Introduction
Colorectal cancer (CRC) is the third most common
type of human malignancy, and the second leading cause of
cancer-associated mortality worldwide (1). The incidence of CRC has been
increasing in Asian countries, including China, Korea, Japan, and
Singapore. The reason for this increasing incidence is currently
unclear; however, demographic trends and adaptation to a
westernized lifestyle, particularly with regards to factors such as
diet, may have a role in the increased incidence of CRC in these
countries (2). During the past few
decades, there have been numerous advances in CRC research,
including early detection, prevention by adaptation to a healthier
lifestyle, and improved treatment strategies; however, a high
percentage of patients with CRC still develop advanced stages of
the disease and tumor metastasis (3). Therefore, the identification and
evaluation of biomarkers for the prediction of disease progression,
prognosis, and treatment response may help physicians effectively
treat and control the spread of cancer in these patients. To date,
clinicopathological characteristics, including CRC tumor-lymph
node-metastasis (TNM) stage, differentiation grade, lymph node
metastasis, invasion to other tissue or structures, and tumor size,
have been used to evaluate treatment selection and prognosis of
patients with CRC. Various biomarkers, such as carcinoembryonic
antigen (CEA) and carbohydrate antigen (CA)19–9, have previously
been identified and evaluated with regards to the diagnosis, and
prediction of tumor recurrence or treatment response (4–6).
However, these biomarkers are not without fault, due to the lack of
specificity and sensitivity; therefore, more research is required
to identify novel serum-based tumor markers that may be used screen
the general or high-risk population for early diagnosis of CRC, or
predict CRC prognosis and treatment response (4).
Kallikrein-related peptidase 5 (KLK5), which is one
of 15 kallikrein subfamily members, is a serine protease located on
chromosome 19 (7). Functionally,
KLK5 is usually expressed in the epidermis and regulates cell
shedding in conjunction with KLK7 and KLK14 (7–9).
KLK5 can become activated from the secreted pro-KLK5, in order to
activate several other KLKs, such as KLK2, -3, -6, -7, -11, -12 and
-14 (10). In addition, KLK5
proteolytic activity is able to target metal-loproteases and
extracellular matrix components, including collagens, fibronectin,
and laminin (11,12). Previous studies have demonstrated
that KLK5 is upregulated in numerous types of human cancer
(7,13–19).
Indeed, kallikrein-mediated extracellular proteolysis is important
in several facets of cancer development, including regulation of
tumor growth, invasion, metastasis, and angiogenesis (20). A previous study demonstrated that
altered expression of KLKs is associated with cancer development
and diagnosis, and may therefore be considered a useful prognostic
marker for prostate, breast, and ovarian cancer (13). In CRC, KLK5 protease accumulates in
tumor tissues and is associated with an unfavorable outcome for
patients (21). The present study
further assessed KLK5 expression in CRC tissue specimens and serum
samples from patients with CRC using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
immunohistochemistry, and enzyme-linked immunosorbent assay
(ELISA). In addition, the association between KLK5 expression and
clinicopathological parameters of the patients was determined.
Materials and methods
Study subjects
The present study collected 40 fresh CRC and paired
normal tissues from patients who had undergone surgical resection
of CRC lesions at the Department of Surgery, General Hospital of
Jinan Military Command (Jinan, China) between November 2006 and May
2012. Among these patients, there were 23 men and 17 women with a
median age of 58 years (range, 31–77 years). CRC was diagnosed and
graded according to the revised World Health Organization grading
system and was staged according to the Dukes' operative staging
system. In particular, 4/40 cases of CRC (10%) were
well-differentiated, 27/40 (67.5%) were moderately differentiated,
and 9/40 (22.5%) were poorly differentiated; whereas 10/40 cases of
CRC (25%) were at stage A, 11/40 (27.5%) were at stage B, 17/40
(42.5%) were at stage C, and 2/40 (5%) were at stage D (Dukes'
stages). A total of 17 patients exhibited regional lymph node tumor
metastasis, and 2 patients exhibited tumor metastasis to the
liver.
In addition, 48 paraffin-embedded CRC tissue samples
were obtained from 28 male and 20 female patients with a median age
of 60 years and a range of 22–80 years., and serum samples were
collected from an additional 70 patients with CRC (Table I), including 38 serum samples taken
pre- and post-surgery, and 53 healthy individuals from the same
hospital. The present study was approved by the Medical Ethics
Committee of the General Hospital of Jinan Military Command (Jinan,
China), and all participants provided written informed consent.
 | Table IAssociation of serum KLK5 levels with
the clinicopathological features of colorectal cancer. |
Table I
Association of serum KLK5 levels with
the clinicopathological features of colorectal cancer.
| Characteristics | Total number | Serum KLK5 level
| P-value |
|---|
| High | Low/negative |
|---|
| Gender | | | | 0.092 |
| Male | 44 | 33 | 11 | |
| Female | 26 | 17 | 9 | |
| Age | | | | 0.720 |
| <60 years
old | 33 | 24 | 9 | |
| ≥60 years old | 37 | 26 | 11 | |
| Tumor site | | | | 0.700 |
| Rectum | 49 | 35 | 14 | |
| Colon | 21 | 15 | 6 | |
| Differentiation | | | | 0.890 |
| Well/moderate | 51 | 36 | 15 | |
| Poor | 11 | 6 | 5 | |
| Tumor size | | | | 0.874 |
| <5 cm | 32 | 24 | 8 | |
| ≥5cm | 30 | 23 | 7 | |
| Dukes' stage | | | | 0.005 |
| A/B | 37 | 23 | 14 | |
| C/D | 28 | 25 | 3 | |
| TNM stage | | | | 0.004 |
| I/II | 40 | 26 | 14 | |
| III/IV | 26 | 23 | 3 | |
| Metastasis | | | | 0.003 |
| Absent | 37 | 23 | 14 | |
| Present | 32 | 23 | 9 | |
RT-qPCR
Total RNA was isolated from the 40 paired normal and
tumor frozen tissues using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. Frozen samples (100 mg) were homogenated
in 1 ml TRIzol, RNA quality and quantity were measured using a
LAMBDA Bio UV/VIS Spectrophotometer (PerkinElmer, Inc., Waltham,
MA, USA), and RNA was reverse transcribed into cDNA using a
real-time qPCR kit (Takara Biotechnology Co., Ltd., Dalian, China),
according to the manufacturer's protocol. The SYBR Green qPCR assay
was performed to amplify the cDNA samples using a Roche LightCycler
480 II (Roche Diagnostics, Basel, Switzerland). The primers used
were as follows: KLK5, forward 5′-AAGGTCCTCCAGTGCTTGAA-3′, reverse
5′-CCAACAGACCGGGTGTCTAC-3′ (synthesized in our laboratory); and
GAPDH, forward 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse
5′-GAAGATGGTGATGGGATTTC-3′ (Sango Biotech, Shanghai, China). The
qPCR 25-µl reaction mixture consisted of 12.5 µl 2X
SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.), 1
µl each primer (100 nM), 1 µl cDNA and RNase free
H2O up to 25 µl. The qPCR conditions were set as
follows: Initial denaturation at 95°C for 5 min, followed by
40 cycles at 95°C for 5 sec and 60°C for 10 sec, the
temperature was then increased from 65 to 92°C to obtain the
melting curve, which was used to distinguish specific products from
non-specific products or primer dimers. The relative mRNA
expression levels of KLK5 were determined by normalization to the
endogenous control, GAPDH mRNA, and were calculated using the
2−ΔΔCq method as follows: ΔCq = Cq (mRNA of KLK5) − Cq
(mRNA of GAPDH). The Cq value was the threshold cycle at which
fluorescence was detected. Each sample was measured in duplicate
and repeated at least once. The KLK5 and GAPDH qPCR products
subsequently underwent 1.5% agarose gel electrophoresis, and were
visualized by ethidium bromide staining in order to confirm product
size.
Immunohistochemistry
The indirect immunoperoxidase method was used to
analyze the KLK5 expression in archived formalin-fixed,
paraffin-embedded samples from 48 CRC and matched tumor-free
tissues. These tissue samples were used to confirm the RT-qPCR
results. For the immunohistochemistry experiments, tissue sections
(4 µm) were cut from the paraffin blocks, deparaffinized
twice in warm xylene (5 min each), and rehydrated through a series
of graded alcohol solutions. Endogenous peroxidase activity was
blocked with 3% H2O2 in methanol for 10 min,
and with normal serum for 30 min at room temperature. Subsequently,
tissue sections were incubated with 200 µl KLK5 primary
antibody at a dilution of 1:400 in phosphate-buffered saline (PBS)
at 4°C overnight. This KLK5 rabbit polyclonal antibody was
generated in our laboratory using recombinant KLK5 protein
(22). Immunohistochemical
staining of antibodies was performed using the Dako K5007 Envision
Plus System (Dako, Glostrup, Denmark). The antibody binding was
visualized with a 3,3′-Diaminobenzidine staining prior to a brief
counterstaining with Mayer's hematoxylin. In the negative control
experiments, the primary antibody was replaced with antibody
diluent. The stained tissue sections were reviewed and scored under
an Olympus BX53 microscope (Olympus Corporation, Tokyo, Japan)
using a semi-quantitative scoring system for both the intensity of
the stain and the percentage of positive neoplastic cells (23).
ELISA
A monoclonal anti-KLK5 antibody generated in our
laboratory (22) was used to coat
96-well white polystyrene plates in PBS (750 ng/well) overnight at
4°C. Subsequently, the plates were washed three times with
PBS plus 0.5% Tween 20 (PBS-T), and diluted serum samples or
calibrators (100 µl/well; Sino Biological, Inc., Beijing,
China) were added to each well and incubated at 37°C for 1
h. The plate was then washed a further six times with PBS-T.
Subsequently, 100 µl horseradish peroxidase-conjugated
rabbit anti-KLK5 antibody (1 mg/ml, diluted 1,000-fold in PBS;
generated in our laboratory) was added to each well and incubated
at 37°C for 40 min. After washing six times with PBST, 100
µl chromogen (Tetramethylbenzidine) was added to each well
and the plates were incubated at 37°C for 10 min. After
quenching the reaction with 50 µl 2 M
H2SO4, the absorbance (optical density) was
measured at 450 nm using a SpectraMax M2 Multi-Mode microplate
reader (Molecular Devices LLC, Sunnyvale, CA, USA).
Statistical analysis
All statistical analyses were performed using the
SPSS software package, version 17.0 (SPSS Inc., Chicago, IL, USA).
The serum KLK5 levels in healthy individuals compared with patients
with CRC and the differences in various stages were analyzed with
the Mann-Whitney U test. The serum KLK5 levels were analyzed by
Wilcoxon test in patients prior to surgery and were compared with
results following the surgery. The differences in expression of
KLK5 mRNA in CRC and normal tissues were analyzed by Wilcoxon test.
Receiver operating characteristic (ROC) curves were performed to
analyze the data of CEA and serum KLK5 levels. The ROC curves were
delineated using Sigmaplot 11.0 (Systat Software Inc., Chicago, IL,
USA). Data are expressed as the mean ± standard deviation.
P<0.05 (two-sided) was considered to indicate a statistically
significant difference.
Results
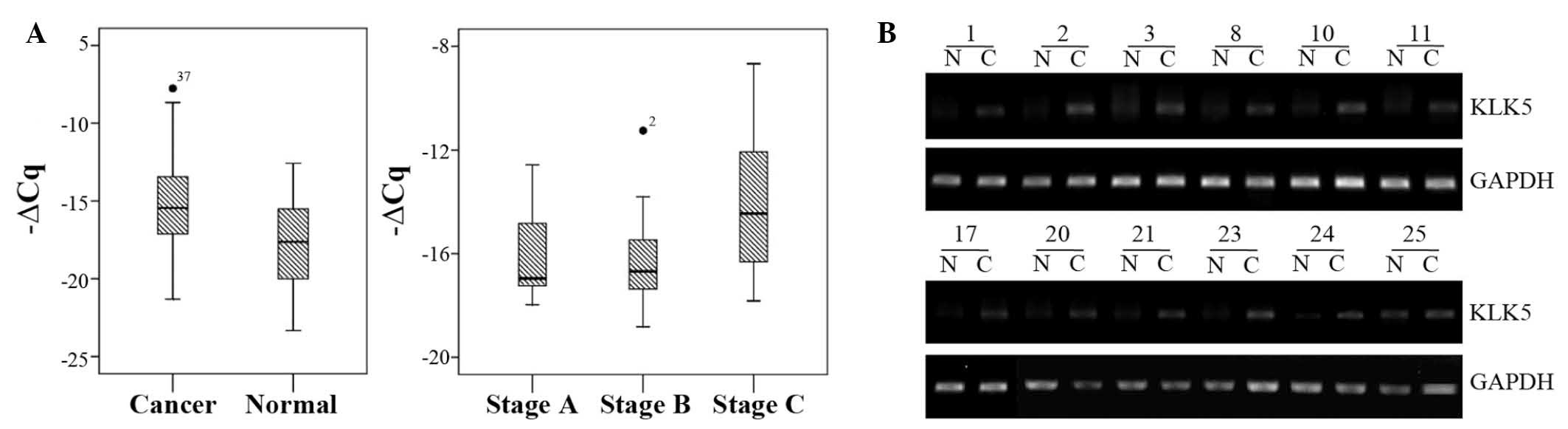
KLK5 mRNA expression is upregulated in
CRC tissues
The present study initially assessed the mRNA
expression levels of KLK5 in 40 tumor and paired normal tissue
samples from patients with CRC. The mRNA expression levels of KLK5
were upregulated in CRC tissues, as compared with in the paired
normal controls (Fig. 1), and
32/40 CRC tissue samples (80%) exhibited increased KLK5 expression.
In addition, the upregulated KLK5 levels were associated with
advanced tumor stages (stage C/D vs. stage A/B; P<0.001;
Fig. 1).
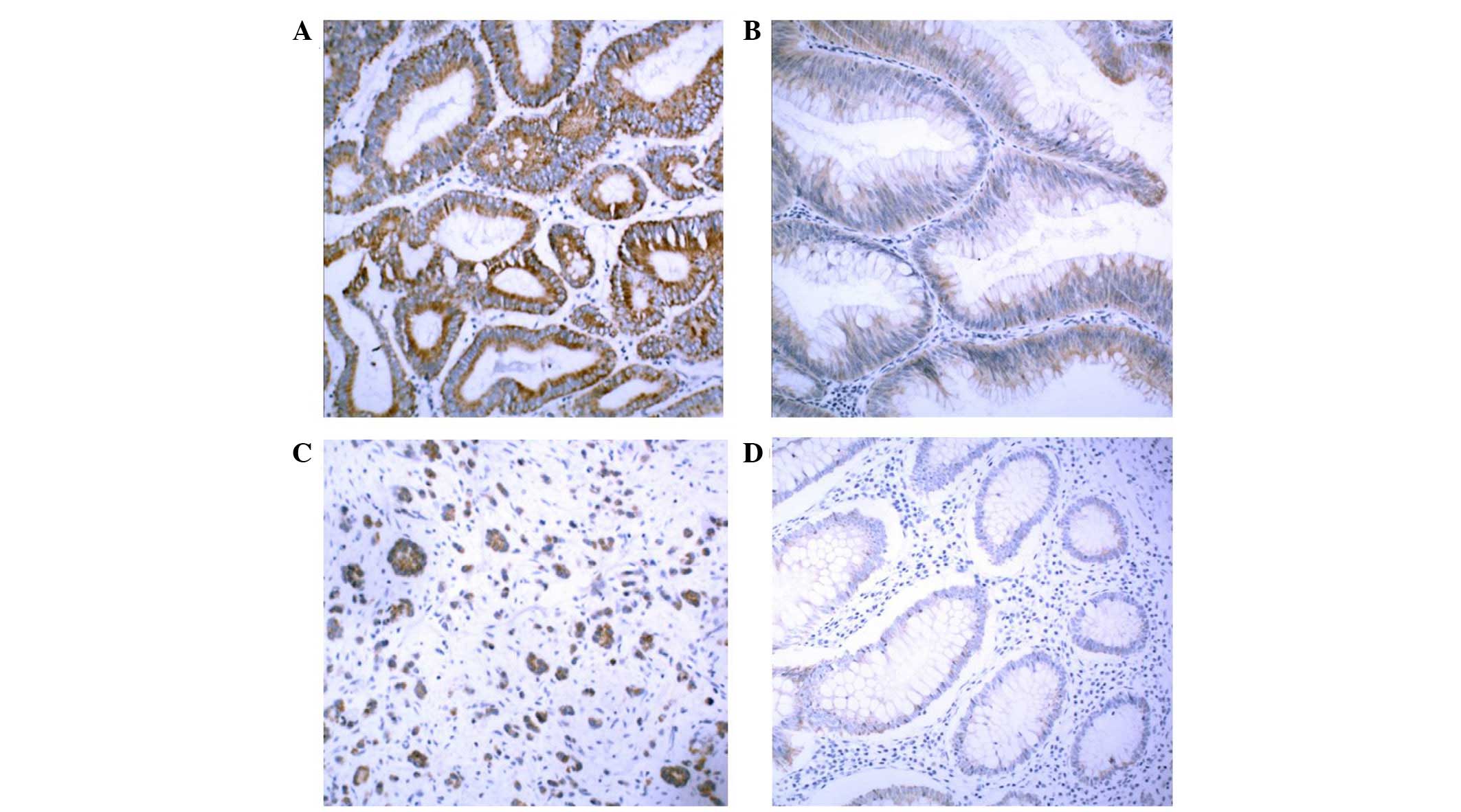
KLK5 protein expression is upregulated in
CRC tissues and sera
The present study also assessed the protein
expression levels of KLK5, since mRNA expression does not
necessarily correlate with protein abundance. Immunohistochemical
staining of KLK5 was performed in 48 CRC samples. KLK5 protein was
highly expressed in cancerous lesions but not in normal tissues
(Fig. 2A–D).
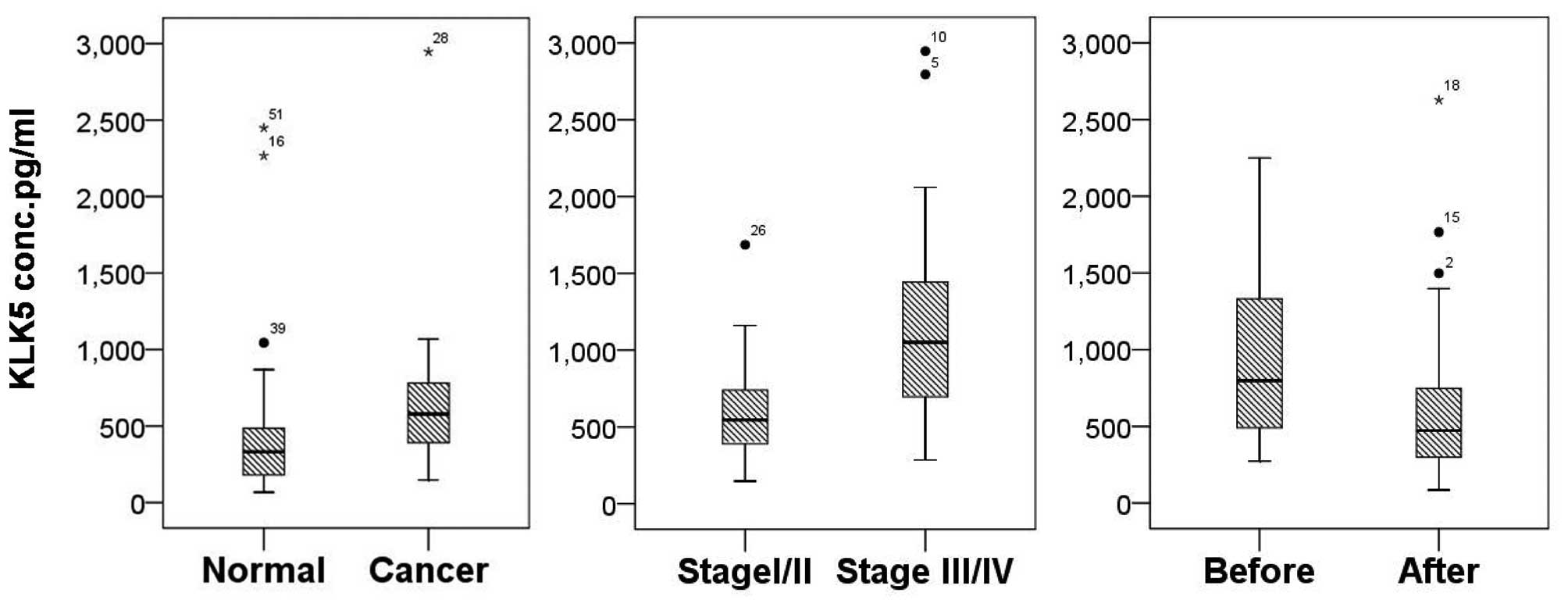
The present study also analyzed the KLK5 levels in
serum samples from 70 patients with CRC and 53 healthy individuals.
Among the 70 CRC serum samples, there were 38 paired pre- and
post-surgery serum samples taken from patients with CRC. The serum
levels of KLK5 were upregulated in patients with CRC, as compared
with in the healthy controls (878.02±602.02 vs. 391.07±331.13
pg/ml; P<0.001; Fig. 3). The
serum KLK5 levels were also significantly higher in patients prior
to surgery compared with after surgery (909.48±536.72 vs.
644.00±522.87 pg/ml; P<0.001). Furthermore, the serum levels of
KLK5 were higher in stage III/IV CRC than in stage I/II CRC
(1153.56±679.97 vs. 737.91.23±500.53 pg/ml; P=0.004).
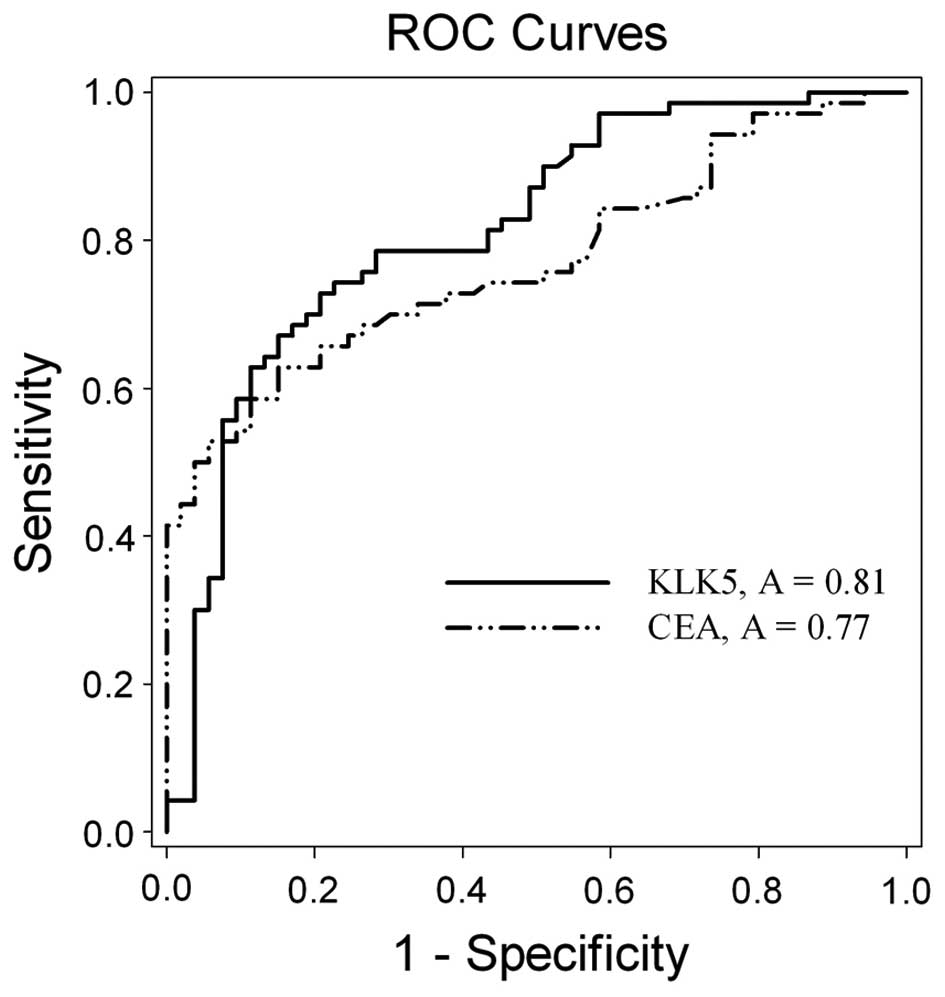
Finally, ROC curve analysis revealed the significant
and the independent value of the KLK5 expression, for the
discrimination of the CRC from the normal individuals. The curve
demonstrated that serum KLK5 levels [area under the curve (AUC),
0.81; 95% confidence interval (CI), 0.7375–0.8919; P<0.001]
exhibited a significant discriminatory value in the whole
population (Fig. 4), compared to
that of CEA (AUC, 0.77; 95% CI, 0.6901–0.8533; P<0.001). No
significant difference was observed between the KLK5 and CEA curves
(P=0.462).
Association of KLK5 expression with
clinicopathological features of patients with CRC
The association between KLK5 expression and
clinicopathological characteristics is summarized in Tables I and II. There were significant associations
between serum KLK5 levels and CRC lymph node or distant metastasis
(P=0.003), TNM stage (P=0.004), and Dukes' stage (P=0.005)
(Table I); whereas the mRNA
expression levels of KLK5 in CRC tissues were associated with a
high Dukes' stage (P<0.001; Table
II).
 | Table IIAssociation of KLK5 mRNA expression
with the clinicopathological features of colorectal cancer. |
Table II
Association of KLK5 mRNA expression
with the clinicopathological features of colorectal cancer.
|
Characteristics | N | KLK5 mRNA level
(ΔCq) | P-value |
|---|
| Gender | | | 0.436 |
| Male | 23 | −15.2±2.48 | |
| Female | 17 | −14.4±3.08 | |
| Age | | | 0.224 |
| <60 years
old | 20 | −14.40±2.82 | |
| ≥60 years old | 20 | −15.30±2.64 | |
| Tumor site | | | 0.913 |
| Rectum | 22 | −14.79±3.02 | |
| Colon | 18 | −14.94±2.43 | |
|
Differentiation | | | 0.295 |
| Well/moderate | 32 | −15.00±2.84 | |
| Poor | 8 | −14.27±2.39 | |
| Tumor size | | | 0.699 |
| <5 cm | 16 | −14.81±3.34 | |
| ≥5 cm | 24 | −14.88±2.33 | |
| Dukes' stage | | | <0.001 |
| A/B | 21 | −16.23±1.94 | |
| C/D | 19 | −13.34±2.73 | |
Discussion
Early clinical detection of CRC typically relies on
endoscopy and pathological analysis of tissue samples, which is
expensive and invasive. Studies regarding CRC biomarker discovery
have made little progress, possibly due to a lack of sensitive and
specific tumor markers (3,4,6). To
date, CEA and CA19–9 remain the only widely used serum tumor
markers in the early detection or prediction of treatment response
in CRC (24). The present study
assessed KLK5 expression in CRC tissue and serum samples using
RT-qPCR, immunohistochemistry, and ELISA using our own anti-KLK5
antibody. The results demonstrated that the mRNA and protein
expression levels of KLK5 were significantly upregulated in
independent CRC tissue and serum samples, as compared with in
normal samples. As a biomarker, KLK5 exhibited a better ROC curve
than CEA. In addition, KLK5 expression was associated with
malignant CRC behavior. The results of the present study indicated
that KLK5 may be considered a useful biomarker for CRC. However,
further studies are required to confirm these findings.
Dysregulated expression of various KLKs has been
detected in a large number of human malignancies during development
and progression of the disease. Therefore, the detection of KLK
levels may serve as a novel and useful tumor biomarker for the
early detection and monitoring of progression in patients with
cancer (25,26). Among these KLKs, KLK5 is important
since it can activate other KLKs. A previous study reported that
upregulation of KLK5 in ovarian cancer tissues was associated with
advanced stages and grades of the disease, and shorter disease-free
and overall survival rates of patients (27). In addition, another previous study
identified a novel variant of the KLK5 5′-untranslated region due
to alternative splicing, which was able to contribute to
differential KLK5 expression in prostate and ovarian cancer
(28). This finding indicated that
KLK5-SV1 may have clinical use in various malignancies, and should
be further explored as a potential novel biomarker for prostate and
ovarian cancer (28). It has
previously been shown that KLK6 and KLK10 are significantly
upregulated in patients with CRC, and that their upregulation is
associated with a poor prognosis (29,30).
The present study detected the upregulation of KLK5 mRNA and
protein in CRC tissues and in sera from patients with CRC, thus
indicating that KLK5 may be a useful biomarker for the early
diagnosis of CRC.
However, the potential mechanism underlying KLK5
activity in CRC development and progression remains to be
elucidated. KLK5 was originally discovered in the epidermis and has
functions related to keratinocyte turnover (desquamation), during
which this serine protease can digest the extracellular matrix and
increase cell mobility (8).
Nevertheless, it is currently unknown why and how KLK5 is
upregulated in CRC tissues and in sera from patients with CRC.
Previous studies have indicated that altered microRNA expression
may lead to the upregulation of KLK5 (31,32),
and our unpublished data support this notion.
The present study demonstrated that upregulated KLK5
levels may promote CRC development and progression. In addition,
after surgery, serum KLK5 levels were downregulated, thus
suggesting that KLK5 may be used to monitor CRC recurrence,
metastasis, and treatment response. However, the present study does
have some limitations; for example, survival or treatment data was
not collected from all of the patients with CRC, in order to
determine the association with prognosis or treatment response.
Furthermore, the sample size was relatively small and the
conclusion needs to be verified in a study with a larger sample
size. Future studies will focus on the following: Function of KLK5
in CRC cells and the underlying molecular mechanism; evaluation of
KLK5 as a novel biomarker for early detection, tumor progression,
and treatment response of patients with CRC; and the mechanisms
underlying KLK5 upregulation in patients with CRC.
Acknowledgments
The authors would like to thank the patients and
healthy controls that participated in the present study. This study
was supported in part by a grant from Medjaden Bioscience
Limited.
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McCracken M, Olsen M, Chen MS Jr, Jemal A,
Thun M, Cokkinides V, Deapen D and Ward E: Cancer incidence,
mortality, and associated risk factors among asian americans of
chinese, filipino, vietnamese, korean, and Japanese ethnicities. CA
Cancer J Clin. 57:190–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sarela AI: Systematic review of genetic
influences on the prognosis of colorectal cancer (Br J Surg 2004;
91: 1275–1291). Brit J Surg. 91:1275–1291. 2004. View Article : Google Scholar
|
|
4
|
Duffy MJ, van Dalen A, Haglund C, Hansson
L, Klapdor R, Lamerz R, Nilsson O, Sturgeon C and Topolcan O:
Clinical utility of biochemical markers in colorectal cancer:
European Group on Tumor Markers (EGTM) guidelines. Eur J Cancer.
39:718–727. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thomas SN, Tong Z, Stebe KJ and
Konstantopoulos K: Identification, characterization and utilization
of tumor cell selectin ligands in the design of colon cancer
diagnostics. Biorheology. 46:207–225. 2009.
|
|
6
|
Hammarström S: The carcinoembryonic
antigen (CEA) family: Structures, suggested functions and
expression in normal and malignant tissues. Semin Cancer Biol.
9:67–81. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Borgoño CA, Michael IP and Diamandis EP:
Human tissue kallikreins: Physiologic roles and applications in
cancer. Mol Cancer Res. 2:257–280. 2004.PubMed/NCBI
|
|
8
|
Yousef GM and Diamandis EP: The new
kallikrein-like gene, KLK-L2. Molecular characterization, mapping,
tissue expression, and hormonal regulation. J Biol Chem.
274:37511–37516. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brattsand M and Egelrud T: Purification,
molecular cloning, and expression of a human stratum corneum
trypsin-like serine protease with possible function in
desquamation. J Biol Chem. 274:30033–30040. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Michael IP, Pampalakis G, Mikolajczyk SD,
Malm J, Sotiropoulou G and Diamandis EP: Human tissue kallikrein 5
is a member of a proteolytic cascade pathway involved in seminal
clot liquefaction and potentially in prostate cancer progression. J
Biol Chem. 281:12743–12750. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Michael IP, Sotiropoulou G, Pampalakis G,
Magklara A, Ghosh M, Wasney G and Diamandis EP: Biochemical and
enzymatic characterization of human kallikrein 5 (hK5), a novel
serine protease potentially involved in cancer progression. J Biol
Chem. 280:14628–14635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ohler A, Debela M, Wagner S, Magdolen V
and Becker-Pauly C: Analyzing the protease web in skin: Meprin
metalloproteases are activated specifically by KLK4, 5 and 8 vice
versa leading to processing of proKLK7 thereby triggering its
activation. Biol Chem. 391:455–460. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diamandis EP and Yousef GM: Human tissue
kallikreins: A family of new cancer biomarkers. Clin Chem.
48:1198–1205. 2002.PubMed/NCBI
|
|
14
|
Kim H, Scorilas A, Katsaros D, Yousef GM,
Massobrio M, Fracchioli S, Piccinno R, Gordini G and Diamandis EP:
Human kallikrein gene 5 (KLK5) expression is an indicator of poor
prognosis in ovarian cancer. Br J Cancer. 84:643–650. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yousef GM, Scorilas A, Kyriakopoulou LG,
Rendl L, Diamandis M, Ponzone R, Biglia N, Giai M, Roagna R,
Sismondi P and Diamandis EP: Human kallikrein gene 5 (KLK5)
expression by quantitative PCR: An independent indicator of poor
prognosis in breast cancer. Clin Chem. 48:1241–1250.
2002.PubMed/NCBI
|
|
16
|
Avgeris M, Papachristopoulou G,
Polychronis A and Scorilas A: Down-regulation of kallikrein-related
peptidase 5 (KLK5) expression in breast cancer patients: A
biomarker for the differential diagnosis of breast lesions. Clin
Proteomics. 8:52011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yousef GM, Obiezu CV, Jung K, Stephan C,
Scorilas A and Diamandis EP: Differetial expression of kallikrein
gene 5 in cancerous and normal testicular tissues. Urology.
60:714–718. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yousef GM, Polymeris ME, Grass L,
Soosaipillai A, Chan PC, Scorilas A, Borgoño C, Harbeck N,
Schmalfeldt B, Dorn J, et al: Human kallikrein 5: A potential novel
serum biomarker for breast and ovarian cancer. Cancer Res.
63:3958–3965. 2003.PubMed/NCBI
|
|
19
|
Korbakis D, Gregorakis AK and Scorilas A:
Quantitative analysis of human kallikrein 5 (KLK5) expression in
prostate needle biopsies: An independent cancer biomarker. Clin
Chem. 55:904–913. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sotiropoulou G, Pampalakis G and Diamandis
EP: Functional roles of human kallikrein-related peptidases. J Biol
Chem. 284:32989–32994. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Talieri M, Li L, Zheng Y, Alexopoulou DK,
Soosaipillai A, Scorilas A, Xynopoulos D and Diamandis EP: The use
of kallikrein-related peptidases as adjuvant prognostic markers in
colorectal cancer. Br J Cancer. 100:1659–1665. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Y, Liu X, Chen Y and Hu C: Development
and evaluation of an ELISA method for the measurement of
kallikrein-related peptidase 5 (KLK5) in human serum. Open J Clin
Diagn. 3:159–166. 2013. View Article : Google Scholar
|
|
23
|
Fan X, Liu B, Xu H, Yu B, Shi S, Zhang J,
Wang X, Wang J, Lu Z, Ma H and Zhou X: Immunostaining with EGFR
mutation-specific antibodies: a reliable screening method for lung
adenocarcinomas harboring EGFR mutation in biopsy and resection
samples. Human Pathology. 44:1499–1507. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Anthony T, Simmang C, Hyman N, Buie D, Kim
D, Cataldo P, Orsay C, Church J, Otchy D, Cohen J, et al Standards
Practice Task Force, The American Society of Colon and Rectal
Surgeons: Practice parameters for the surveillance and follow-up of
patients with colon and rectal cancer. Dis Colon Rectum.
47:807–817. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Avgeris M, Mavridis K and Scorilas A:
Kallikrein-related peptidase genes as promising biomarkers for
prognosis and monitoring of human malignancies. Biol Chem.
391:505–511. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mavridis K and Scorilas A: Prognostic
value and biological role of the kallikrein-related peptidases in
human malignancies. Future Oncol. 6:269–285. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Diamandis EP, Borgoño CA, Scorilas A,
Yousef GM, Harbeck N, Dorn J, Schmalfeldt B and Schmitt M:
Immunofluorometric quantification of human kallikrein 5 expression
in ovarian cancer cytosols and its association with unfavorable
patient prognosis. Tumour Biol. 24:299–309. 2003. View Article : Google Scholar
|
|
28
|
Kurlender L, Yousef GM, Memari N, Robb JD,
Michael IP, Borgoño C, Katsaros D, Stephan C, Jung K and Diamandis
EP: Differential expression of a human kallikrein 5 (KLK5) splice
variant in ovarian and prostate cancer. Tumor Biol. 25:149–156.
2004. View Article : Google Scholar
|
|
29
|
Kim JT, Song EY, Chung KS, Kang MA, Kim
JW, Kim SJ, Yeom YI, Kim JH, Kim KH and Lee HG: Up-regulation and
clinical significance of serine protease kallikrein 6 in colon
cancer. Cancer. 117:2608–2619. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Talieri M, Alexopoulou DK, Scorilas A,
Kypraios D, Arnogiannaki N, Devetzi M, Patsavela M and Xynopoulos
D: Expression analysis and clinical evaluation of
kallikrein-related peptidase 10 (KLK10) in colorectal cancer. Tumor
Biol. 32:737–744. 2011. View Article : Google Scholar
|
|
31
|
White NM, Bui A, Mejia-Guerrero S, Chao J,
Soosaipillai A, Youssef Y, Mankaruos M, Honey RJ, Stewart R, Pace
KT, et al: Dysregulation of kallikrein-related peptidases in renal
cell carcinoma: Potential targets of miRNAs. Biol Chem.
391:411–423. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
White NM, Chow TF, Mejia-Guerrero S,
Diamandis M, Rofael Y, Faragalla H, Mankaruous M, Gabril M, Girgis
A and Yousef GM: Three dysregulated miRNAs control kallikrein 10
expression and cell proliferation in ovarian cancer. Br J Cancer.
102:1244–1253. 2010. View Article : Google Scholar : PubMed/NCBI
|