Introduction
Mammalian genomes contain 23 members of the
fibroblast growth factor (FGF) family (1), which are essential for metabolism and
development. FGFs have been identified to be involved in the
processes of embryogenesis, gastrulation, somitogenesis, body plan
formation, organogenesis and skin wound healing (2–7).
FGF21 is the most studied family member, and has been reported to
be preferentially expressed in the liver early in development
(8). However, recent studies have
identified that FGF21 production is inducible by starvation or drug
administration, and revealed its diverse functions in glucose
homeostasis and protection of the liver and heart from injury
(9–11). FGF19, 21 and 23 belong to the FGF19
subfamily. FGF21 primarily binds to the FGF receptor 1c isoform,
the activation of which requires the presence of the cofactor
β-klotho (12,13). A recent study reported that FGF1
regulated insulin sensitivity in order to maintain blood sugar
homeostasis (14).
Skin wound repair requires the cooperation of
various cell types, including keratinocytes, fibroblasts,
endothelial cells, macrophages and platelets. Fibroblast cell
proliferation and migration, collagen deposition and remodeling,
wound contraction and angiogenesis are important steps of this
process (15,16). Extracellular matrix (ECM) forms the
largest component of the dermal skin layer; therefore, repair of
ECM is key to wound healing (15).
Fibroblasts form a critical cell layer that participates in the
production and remodeling of the ECM, and their proliferation and
migration is important for the formation of granulation tissue and
skin repair (16). FGF2/basic
(b)FGF is well-known for its efficacy in skin wound healing, via
effects on cell proliferation and migration (16). However, the role of other FGFs in
this process remains to be elucidated.
In the present study, mouse heart, liver, skin and
kidney tissues were analyzed to determine the expression of
FGFs. Numerous FGFs were relatively highly expressed
in tissues; however, in skin only four FGFs (FGF2,
7, 10 and 21) were identified. In addition,
the function of four FGFs in fibroblast cell migration was analyzed
and possible roles of FGFs in skin wound healing were
identified.
Materials and methods
Fibroblast cell culture
The mouse NIH3T3 fibroblast cell line (Nanjing
Branch Bai Biological Technology Co., Ltd., Nanjing, China) was
placed into 25-cm2 flasks pretreated with fetal bovine
serum (FBS; Gibco, Thermo Fisher Scientific, Inc., Waltham, MA,
USA), and incubated horizontally for 1 h and then vertically for 3
h in an atmosphere of 5% CO2 at 37°C. The cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) containing
5.5 mM glucose, 10% FBS and 1% penicillin-streptomycin. Medium was
replaced every three days. The cultured cells were digested and
passaged with 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.)
when cell confluence reached ~80%. Cells passaged 3–6 times were
used in the following experiments.
Overexpression of FGF7 in fibroblast
cells
To overexpress FGF7 in NIH3T3 cells,
FGF7 open reading frame sequences (NM_008008.4, NCBI) were
synthesized and N-terminally fused with FLAG-tag coding sequences
(Sangon Biotech Co., Ltd., Shanghai, China). To transduce NIH3T3
cells with lentivirus (Beijing Omega Bio-Technology Co., Ltd,
Beijing, China), NIH3T3 cells were seeded at 2×105/well
into 24-well plates. Following overnight culture, 3, 9 or 12
μl lentivirus (108/ml) was added to the wells in
the presence of 4 mg/ml Polybrene® (Sigma-Aldrich, St.
Louis, MO, USA). The plates were then centrifuged at 800 × g at
room temperature for 1 h and returned to culture in DMEM.
Transduced cells, and mock-treated NIH3T3 cells, were analyzed 24 h
later by a confocal microscope (Olympus, Japan) to detect
expression of the reporter green fluorescent protein (GFP), and
then >20 single transfected cells expressing GFP were collected
and maintained for 5–6 generations to increased the cell number.
The dose of 12 μl lentivirus was selected for subsequent
experiments.
Wound healing assay
Cell migration was determined using the wound
healing scratch assay. Cells were seeded into 6-well plates
(103/plate) and grown overnight at 37°C. Confluent cells
were cultured in DMEM containing 0.5% FBS and 5 μg/ml
mitomycin-C for 24 h at 37°C, and then wounded by a 1-mm linear
scratch from a sterile pipette tip. Images of the wounded cell
monolayers were captured at 0, 12 and 24 h following wounding using
an inverted light microscope (model IX70; Olympus Corporation,
Tokyo, Japan) equipped with a charge-coupled device camera
(CoolSNAP HQ; Nippon Roper K.K., Chiba, Japan) and controlled by
MetaMorph® software version 7 (Universal Imaging, Inc.,
Bedford Hills, NY, USA). All experiments were performed in the
presence of 5 mg/ml of mitomycin-C to inhibit cell proliferation.
Cells were treated prior to wound healing with 100 ng/ml FGF2,
FGF10 or FGF21 (Sigma-Aldrich) for 1 h, or transduced to
overexpress FGF7 as previously described, and wound healing
was measured at 12 and 24 h. A total of 20 cells/experiment at the
edge of wound region were randomly selected from the wound area. At
12 and 24 h following wounding, the distance between the 20
selected cells and the wound edge at 0 h was measured using Image J
Fiji software (National Institutes of Health, Bethesda, MD,
USA).
Western blot analysis
The cells were lysed in an ice-cold lysis solution
[7 M urea, 2 M thiourea, 2% 3-([3-Cholamidopropyl]
dimethylammonio)-1-propanesulfonate, 40 mM Trizma® base,
40 mM dithiothreitol, 1% protease inhibitor] for 10 min. Following
centrifugation at 15,000 × g for 15 min at 4°C, the protein
concentration in the supernatant was measured by the Bradford
protein assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
according to the manufacturer's protocol. The proteins (20
μg) were loaded onto a 10% SDS-PAGE gel, separated by
electrophoresis at 100 V for 2 h and transferred onto Immobilon-P
Transfer Membranes (Merck Millipore, Tokyo, Japan). The membranes
were blocked with Tris-buffered saline containing 5% skim milk and
0.05% Tween-20 for 1 h and then probed with primary antibodies at
4°C overnight. Anti-GAPDH (mouse monoclonal; dilution, 1:2,500;
catalog no., mAbcam 9484; Abcam, Cambridge, UK),
anti-phospho-stress-activated protein kinase (SAPK)/c-Jun
N-terminal kinase (JNK) phosphorylated at Thr183/Tyr185 (rabbit
monoclonal; dilution, 1:1,000; catalog no., 4668; Cell Signaling
Technology, Inc., Danvers, MA, USA), anti-JNK (rabbit monoclonal;
dilution, 1:1,000; catalog no., ab179461) and anti-FLAG (mouse
monoclonal; dilution, 1:2,000; catalog no., ab49763; Abcam) were
used as primary antibodies. The membranes were then incubated for 1
h with an anti-mouse (dilution, 1:2,000; ab131368; Abcam) or
anti-rabbit horseradish peroxidase-conjugated secondary antibody
(dilution, 1:2,000; catalog no., 7074; Cell Signaling Technology,
Inc.), and visualized using an electrochemiluminescence kit (GE
Healthcare Life Sciences, Chalfont, UK). Images of western blots
were captured using an ImageQuant LAS 4000 Mini (GE Healthcare Life
Sciences).
Total RNA extraction, complementary DNA
synthesis and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR)
Male C57/BL6J mice, aged 3 months and weighing 2835
g, were obtained from the Laboratory Animal Centre of Wenzhou
Medical University (Wenzhou, China). The mice were housed at 22°C
and 50% humidity, with a 12 h light/dark cycle. Mice had free
access to food and water. Approval was given for the use of mice in
the present study by the Ethics Committee of Wenzhou Medical
University (Wenzhou, China). All mice were anesthetized via
intraperitoneal injection of 3% sodium pentobarbital (45 mg/kg) and
their dorsal areas were completely depilated using sodium sulfide
(8.0%; w/v; both SigmaAldrich) prior to tissue extraction during
surgery. Total RNA was extracted from liver, heart, kidney and skin
tissues of 9 mice, which were randomly divided into three groups (3
mice/group) for biological duplication (the nature of each group
was the same, and simply served as a replication), using
TRIzol® Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA (1 μg) was reverse-transcribed using a
GoScript Reverse Transcription kit (Promega Corporation, Madison,
WI, USA) according to the manufacturer's instructions. PCR was
performed using the SapphireAmp® Fast PCR Master Mix
(Takara Biotechnology Co., Ltd., Dalian, China) on a T100 thermal
cycler (BioRad Laboratories, Inc.), with the following cycling
conditions: 95°C for 5 min, followed by 35 cycles at 94°C for 30
sec and 58°C for 30 sec, 72°C for 30 sec, elongation at 72°C for 5
min and maintenance at 10°C. Gene expression levels were quantified
by the ΔΔCq method as described previously (17). mRNA levels were normalized against
those of GAPDH using Image J2x version 2.1.4.7 software and the
2−ΔΔCq method (18).
Primers used for RT-qPCR are listed in Table I.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Primer | Forward
sequence | Reverse
sequence |
|---|
| FGF1 |
TGCTCTACTGCAGCAACG |
CTAGTCAGAAGACACCGG |
| FGF2 |
CAAGAACGGCGGCTTCTTC |
GGAAGAAACAGTATGGCCT |
| FGF3 |
CAAGCTCTACTGCGCTACC |
GTCCACCTGTATGCAGCT |
| FGF4 |
TACTGCAACGTGGGCATC |
GGAAGTGGGTTACCTTCA |
| FGF5 |
GAAGTAGCGCGACGTTTTC |
GGCTTAACACACTGGCTTC |
| FGF6 |
CTCTACTGCAACGTGGGC |
GGAAGTGAGTGACAGTCA |
| FGF7 |
AGACTGTTCTGTCGCACC |
CCGCTGTGTGTCCATTTAG |
| FGF8 |
ACCTACCAGCTCTACAGCC |
GGCGGGTAGTTGAGGAACT |
| FGF9 |
CTGCAGGACTGGATTTCATTT |
GTTCAGGTACTTTGTCAGGG |
| FGF10 |
TGTCCGCTGGAGAAGGCTGTTC |
CTATGTTTGGATCGTCATGG |
| FGF11 |
ATCGTCACCAAACTGTTCTG |
CAGGAACACTGTGGAGAGAA |
| FGF12 |
TCAGCCAGCAGGGATATTTC |
CACGACTTTGCCTCCATTCA |
| FGF13 |
TAACCTCATCCCTGTGGG |
GAGAACTCCGTGAGATCG |
| FGF14 |
CAACCTCATCCCAGTGGGA |
GGGACTGTTTCACCAACATC |
| FGF15 |
ACTCCGCTGGTCCCTATGTC |
CTACATCCTCCACCATCCT |
| FGF16 |
GCTTCCACCTTGAGATCTTC |
GAGATCTCTGGACATGGAG |
| FGF17 |
CCAGCTCTACAGCCGGAC |
GGGGCGGAGCCCACAAAT |
| FGF18 |
CCAGCTCTATAGCAGGAC |
GCTTGGTGACTGTGGTGT |
| FGF19 |
AACTTTATCCCCATATTTCACC |
GAAGCTGGGACTCTTCACT |
| FGF20 |
TCAGAGAAATTGACTTCTG |
GTGTACATCAGTAGGTCTT |
| FGF21 |
GATGACGACCAAGACACTG |
CGGCCCTGTAAAGGCTCT |
| FGF22 |
GCCTCTTCTCCTCCACTC |
CGAGACCAAGACTGGCAG |
| FGF23 |
ACAGCCAGGACCAGCTATC |
CTCGCGAGAGCAGGATACA |
| GAPDH |
GCCAAGGTCATCCATGACAACT |
GAGGGGCCATCCACAGTCTT |
Phylogenetic analysis
FGF sequences used for similarity searches were
collected from the National Center for Biotechnology Information
website (http://www.ncbi.nlm.nih.gov/gene/?term=FGF). ClustalW
(http://align.genome.jp) was used to perform a
homologous sequence alignment of the amino acid sequences of FGF
family proteins using default settings. Based on the results of
sequence alignment, an unrooted phylogenetic tree of the FGF gene
family was constructed using Mega software version 6.0 (http://www.megasoftware.net). The neighbor-joining
method was applied to construct a phylogenetic tree (19), in which Poisson correction,
pairwise deletion and bootstrapping (1000 replicates; random seeds)
served as default values to evaluate the reliability of the
tree.
Statistical analysis
Statistical analyses were performed in GraphPad
Prism version 5 (GraphPad Software, Inc., La Jolla, CA, USA). Data
are presented as the mean ± standard error. Comparisons between
groups were performed using Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
FGF expression patterns in mouse
tissues
The expression patterns of 23 FGF members in mouse
tissues was dissected by analyzing their transcript levels in
liver, heart, kidney and skin tissue samples from 9 C57/BL6J male
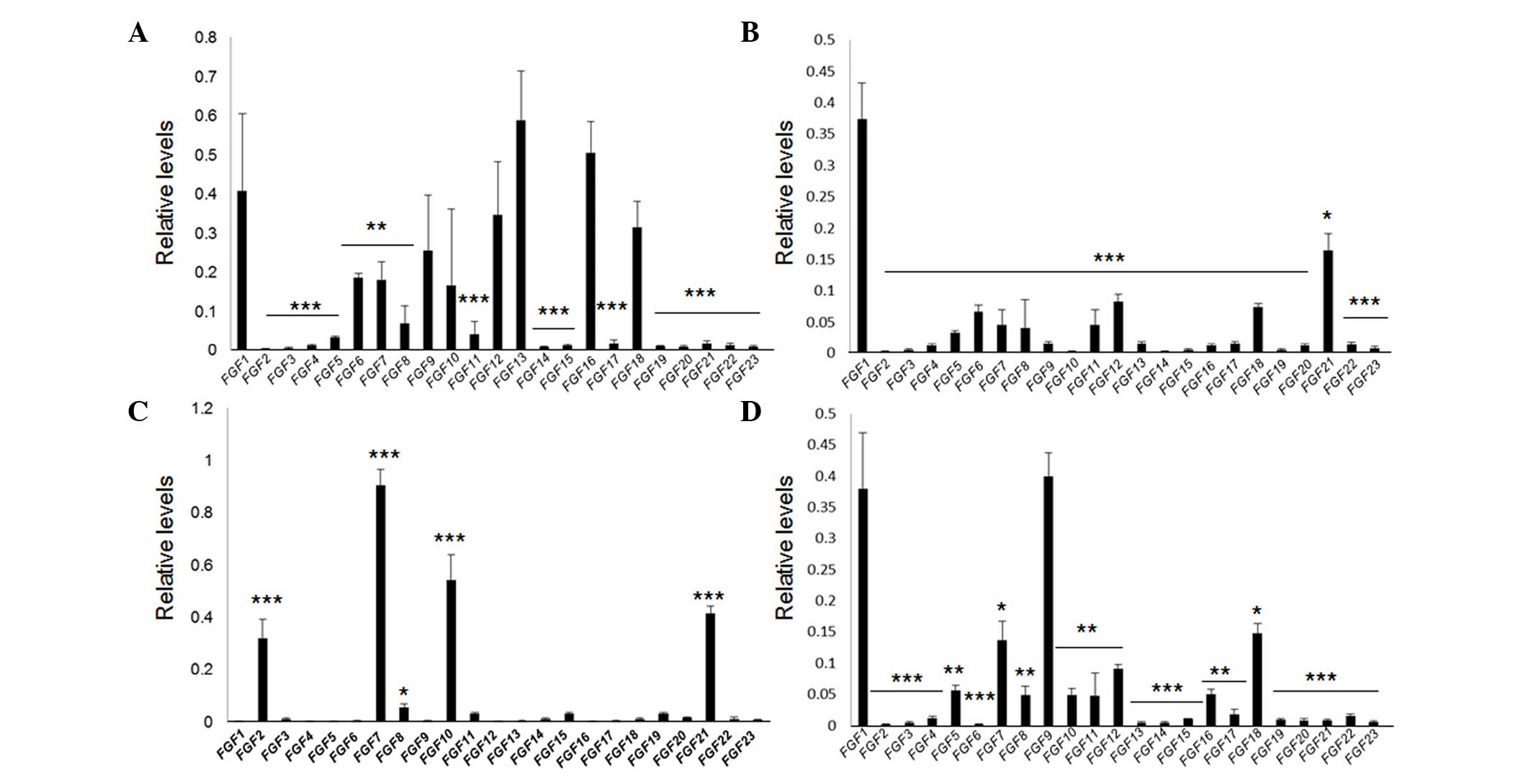
mice. RT-qPCR results revealed that FGF1, 6,
7, 9, 10, 12, 13, 16 and
18 were highly expressed in the heart (Fig. 1A). In the liver, expression of
FGF1, 5, 6, 7, 8, 11,
12, 18 and 21 were relatively high, with FGF1
having the greatest expression (Fig.
1B). Notably, the expression of FGFs was relatively less
complex in skin compared with other tissues. A total of four
FGFs, FGF2, 7, 10 and 21 were
significantly highly expressed in the skin (Fig. 1C). In the kidney, levels of
FGF1, 5, 7, 8, 9, 10,
11, 12, 16 and 18 were relatively high
(Fig. 1D). FGF2/bFGF is widely
known for its efficacy in wound healing (7,20),
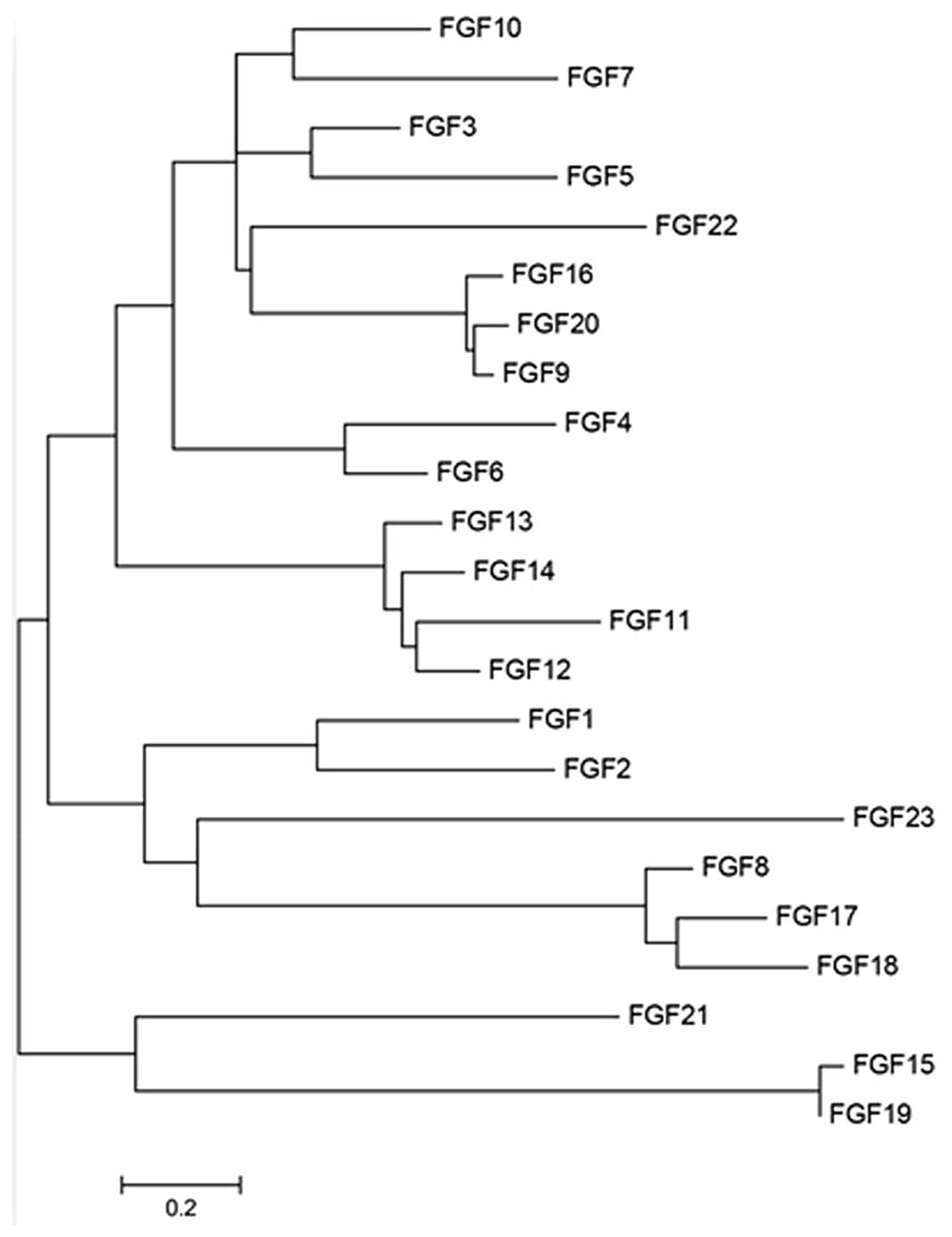
and as the profile of FGFs is relatively simple in skin
tissue, a phylogenetic tree was generated to understand the
associations between the four FGF proteins. The data revealed that
FGF7 and FGF10 belong to the same sub-group, while FGF2 shares a
sub-group with FGF1. FGF21 is on a separate branch of the tree
(Fig. 2), indicating that the four
FGFs are not highly correlated.
FGF2, 10 and 21 promote fibroblast cell
migration
FGF2, 7, 10 and 21 are
highly expressed in skin tissue (Fig.
1C). Therefore, the function of these FGFs in wound healing was
examined. Wound healing involves multiple steps, including cell
proliferation and migration. To investigate the effect of FGFs on
the cell migration process, mouse NIH3T3 foreskin fibroblast cells
were grown in a low glucose medium (5.5 mM) containing 5 mg/ml of
mitomycin-C, which prevents cell proliferation, for 24 h at 37°C
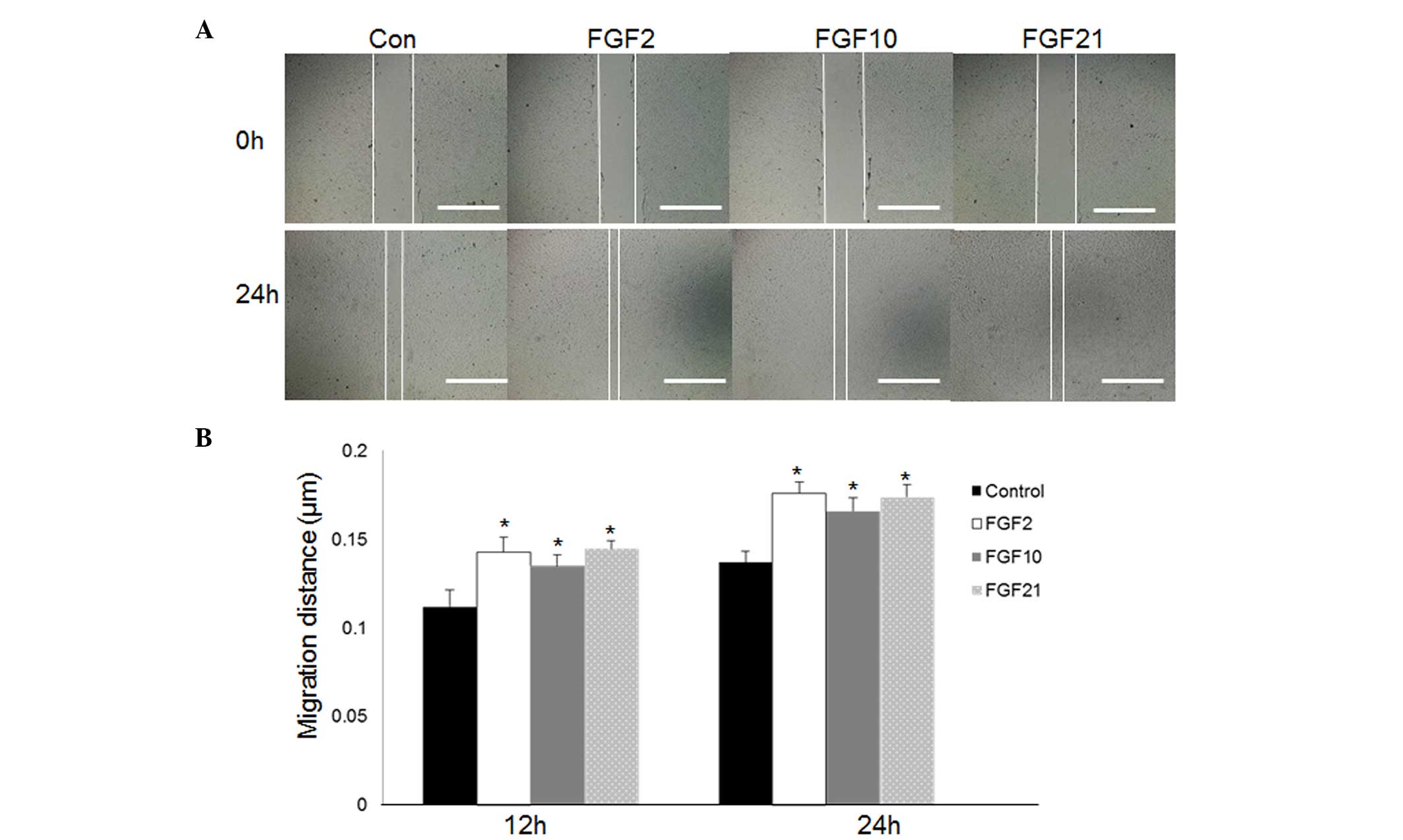
prior to treatment with FGFs. As presented in Fig. 3, the cells treated with
commercially purified FGF2 (P=0.023), 10 (P=0.027) or 21 (P=0.018)
at 100 ng/ml exhibited accelerated cell migration 24 h following
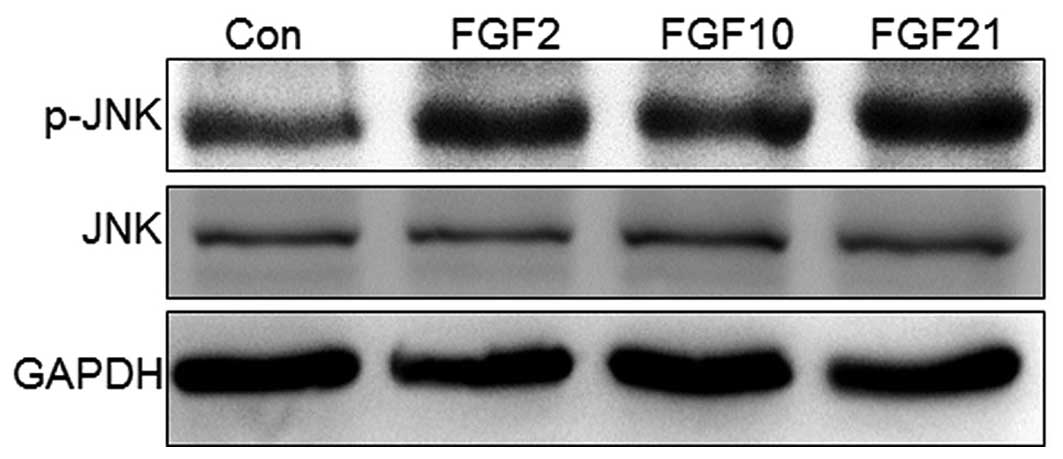
scratching as compared with control cells. JNK is activated via
phosphorylation, and is important for cell migration in wound
healing (16); therefore,
JNK-phosphorylation was examined following FGF-treatment of NIH3T3
cells for 1 h. The data revealed that treatment with the three FGFs
(FGF2, 10 and 21) increased p-JNK levels, while the total JNK level
remained unchanged (Fig. 4).
As commercially produced FGF7 is not available,
FGF7-FLAG fusion protein was overexpressed in NIH3T3 cells via a
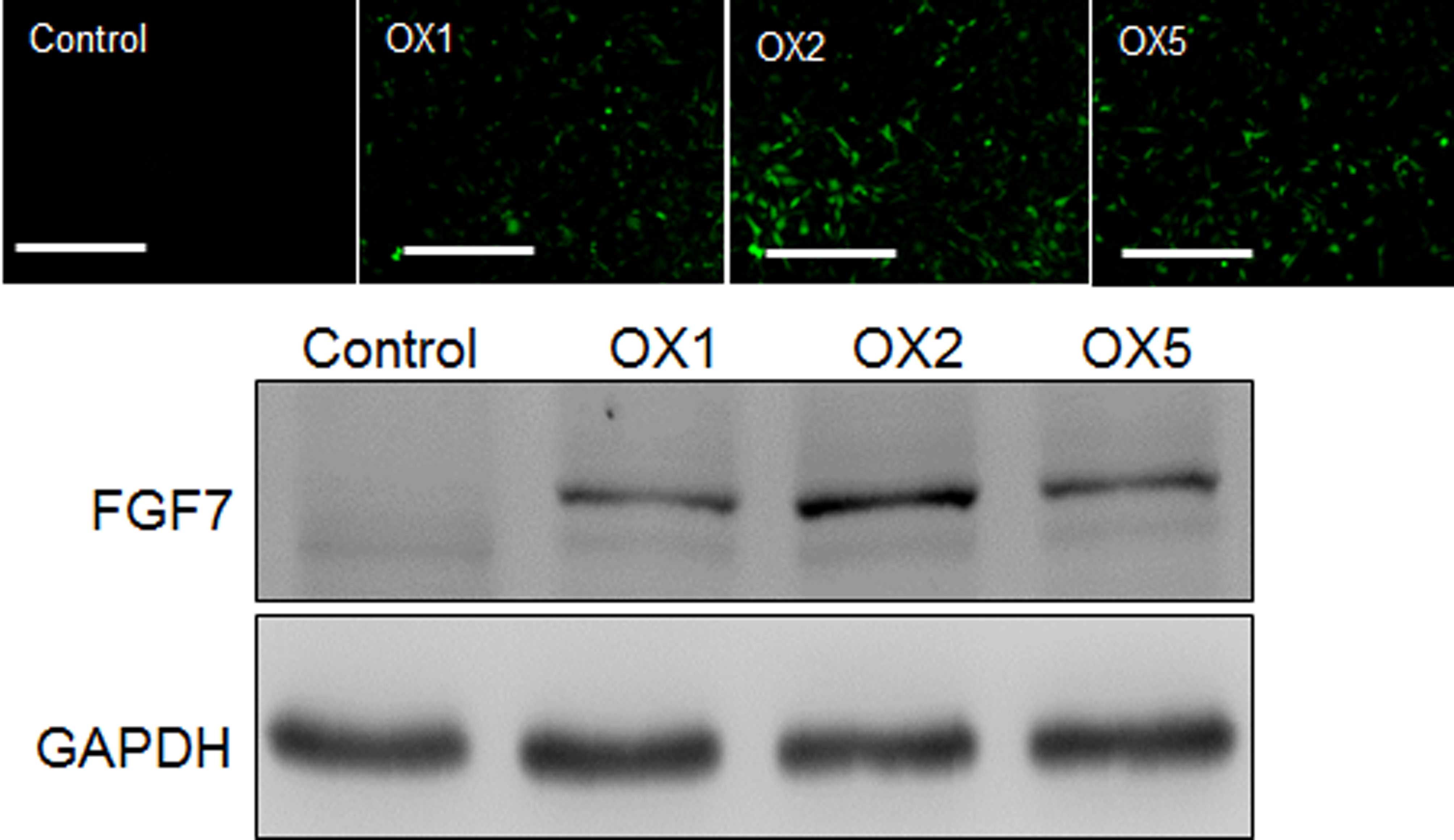
lentivirus system. Transduction efficiency was analyzed by
monitoring GFP expression, and FGF7 expression was evaluated by
western blotting using an anti-FLAG antibody (Fig. 5). In addition, FGF7
transcripts were analyzed in transduced and non-transduced NIH3T3
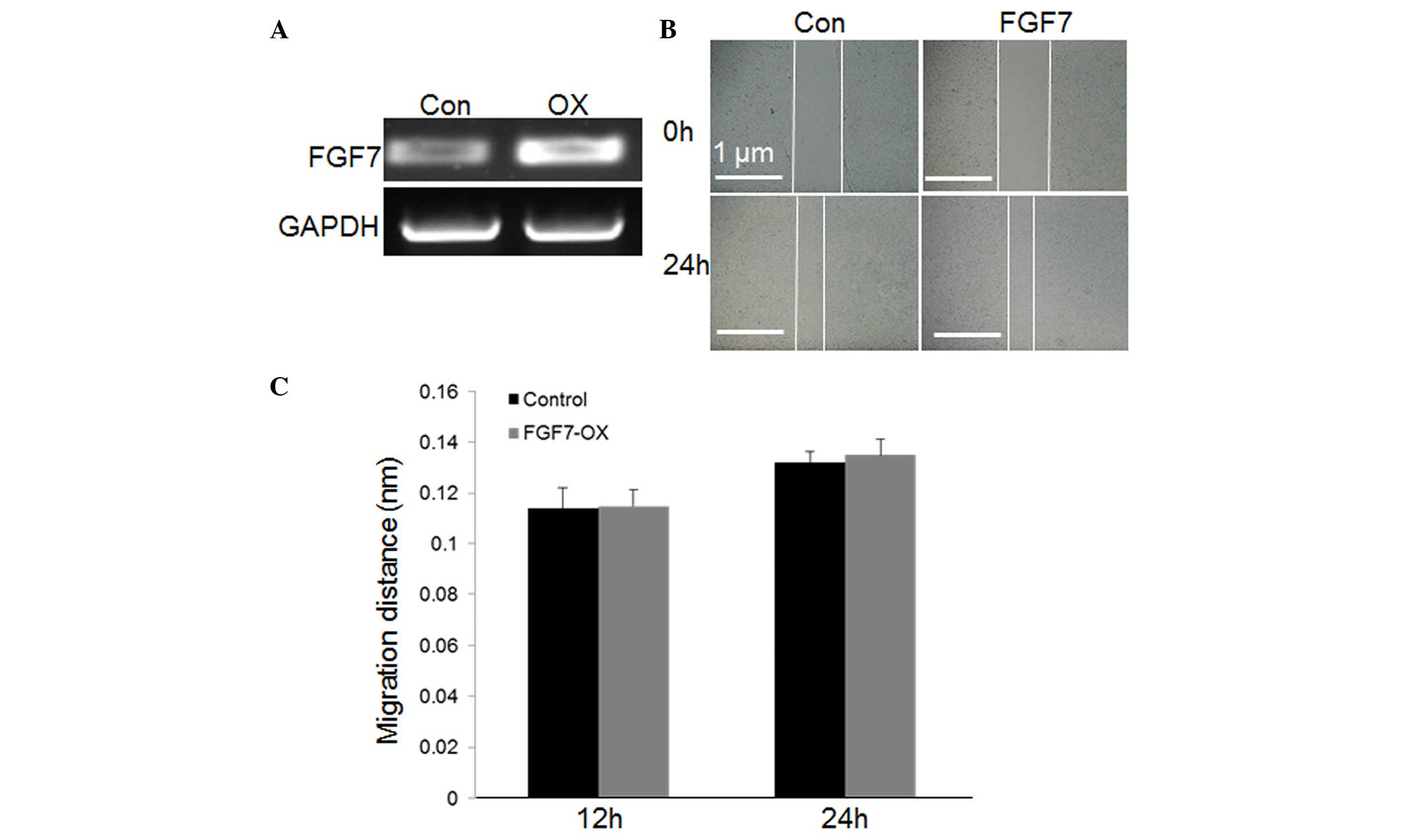
cells. RT-qPCR revealed that FGF7 levels were much higher in
transduced, (FGF7 OX) compared with non-transduced, cells (Fig. 6A). Furthermore, cell migration was
evaluated in FGF7 OX and non-transduced cells. However, FGF7
overexpression did not significantly alter the speed of cell
migration (Fig. 6B and C;
P=0.094). These results indicate that FGF2, 10 and 21 are highly
expressed in skin tissue, and are involved in the fibroblast cell
migration process.
Discussion
Skin wound healing is a complex process that
requires keratinocytes, fibroblasts, endothelial cells, macrophages
and platelets. These cells undergo multiple steps including
proliferation and migration to rebuild the skin (21). Fibroblasts are critical in wound
contraction. Fibroblast cell migration, considered to be a
fundamental step in wound healing, involves a multi-step cyclic
process, including extension of a protrusion, stable attachment
close to the leading edge of the protrusion, forward movement of
the cell body, and release of adhesions and retraction at the rear
of the cell (22).
FGF2/bFGF is a member of FGF family, and its
efficacy in the promotion of fibroblast cell migration is
well-understood (7). In the
present study, it was observed that besides FGF2,
FGF7, 10 and 21 are highly expressed in skin
tissue. Previously, the role of FGF21 in glucose homeostasis has
been well-characterized, and its production is induced by stress in
the liver and heart (9–11). However, in the present study,
FGF21 expression was revealed to be greatest in the skin,
where expression was even higher than in the liver (data not
shown). In addition, FGF21 was demonstrated to accelerate the
migration of mouse fibroblast cells, similar to FGF2. Furthermore,
FGF10 was identified as predominant in skin, and also accelerated
cell migration. FGF2 has previously been revealed to accelerate
fibroblast cell migration via activation of the phosphoinositide
3-kinase-Ras-related C3 botulinum toxin substrate 1-JNK signaling
pathway (7). In the present study,
FGF2, 10 and 21 treatment increased JNK phosphorylation levels.
FGF7 has the greatest expression of all FGFs in skin;
however, overexpression of FGF7 in fibroblast cells did not
alter cell migration speed, implying that FGF7 may have alternative
roles in skin tissue. A previous study demonstrated that induction
of FGF7 expression in the dermal papilla cells of
adenosine-stimulated hair, and treatment with exogenous FGF7,
stimulated hair fiber elongation in human scalp hair follicle organ
cultures (23).
Numerous FGFs were relatively highly expressed in
the heart, liver and kidney, suggesting potential, although as yet
uncharacterized, roles of these FGFs in various tissues. Notably,
FGF1 was one of the predominant FGFs in all the tissues
examined except the skin, and was dominant in the liver. Recently,
FGF1 was reported to regulate insulin sensitivity (14), therefore, it may be beneficial to
analyze the role of FGF1, particularly in the liver. In conclusion,
the findings of the present study indicate that FGF family members
FGF2, 10 and 21 coordinate to activate the FGF signaling pathway,
which is important in the promotion of wound repair. In addition,
this experimental approach may provide a basis for isolating and
analyzing FGF functions in various tissues.
Acknowledgments
The present study was supported by initiative
funding from the second Affiliated Hospital of Wenzhou Medical
University (grant no. GC130798).
References
|
1
|
Mohammadi M, Olsen SK and Ibrahimi OA:
Structural basis for fibroblast growth factor receptor activation.
Cytokine Growth Factor Rev. 16:107–137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feldman B, Poueymirou W, Papaioannou VE,
DeChiara TM and Goldfarb M: Requirement of FGF-4 for
postimplantation mouse development. Science. 267:246–249. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dubrulle J and Pourquié O: fgf8 mRNA decay
establishes a gradient that couples axial elongation to patterning
in the vertebrate embryo. Nature. 427:419–422. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun X, Meyers EN, Lewandoski M and Martin
GR: Targeted disruption of Fgf8 causes failure of cell migration in
the gastrulating mouse embryo. Genes Dev. 13:1834–1846. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martin GR: The roles of FGFs in the early
development of vertebrate limbs. Genes Dev. 12:1571–1586. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goldfarb M: Functions of fibroblast growth
factors in vertebrate development. Cytokine Growth Factor Rev.
7:311–325. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kanazawa S, Fujiwara T, Matsuzaki S,
Shingaki K, Taniguchi M, Miyata S, Tohyama M, Sakai Y, Yano K,
Hosokawa K and Kubo T: bFGF regulates PI3-kinase-Rac1-JNK pathway
and promotes fibroblast migration in wound healing. PloS One.
5:e122282010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishimura T, Nakatake Y, Konishi M and
Itoh N: Identification of a novel FGF, FGF-21, preferentially
expressed in the liver. Biochim Biophys Acta. 1492:203–206. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang Q, Zhong L, Zhang J, Wang Y,
Bornstein SR, Triggle CR, Ding H, Lam KS and Xu A: FGF21 maintains
glucose homeostasis by mediating the cross talk between liver and
brain during prolonged fasting. Diabetes. 63:4064–4075. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin Z, Tian H, Lam KS, Lin S, Hoo RC,
Konishi M, Itoh N, Wang Y, Bornstein SR, Xu A and Li X: Adiponectin
mediates the metabolic effects of FGF21 on glucose homeostasis and
insulin sensitivity in mice. Cell Metab. 17:779–789. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin Z, Wu F, Lin S, Pan X, Jin L, Lu T,
Shi L, Wang Y, Xu A and Li X: Adiponectin protects against
acetaminophen-induced mitochondrial dysfunction and acute liver
injury by promoting autophagy in mice. J Hepatol. 61:825–831. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yie J, Wang W, Deng L, Tam LT, Stevens J,
Chen MM, Li Y, Xu J, Lindberg R, Hecht R, et al: Understanding the
physical interactions in the FGF21/FGFR/beta-Klotho complex:
structural requirements and implications in FGF21 signaling. Chem
Biol Drug Des. 79:398–410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Belov AA and Mohammadi M: Molecular
mechanisms of fibroblast growth factor signaling in physiology and
pathology. Cold Spring Harb Perspect Biol. 5:pii: a015958. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Suh JM, Jonker JW, Ahmadian M, Goetz R,
Lackey D, Osborn O, Huang Z, Liu W, Yoshihara E, van Dijk TH, et
al: Endocrinization of FGF1 produces a neomorphic and potent
insulin sensitizer. Nature. 513:436–439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brem H and Tomic-Canic M: Cellular and
molecular basis of wound healing in diabetes. J Clin Invest.
117:1219–1222. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xuan YH, Huang BB, Tian HS, Chi LS, Duan
YM, Wang X, Zhu ZX, Cai WH, Zhu YT, Wei TM, et al: High-glucose
inhibits human fibroblast cell migration in wound healing via
repression of bFGF-regulating JNK phosphorylation. PloS One.
9:e1081822014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zittermann SI and Issekutz AC: Basic
fibroblast growth factor (bFGF, FGF-2) potentiates leukocyte
recruitment to inflammation by enhancing endothelial adhesion
molecule expression. Am J Pathol. 168:835–846. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using realtime quantitative PCR and
the 2ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
19
|
Saitou N and Nei M: The neighbor-joining
method: a new method for reconstructing phylogenetic trees. Mol
Biol Evol. 4:406–425. 1987.PubMed/NCBI
|
|
20
|
Iyer VR, Eisen MB, Ross DT, Schuler G,
Moore T, Lee JC, Trent JM, Staudt LM, Hudson J Jr, Boguski MS, et
al: The transcriptional program in the response of human
fibroblasts to serum. Science. 283:83–87. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martin P: Wound healing - aiming for
perfect skin regeneration. Science. 276:75–81. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao C, Sun Y, Healey S, Bi Z, Hu G, Wan S,
Kouttab N, Chu W and Wan Y: EGFR-mediated expression of aquaporin-3
is involved in human skin fibroblast migration. Biochem J.
400:225–234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iino M, Ehama R, Nakazawa Y, Iwabuchi T,
Ogo M, Tajima M and Arase S: Adenosine stimulates fibroblast growth
factor-7 gene expression via adenosine A2b receptor signaling in
dermal papilla cells. J Invest Dermatol. 127:1318–1325. 2007.
View Article : Google Scholar : PubMed/NCBI
|