitation assay buffer (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China) supplemented
with a protease inhibitor cocktail (Pierce; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and protein concentration was
determined by Bradford assay. Protein lysates (20 μg) were
separated by sodium dodecyl sulfate polyacrylamide gel
electrophoresis and resulting gels were transferred to a
polyvinylidene difluoride membrane (EMD Millipore). Indicated
primary antibodies were incubated with the membranes overnight at
4°C following blocking with skimmed milk powder for 2 h at room
temperature. Protein bands were visualized using Pierce ECL Western
Blotting Substrate (Pierce; Thermo Fisher Scientific, Inc.)
following incubation with corresponding horseradish peroxidase
(HRP)-conjugated antibodies. An automated chemiluminescence image
analysis system (Tanon 6200; Tanon Science and Technology Co.,
Ltd., Shanghai, China) was used for protein visualization and to
measure the optical density of protein bands. The primary
antibodies used for experiments were as follows: anti-GAPDH (cat.
no. KC-5G4; Kangchen Biotech), polyclonal rabbit anti-PRMT1 (cat.
no. ab73246; Abcam, Cambridge, UK) and monoclonal rabbit
anti-activated leukocyte cell adhesion molecule (ALCAM; cat. no.
ab109215; Abcam). The corresponding anti-rabbit-HRP (cat. no.
AP307P) and anti-mouse-HRP (cat. no. AP181P) antibodies (EMD
Millipore) were used as secondary antibodies. All antibodies were
diluted according to the manufacturer's instructions.
Cell proliferation assay
For the cell proliferation assay, cells were plated
in 96-well plates at a density of 800 cells/well. Cell Counting
Kit-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan)
was used to measure cell proliferation according to the
manufacturer's protocol, and an Epoch microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA) was used to detect optical
density values at 450 nm at the same time point of each day for 6
days.
In vivo tumor growth assay
A375/shNC or A375/shPRMT1 cells (1×106 in
0.2 ml sterilized phosphate-buffered saline) were implanted
subcutaneously into the right flank of 4-week-old male BALB/c nude
mice (five mice per group; weight, ~20 g). The mice were maintained
in a specific pathogen-free environment at 20–22°C (12-h light/dark
cycle) and provided with ad libitum access to food and
autoclaved water. Tumor volume was calculated every 7 days using
the following formula: V=(length) × (width) × (width)/2. The
subcutaneous tumors were weighed following excision. The mice were
housed in a specific pathogen-free environment and all experiments
were performed in accordance with the official recommendations of
the Chinese animal community.
Colony formation assay
For colony formation assays, cells from each
transfection group in the logarithmic growth phase were incubated
in 6-well plates at a density of 1,000 cells/well for 14 days and
clones were stained with 1% crystal violet for counting.
Cell migration assay
Cell migration ability was assessed by Transwell
assay (Corning Incorporated, Corning, NY, USA). Briefly,
1×105 cells from each group were added into the upper
chamber of the Transwell containing 200 μl serum-free
RPMI-1640 medium with tumor supernatant of each group and 600
μl DMEM containing 10% FBS was added into lower chamber.
Following incubation at 37°C and 5% CO2 for 24 h, cells
that migrated to the underside of upper chamber were fixed with 1%
methanol for 1 h at room temperature, then stained by 0.1% crystal
violet, and counted in 5 random fields using a light
microscope.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and qPCR array
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for total RNA extraction from cell
lines. cDNA was synthesized from 1 μg of total RNA using a
two-step method with a ReverTra Ace qPCR RT kit (Toyobo Co., Ltd.,
Osaka, Japan) according to the manufacturer's protocol. SYBR Green
PCR Core reagent kit and ABI 7900 real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) were used for qPCR
reactions. The primers used are as follows: PRMT1, forward
5′-CTTTGACTCCTACGCACACTT-3′, and reverse
5′-GTGCCGGTTATGAAACATGGA-3′; GAPDH, forward
5′-AGCCACATCGCTCAGACAC-3′, and reverse 5′-GCCCAATACGACCAAATCC-3′.
Total RNA from the A375, A375/shNC and A375/shPRMT1 cells were used
to perform a qPCR array by wcgene (Shanghai, China) according to
the manufacturer's protocol. The relative mRNA levels were
evaluated using the 2−ΔΔCq method (17).
Transient transfection
pcDNA3.1-EGFP-ALCAM or control plasmids (GeneChem,
Inc., Daejeon, Korea) were transiently transfected into
A375/shPRMT1 or Hs294T/shPRMT1 cells. Small interfering RNA (siRNA)
targeting ALCAM (Shanghai GenePharma Co., Ltd., Shanghai, China)
was employed for RNA interference (RNAi) experiments. The sequence
of the ALCAM targeting siRNA was 5′-GUAAUUGUCCACUGAAUGGCUGGC-3′
(sense). Briefly, cells were seeded in 6-well plates at a density
of 3×105 cells/well and transfected with the
corresponding plasmids/siRNA using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Western blotting was performed to determine the
transfection efficiency. All functional studies were performed
following transfection for 48 h.
Statistical analysis
Student's t-test and one-way analysis of variance
were used where appropriate. Data are presented as the mean ±
standard deviation from three independent experiments. SPSS
software (version 15.0; SPSS, Inc., Chicago, IL, USA) was used to
perform statistical analyses. P<0.05 was considered to indicate
a statistically significant difference.
Results
Identification of PRMT1 as a potential
biomarker by screening the Oncomine database
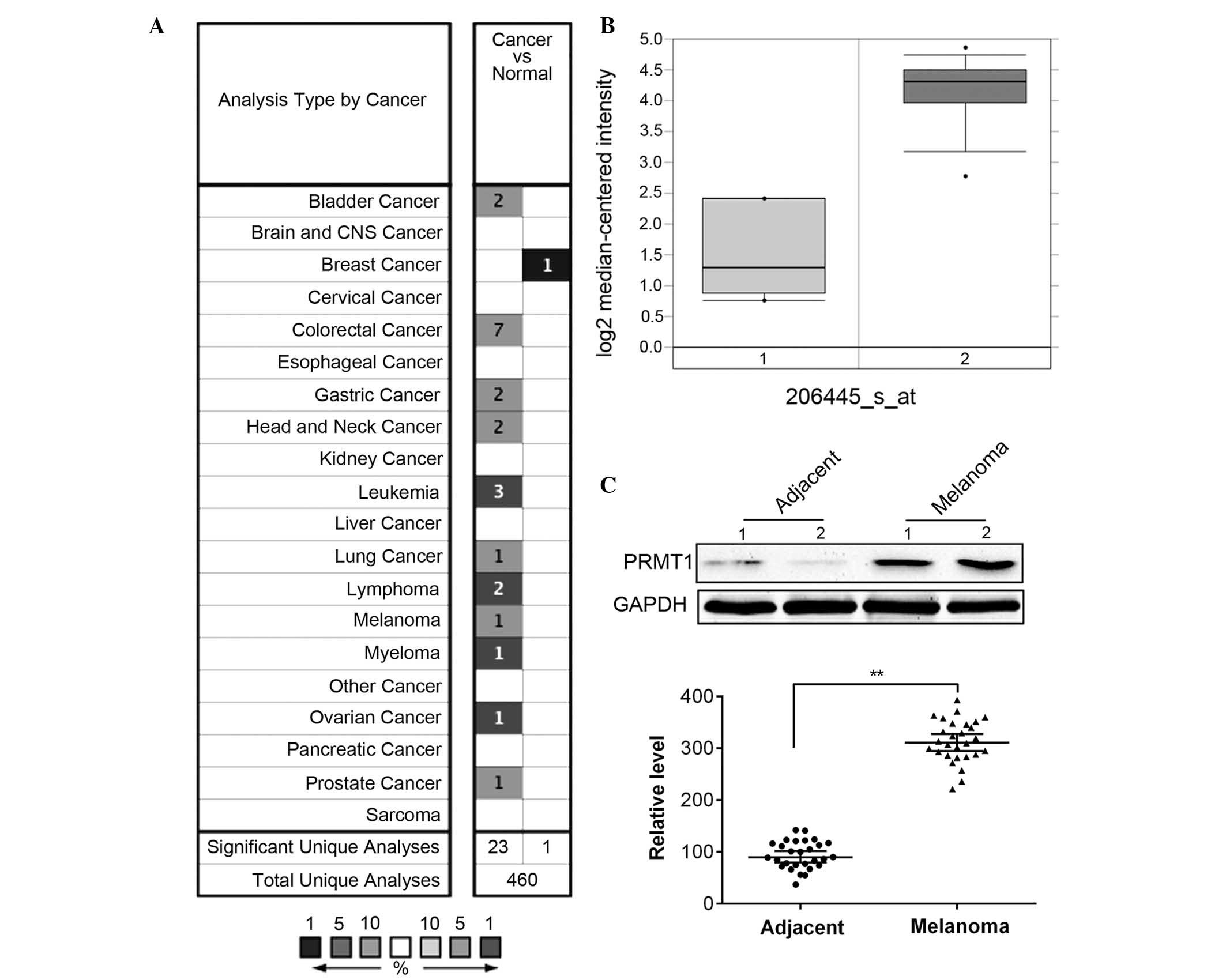
Using the Oncomine database (www.oncomine.org), overexpression of PRMT1 was
demonstrated in 11 out of 20 types of cancer types compared with
normal tissue, whereas downregulation of PRMT1 was only
demonstrated in breast cancer (Fig.
1A). In melanoma, PRMT1 was significantly overexpressed in
Talantov's dataset (Fig. 1B).
Furthermore, the protein level of PRMT1 in 28 melanoma tissues and
paired non-tumor tissues was determined. The present study
demonstrated that the protein expression level of PRMT1 was
significantly upregulated in melanoma compared with adjacent normal
tissues (P<0.01; Fig. 1C).
PRMT1 regulates cell proliferation in
vitro and in vivo
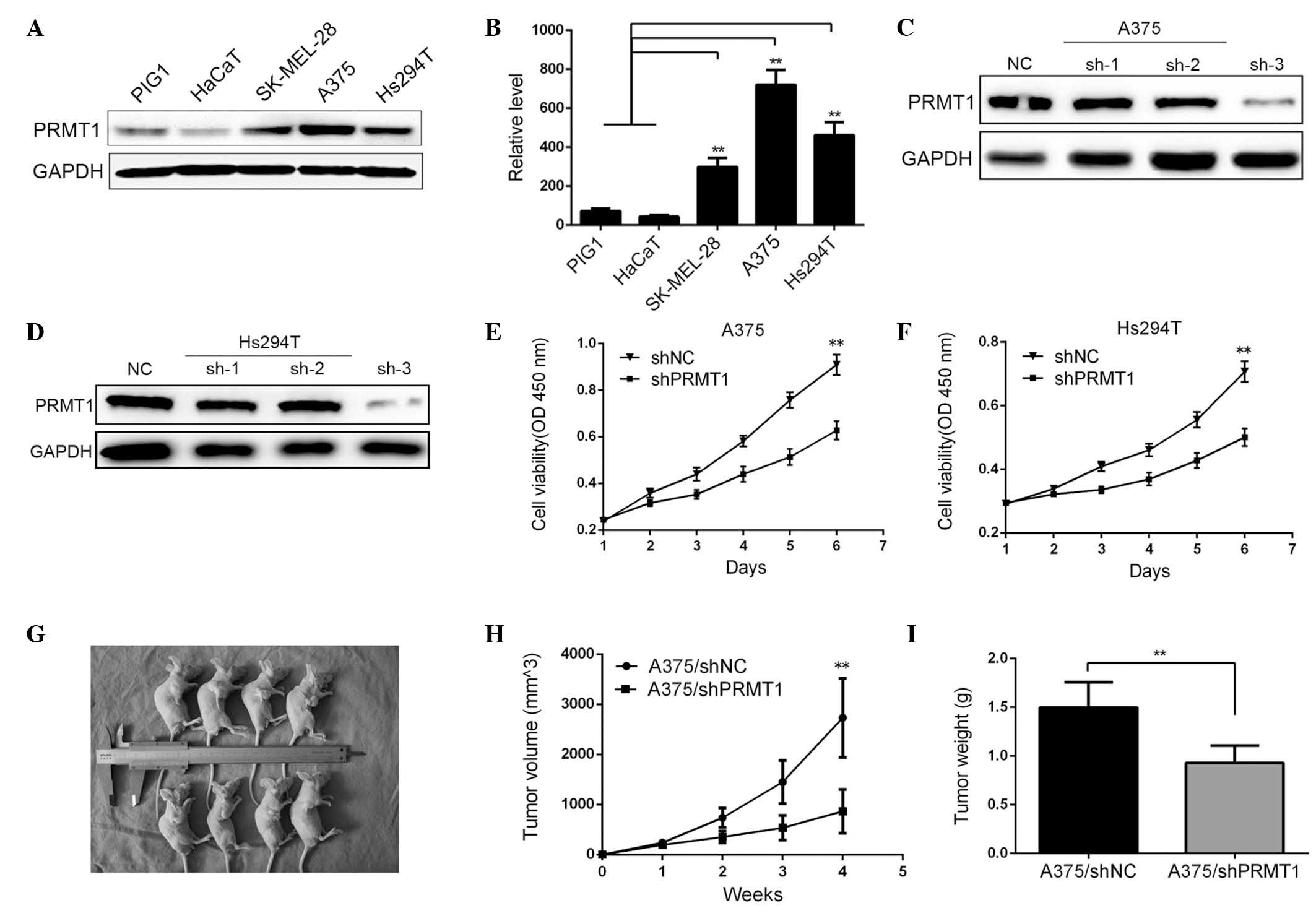
The protein expression level of PRMT1 was determined
by western blotting in several melanoma cell lines. It was
demonstrated that the PRMT1 level was significantly higher in three
melanoma cell lines (SK-MEL-28, A375 and Hs294T) compared with
HaCaTs or immortalized human melanocyte cell line, PIG1 (P<0.01;
Fig. 2A and B). Additionally, the
effect of PRMT1 silencing on cell proliferation was analyzed. Of
the three different shRNAs used, western blotting demonstrated that
sh-3 yielded an efficient silencing effect on PRMT levels (Fig. 2C and D). The proliferative ability
of A375 and Hs294T cells were determined at the same point on each
day for 6 days. As indicated in Fig.
2E and F, silencing of PRMT1 significantly inhibited the
proliferation of the cell lines (P<0.01). As PRMT1 was
demonstrated to regulate in vitro cell proliferation, the
effect of PRMT1 on tumor growth in vivo was investigated.
A375 cells of each group were subcutaneously injected into nude
mice. As demonstrated in Fig.
2G–I, knockdown of PRMT1 in A375 cells reduced tumor size and
tumor growth compared with A375/shNC cells.
PRMT1 regulates colony formation,
metastasis and multiple downstream cancer-associated genes
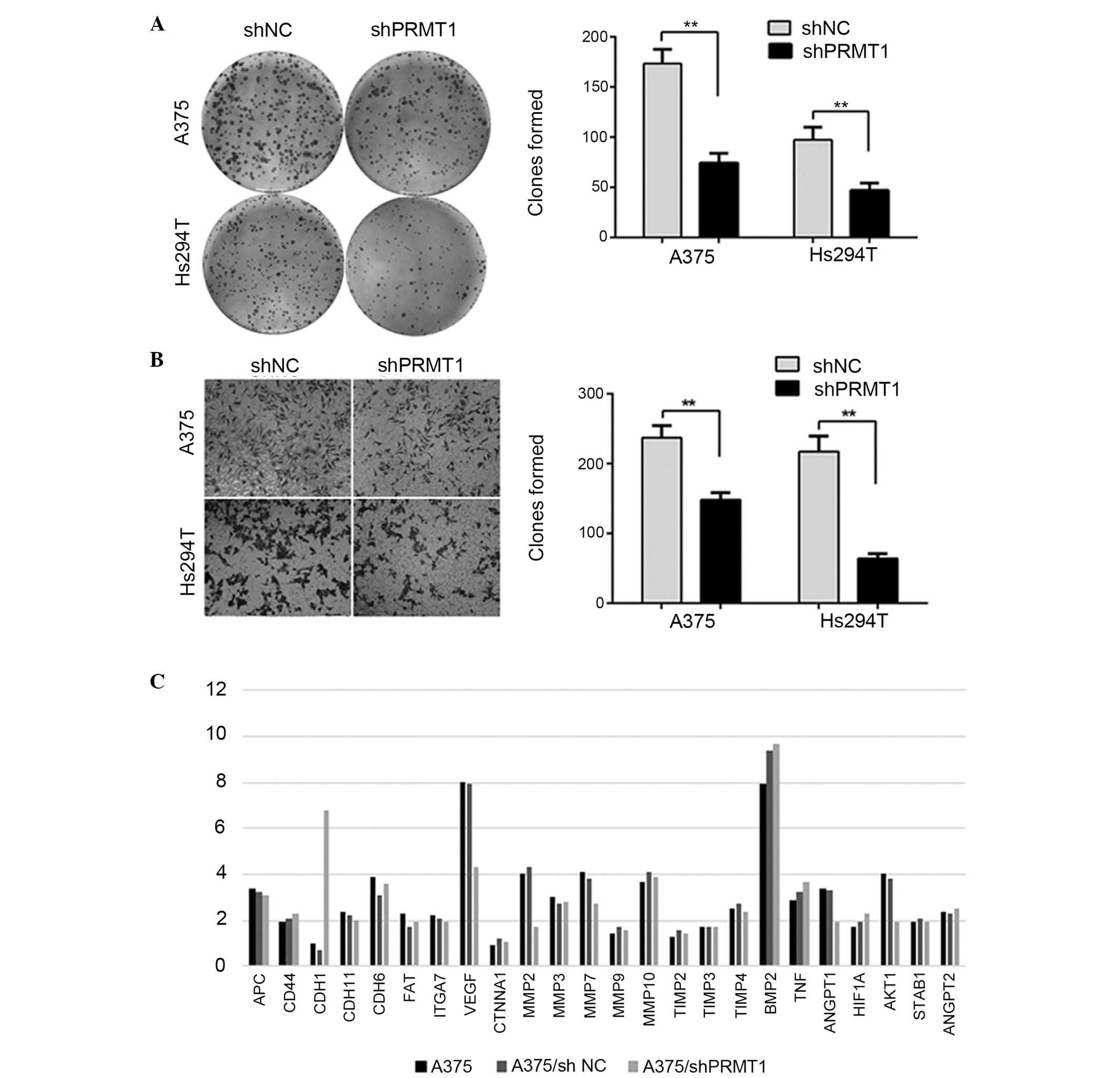
Plate colony formation assay was performed to
determine the regulatory effect of PRMT1 on colony formation. As
demonstrated in Fig. 3A, cells
transfected with shNC exhibited significantly increased colony
formation compared with the shPRMT1 group (P<0.01). Furthermore,
the effect of PRMT1 on migration was investigated. A migration
assay was performed using A375 and Hs294T cells transfected with
shPRMT1 or shNC. As demonstrated in Fig. 3B, knockdown of PRMT1 significantly
reduced cell migration through Transwell chambers (P<0.01). To
understand the potential mechanisms that mediate the effect of
PRMT1 on tumor growth and metastasis, a qPCR array including 24
genes was performed to investigate the changes that occur following
PRMT1 silencing. Compared with shNC cells, A375/shPRMT1 cells
exhibited significantly increased mRNA levels of cadherin 1 (CDH1),
which is a well-known metastatic inhibitor. However, the mRNA
levels of vascular endothelial growth factor (VEGF), matrix
metalloproteinase 2 (MMP2), v-akt murine thymoma viral
oncogene homolog 1 (AKT1) and angiopoietin 1 (ANGPT1) were
downregulated in A375/shPRMT1 cells compared with A375/shNC cells
(fold change, >1.5; Fig. 3C).
VEGF and ANGPT1 are important angiogenic factors involved in tumor
growth and metastasis of human melanoma. MMP2 is a member of the
MMP protein family involved in angiogenesis, wound healing, tumor
invasion and fibrosis. AKT1 is an important protein of the
phosphatidylinositol-4,5-bisphosphate 3-kinase/AKT signaling
pathway. Taken together, the changes in the mRNA levels of these
genes provided a preliminary mechanism of PRMT1-induced tumor
growth.
PRMT1 is a direct regulator of ALCAM and
influence multiple aggressive behaviors
The current study demonstrated that PRMT1 regulates
tumor progression and multiple downstream metastasis-associated
genes; thus, proteins that interact with PRMT1 were further
investigated using LC-MS and co-IP. The results of the present
study indicated that ALCAM, a well-established oncogene in multiple
human tumors, including human melanoma, interacts with PRMT1, as
demonstrated by LC-MS and validated by co-IP (Fig. 4A and B). Additionally the current
study investigated whether ALCAM is an upstream regulator of PRMT1
or a downstream co-effector. Silencing of PRMT1 expression in A375
and Hs294T cells downregulated the protein level of ALCAM compared
with shNC (Fig. 4C). However,
silencing of ALCAM using siRNA did not observably affect PRMT1
expression in A375 and Hs294T cells (Fig. 4D). These results indicated that
ALCAM is a downstream target of PRMT1. Furthermore, silencing ALCAM
inhibited cell proliferation (data not shown), which was in
accordance with the phenotypes induced by silencing of PRMT1. To
explore whether ALCAM was involved in PRMT1-induced tumor growth
and metastasis, an ALCAM overexpression vector,
pcDNA3.1-EGFP-ALCAM, was transfected into A375/shPRMT1 and
Hs294T/shPRMT1 cells. ALCAM was fused with enhanced green
fluorescent protein and expressed as a fusion protein. Western blot
analysis demonstrated that transfection of A375/shPRMT1 with
pcDNA3.1-EGFP-ALCAM resulted in overexpression of ALCAM (Fig. 4E). The functional analysis
indicated that re-expression of ALCAM in shPRMT1 cells reversed the
effects of PRMT1 silencing on the colony formation and migration
ability compared with cells not transfected with the ALCAM
expression plasmid (Fig. 4F–I).
These results indicated that PRMT1 regulates tumor progression and
metastasis via directly targeting ALCAM.
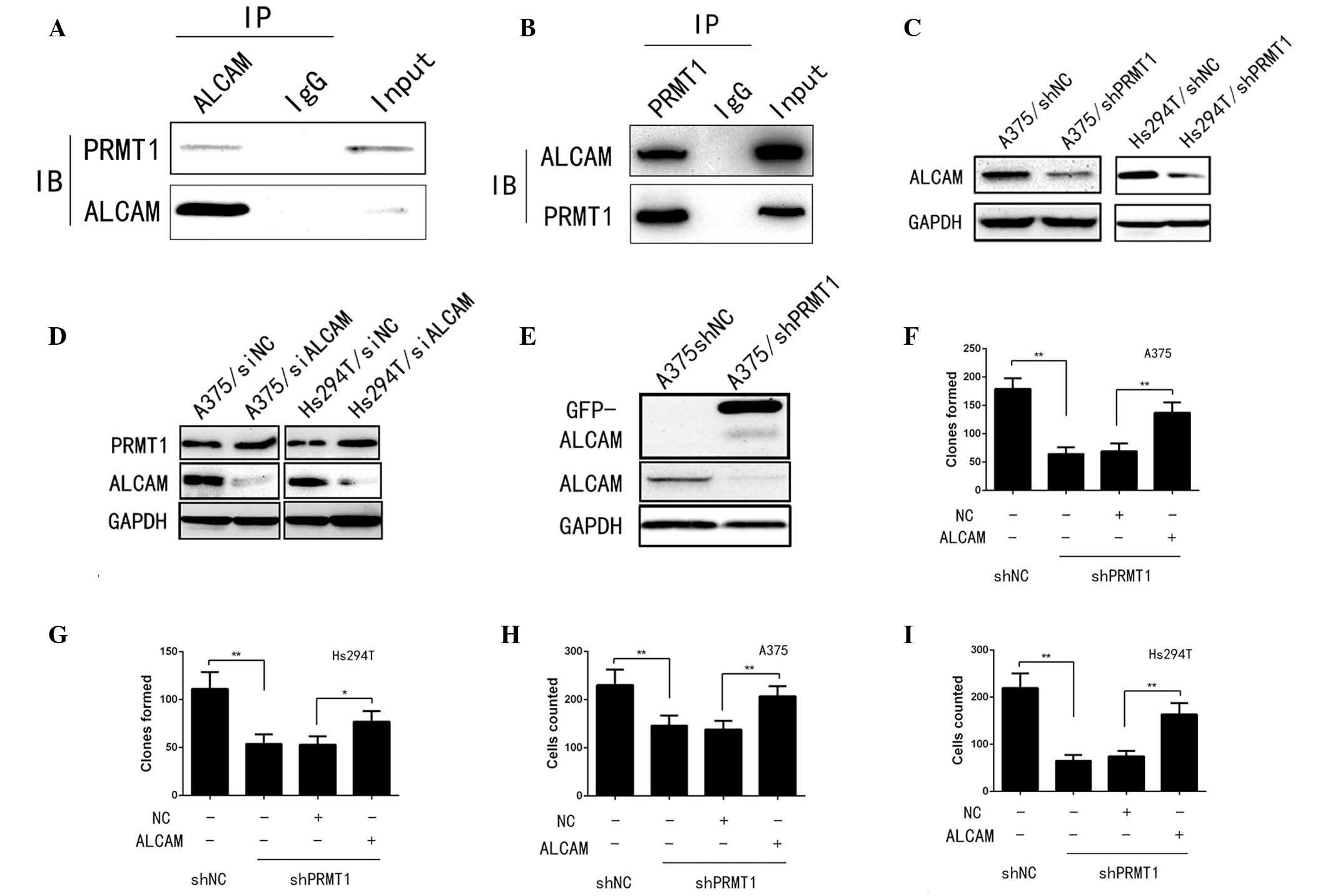 | Figure 4ALCAM was a direct target of PRMT1 and
mediated multiple aggressive behaviors induced by PRMT1. (A and B)
The interaction between PRMT1 and ALCAM was confirmed by co-IP. (C
and D) Silencing PRMT1 in A375 and Hs294T cells could significantly
downregulate ALCAM level while silencing ALCAM in A375 and Hs294T
cells did not effect PRMT1 level. (E) pcDNA3.1-EGFP-ALCAM was
transfected into A375/shPRMT1 cells and the transfection effect was
confirmed by western blotting. (F and G) Colony formation ability
of A375/shPRMT1 and Hs294T/shPRMT1 cells was significantly
increased after ALCAM restoration. *P<0.05,
**P<0.01, comparison indicated by brackets. (H and I)
Migration ability of A375/shPRMT1 and Hs294T/shPRMT1 cells was
significantly reversed after ALCAM re-expression.
**P<0.01, comparison indicated by brackets. IP,
immunoprecipitation; ALCAM, activated leukocyte cell adhesion
molecule; IgG, immunoglobulin G; IB, immunoblot; PRMT, protein
arginine methyltransferase; sh, short hairpin RNA; NC, negative
control; si, small interfering RNA. |
Discussion
The carcinogenesis of human melanoma involves
complex genomic alterations and molecular processes (18). Typically, a malignant tumor
undergoes the following stages during development: Precancerous
stage; carcinoma in situ; and metastasis. For human
melanoma, lymphatic vessel density in primary melanomas is
correlated with the risk of metastasis in patients (19,20).
Furthermore, previous studies have preliminarily investigated the
mechanisms that regulate the aggressive behavior of human melanoma,
particularly the oncogenic function of several epigenetic
regulators, including PRMTs (21–23).
However, accurate molecular markers, and effective targets to
clarify the clinical and biological features of melanoma, remain an
urgent requirement.
Overexpression of PRMTs had been demonstrated in
various types of human tumor (24). In the present study, screening of
an online database (Oncomine) demonstrated that PRMT1 is
overexpressed in human melanoma, which subsequently was confirmed
by the experimental results. Thus, the biological function of PRMT1
in A374 and Hs294T cells was investigated. The current study
demonstrated that silencing PRMT1 significantly inhibited tumor
growth in vitro and in vivo, which is in accordance
with the results of colony formation assays. Furthermore, PRMT1
silencing inhibited the metastatic potential of A375 and Hs294T
cells, demonstrated by migration assay. To preliminarily
investigate the altered phenotype following PRMT silencing, a qPCR
array containing 26 genes was performed to detect potential
downstream metastasis-associated genes. The mRNA level of CDH1, a
metastatic inhibitor, was downregulated, whereas the mRNA level of
VEGF, MMP2, AKT1 and ANGPT1 were upregulated following silencing of
PRMT1 in A375 cells. These results provided direct evidence that
PRMT1 regulates multiple downstream tumorigenesis markers.
Additionally, the mechanisms mediate PRMT1-regulated
tumor growth and metastasis were investigated. Protein was
extracted from A375/shNC and A375/shPRMT1 cells and
immunoprecipitated with anti-PRMT1 antibody for LC-MS analysis.
LC-MS demonstrated that ALCAM may be a potential downstream
co-effector of PRMT1. The negative association between the ALCAM
level and the survival status of patients with colorectal cancer
and esophageal squamous cell carcinoma has been previously reported
(25,26). In human melanoma, the ALCAM level
was previously demonstrated to be correlated with tumor
progression, and regulates melanoma cell clustering and migration
(27–29). The association between PRMT1 and
ALCAM was demonstrated by co-IP. Furthermore, the current study
demonstrated that silencing of PRMT1 in A375 cells significantly
downregulates the ALCAM level. Furthermore, A375/shPRMT1 and
Hs294T/shPRMT1 cells were transfected with pcDNA3.1-EGFP-ALCAM to
determine whether ALCAM restoration affects the phenotypes induced
by PRMT1 silencing. Notably, cells in the shPRMT1 group exhibited
significantly increased colony formation and metastatic ability
following restoration of ALCAM expression. Taken together, the
results of the present study demonstrate that ALCAM is a direct
target of PRMT1 and involved in PRMT1-induced tumorigenesis.
In conclusion, the results of the current study
indicate that PRMT1 is overexpressed in human melanoma, and that
PRMT1 may regulate tumor growth and metastasis via targeting ALCAM.
The PRMT1-ALCAM axis may be a novel anti-cancer target for the
treatment of human melanoma.
Acknowledgments
This study was supported by Medical Science and
Technology Project in Henan province (grant. no. 201403165).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Itakura E, Huang RR, Wen DR, Paul E,
Wünsch PH and Cochran AJ: IL-10 expression by primary tumor cells
correlates with melanoma progression from radial to vertical growth
phase and development of metastatic competence. Mod Pathol.
24:801–809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gandini S, Sera F, Cattaruzza MS, Pasquini
P, Picconi O, Boyle P and Melchi CF: Meta-analysis of risk factors
for cutaneous melanoma: II. Sun exposure. Eur J Cancer. 41:45–60.
2005. View Article : Google Scholar
|
|
4
|
Shi H, Hugo W, Kong X, Hong A, Koya RC,
Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, et al:
Acquired resistance and clonal evolution in melanoma during BRAF
inhibitor therapy. Cancer Discov. 4:80–93. 2014. View Article : Google Scholar :
|
|
5
|
Puig-Butille JA, Escámez MJ, Garcia-Garcia
F, Tell-Marti G, Fabra À, Martínez-Santamaría L, Badenas C,
Aguilera P, Pevida M, Dopazo J, et al: Capturing the biological
impact of CDKN2A and MC1R genes as an early predisposing event in
melanoma and non melanoma skin cancer. Oncotarget. 5:1439–1451.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garbe C, Peris K, Hauschild A, Saiag P,
Middleton M, Spatz A, Grob JJ, Malvehy J, Newton-Bishop J,
Stratigos A, et al: Diagnosis and treatment of melanoma: European
consensus-based interdisciplinary guideline. Eur J Cancer.
46:270–283. 2010. View Article : Google Scholar
|
|
7
|
Loquai C, Schmidtmann I, Beutel M,
Sunderkötter C, Grabbe S, Schiller M and Nashan D: Quality of life
in melanoma patients during adjuvant treatment with pegylated
interferon-α2b: Patients' and doctors' views. Eur J Dermatol.
21:976–984. 2011.PubMed/NCBI
|
|
8
|
Dillman RO, DePriest C, Ellis R and de
Leon C: 5-year survival for patients with metastatic melanoma who
had no evidence of disease at time of treatment with patient
specific tumor stem cell vaccines. Cancer Research. 74:1972014.
View Article : Google Scholar
|
|
9
|
Di Lorenzo A and Bedford MT: Histone
arginine methylation. FEBS Lett. 585:2024–2031. 2011. View Article : Google Scholar
|
|
10
|
Wysocka J, Allis CD and Coonrod S: Histone
arginine methylation and its dynamic regulation. Front Biosci.
11:344–355. 2006. View
Article : Google Scholar
|
|
11
|
Yang Y and Bedford MT: Protein arginine
methyltransferases and cancer. Nat Rev Cancer. 13:37–50. 2013.
View Article : Google Scholar
|
|
12
|
Gu Z, Gao S, Zhang F and Wang Z, Ma W,
Davis RE and Wang Z: Protein arginine methyltransferase 5 is
essential for growth of lung cancer cells. Biochem J. 446:235–241.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoshimoto T, Boehm M, Olive M, Crook MF,
San H, Langenickel T and Nabel EG: The arginine methyltransferase
PRMT2 binds RB and regulates E2F function. Exp Cell Res.
312:2040–2053. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM
and Zhu YJ: Identification of protein arginine methyltransferase 2
as a coactivator for estrogen receptor alpha. J Biol Chem.
277:28624–28630. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nicholas C, Yang J, Peters SB, Bill MA,
Baiocchi RA, Yan F, Sïf S, Tae S, Gaudio E, Wu X, et al: PRMT5 is
upregulated in malignant and metastatic melanoma and regulates
expression of MITF and p27 (Kip1). PLoS One. 8:e747102013.
View Article : Google Scholar
|
|
16
|
Nicholas C, Yan F, Peters SB, Bill MA, Li
P, Li C, Fuchs JR, Baiocchi R and Lesinski GB: The expression of
PRMT5 methyltransferase mediates cell survival and metastatic
phenotype in malignant melanoma. Cancer Research. 71:9332011.
View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Hodis E, Watson IR, Kryukov GV, Arold ST,
Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C,
et al: A landscape of driver mutations in melanoma. Cell.
150:251–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shayan R, Karnezis T, Murali R, Wilmott
JS, Ashton MW, Taylor GI, Thompson JF, Hersey P, Achen MG, Scolyer
RA and Stacker SA: Lymphatic vessel density in primary melanomas
predicts sentinel lymph node status and risk of metastasis.
Histopathology. 61:702–710. 2012.PubMed/NCBI
|
|
20
|
Shields CL, Kaliki S, Furuta M, Fulco E,
Alarcon C and Shields JA: American Joint Committee on Cancer
classification of posterior uveal melanoma (tumor size category)
predicts prognosis in 7731 patients. Ophthalmology. 120:2066–2071.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chapman PB, Hauschild A, Robert C, Haanen
JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et
al: Improved survival with vemurafenib in melanoma with BRAF V600E
mutation. N Engl J Med. 364:2507–2516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin
L and Garraway LA: Highly recurrent TERT promoter mutations in
human melanoma. Science. 339:957–959. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krauthammer M, Kong Y, Ha BH, Evans P,
Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et
al: Exome sequencing identifies recurrent somatic RAC1 mutations in
melanoma. Nat Genet. 44:1006–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bao X, Zhao S, Liu T, Liu Y, Liu Y and
Yang X: Overexpression of PRMT5 promotes tumor cell growth and is
associated with poor disease prognosis in epithelial ovarian
cancer. J Histochem Cytochem. 61:206–217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weichert W, Knösel T, Bellach J, Dietel M
and Kristiansen G: ALCAM/CD166 is overexpressed in colorectal
carcinoma and correlates with shortened patient survival. J Clin
Pathol. 57:1160–1164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Verma A, Shukla NK, Deo SV, Gupta SD and
Ralhan R: MEMD/ALCAM: A potential marker for tumor invasion and
nodal metastasis in esophageal squamous cell carcinoma. Oncology.
68:462–470. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Degen WG, Van Kempen LC, Gijzen EG, van
Groningen JJ, van Kooyk Y, Bloemers HP and Swart GW: MEMD, a new
cell adhesion molecule in metastasizing human melanoma cell lines,
is identical to ALCAM (activated leukocyte cell adhesion molecule).
Am J Pathol. 152:805–813. 1998.PubMed/NCBI
|
|
28
|
Swart GW, Lunter PC, Kilsdonk JW and
Kempen LC: Activated leukocyte cell adhesion molecule
(ALCAM/CD166): Signaling at the divide of melanoma cell clustering
and cell migration? Cancer Metastasis Rev. 24:223–236. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lunter PC, Van Kilsdonk JW, Van Beek H,
Cornelissen IM, Bergers M, Willems PH, van Muijen GN and Swart GW:
Activated leukocyte cell adhesion molecule (ALCAM/CD166/MEMD), a
novel actor in invasive growth, controls matrix metalloproteinase
activity. Cancer Res. 65:8801–8808. 2005. View Article : Google Scholar : PubMed/NCBI
|