Introduction
Increasing evidence has suggested that
single-nucleotide polymorphisms (SNPs) may become a new generation
of genetic markers and valuable indicators for clinical diagnosis
and prognosis (1,2). A polymorphism in Apolipoprotein E
(ApoE) is one of the most widely studied polymorphisms, and has
been considered to be a pre-symptomatic risk predictor for a
variety of diseases, including coronary artery disease and
late-onset Alzheimer's disease (3,4). The
molecular bases of ApoE polymorphism are cysteine (TGC)-arginine
(CGC) interchanges at the one or both of residues 158 and 112 that
determine three major alleles, designated E2, E3 and E4. This
polymorphism leads to the presence of six genotypes in the general
population: E2/E2, E3/E3, E4/E4, E2/E3, E2/E4 and E3/E4 (5).
Several methods have been established for
identifying ApoE polymorphisms. These methods can be divided into
two groups: i) Proteomic analyses using isoelectric focusing
(6) or immunoassay reagents
combined with mass spectrometry (7); and ii) genotyping techniques that
detect sequence differences in the ApoE alleles, including
polymerase chain reaction (PCR) restriction fragment length
polymorphism analysis (8),
quantitative PCR analysis (9),
mass spectrometry (10),
amplification-refractory mutation system (11), denaturing high-performance liquid
chromatography (12), TaqMan
assays (13) and single base
extension genotyping technology (14). Currently, ARMS is generally
considered as a simple and cost-effective method. ARMS for ApoE
genotyping, as previously described (11), requires four separate PCR
reactions. The present study aimed to reduce the complexity of ApoE
genotyping by combining the different primers into two PCR
reactions. However, the method described still requires analysis by
agarose gels, thus, limiting its clinical applications.
Accurate and rapid methods for ApoE genotyping are
in ever-increasing demand. Nanoparticle-associated lateral flow
assays (LFAs) have attracted considerable research interest, as
they provide a promising approach to enable point-of-care nucleic
acid detection (15). In the
emerging and revolutionary diagnostic arena,
Fe3O4/Au nanoparticles, formed of an iron
oxide core with gold coating, are often used to improve the
sensitivity of the immunosensor due to their stability, high
surface-to-volume ratio and biocompatibility (16,17).
Polystyrenesulfonate (PSS) modification has been demonstrated to be
an effective strategy to increase gold nanorods stability and
compatibility for biological interactions (18). Therefore, the
polyelectrolyte-coated gold magnetic (GoldMag) nanoparticle
(PGMN)-mediated conjugates may be a colloidal, monodispersed
particle probe to accurately detect a target with high sensitivity
and specificity, based on LFAs in clinical diagnostics. A GoldMag
based lateral-flow immunoassay was previously reported to rapidly,
specifically and accurately analyze Treponema pallidum
antibody, at the laboratory and clinical level (19). A recent study by our laboratory
proposed a novel approach for the visual detection of MTHFR C677T
polymorphisms via integrating the ARMS-PCR with GoldMag-based LFA
(20); the assay involves two
complementary PCR reactions for each SNP.
The current study describes a PCR-GoldMag LFA for
ApoE genotyping based on the multi-ARMS-PCR and GoldMag LFA, and
analyzes the distribution of ApoE variants in a Han Chinese cohort.
It takes only two multi-ARMS-PCR reactions, using GoldMag-based
LFAs, to distinguish directly the six different ApoE genotypes. The
PCR-GoldMag LFA, as a simple and rapid method, enables visual
identification of SNPs, and avoids complex steps, including
pipetting, incubation, washing and data analysis. This novel method
can also be easily extended to detect SNPs of other
disease-associated genes.
Materials and methods
Materials and reagents
GoldMag nanoparticles (5 mg/ml) with a nanoflower
structure (21) and lateral flow
strips were provided by Xi'an GoldMag Nanobiotech Co., Ltd. (Xi'an,
China). Buffers were prepared according to standard laboratory
procedures. Water (18.2 MΩ cm) purified by the Barnstead Nanopure
Water System (Thermo Fisher Scientific, Inc., Waltham, MA, USA) was
used for all sample preparation. All chemicals listed below were of
analytical grade. Cetyltrimethylammonium bromide (CTAB; 99%
purity), dimethyl sulfoxide (DMSO) and PSS sodium salt (molecular
weight, 70 kDa) were obtained from Sigma-Aldrich (Merck Millipore,
Darmstadt, Germany). A mouse anti-digoxin antibody (catalog no.
MAD53-310C) was purchased from Meridian Life Science, Inc.
(Cincinnati, OH, USA). An anti-fluorescein isothiocyanate (FITC)
antibody (catalog no. bs-0366R) and a goat anti-mouse IgG antibody
(catalog no. bs-0296 G) were obtained from Beijing Biosynthesis
Biotechnology Co., Ltd. (Beijing, China). Streptavidin was obtained
from Promega Corporation (Madison, WI, USA). All labeled
oligonucleotides were synthesized by Invitrogen (Thermo Fisher
Scientific, Inc.). The sequences of each oligonucleotide are listed
in Table I. HotMaster Taq DNA
Polymerase kit was purchased from Tiangen Biotech Co., Ltd.
(Beijing, China), and dNTP and uracil-DNA glycosylase (UDG)
polymerase were from ShineGene Bio-Technologies, Inc. (Shanghai,
China). The Lowry protein assay kit, bovine serum albumin (BSA) and
calf serum were from Sigma-Aldrich (Merck Millipore).
 | Table I.Nucleotide sequences of the primers
used for ApoE genotyping. |
Table I.
Nucleotide sequences of the primers
used for ApoE genotyping.
| Primer | Sequence |
|---|
| ApoE 158C
(wild-type) forward primer |
5′-digoxin-ATGCCGATGACCTGCAGACGC-3′ |
| ApoE 158T (mutant)
forward primer |
5′-FITC-ATGCCGATGACCTGCAGACGT-3′ |
| ApoE 112T
(wild-type) forward primer |
5′-digoxin-CGCGGACATGGAGGACGTTT-3′ |
| ApoE 112C (mutant)
forward primer |
5′-FITC-CGCGGACATGGAGGACGTTC-3′ |
| Common reverse
primer |
5′-biotin-GTTCAGTGATTGTCGCTGGGCA-3′ |
Instruments
The Fourier transform-infrared (FT-IR) spectrum of
particles was recorded using a Nicolet 5700 FT-IR spectrometer
(Thermo Fisher Scientific, Inc.), followed by drying. A Hitachi
H-600 transmission electron microscope (TEM; Hitachi, Ltd., Tokyo,
Japan) was used to acquire images of particles, whereas particles
size and zeta potential were characterized by dynamic light
scattering using Zetasizer Nano ZS (Malvern Instruments Ltd.,
Malvern, UK). A 2550 UV-visible spectrophotometer (Shimadzu
Corporation, Tokyo, Japan) was used to determine the surface
plasmon resonance (SPR).
Surface-modified GMNs with PSS
A pure core/shell of Fe3O4/Au nanoparticle was
obtained by dispersing GoldMag nanoflower (Fe3O4/Au/Fe3O4) with
cationic surfactant, CTAB (22).
After ultrasonic treatment at 45 Hz for 20 min, 10 mg nanoflower
particles were gently mixed with 6 ml CTAB (5 mmol/l) and sonicated
for 40 min. Subsequently, the CTAB-GMNs (GoldMag nanoparticles)
were separated magnetically through the application of an external
permanent magnet, and the supernatant (solution containing uncapped
CTAB) was removed. A PSS solution (6 ml, 0.1 mg/ml) was added to
the CTAB-capped GMNs particles under sonication for 30 min and then
left to stand for 2 h. PSS-GMNs were magnetically separated and the
supernatant was discarded; this step was repeated twice. Then, the
PSS-GMNs were suspended in 7 ml of deionized water.
Conjugation of PSS-GMNs with targeted
moieties
PSS-GMNs (1 mg) were equilibrated in the 600 µl of
phosphate buffer (1X PB, pH 7.2), containing 40 µg of the targeted
moiety (anti-digoxin or anti-FITC antibody). This mixture was
shaken at 180 rpm for 1 h at 22°C. After 1 h of incubation, the
unconjugated antibodies were removed by washing in a magnetic
field. A blocking buffer (1X PB buffer, pH 7.2, containing 3% BSA
and 5% calf serum) was added to the conjugates, and the mixture was
incubated for 1 h. After incubation for 2 h, anti-digoxin antibody
or anti-FITC antibody functionalized PSS-GMNs conjugates, were
magnetically separated and then suspended in buffer (1X PB, pH 7.2,
containing 1% BSA) at 2–8°C prior to use. The conjugation
efficiency was calculated by determining the concentration of
anti-digoxin or anti-FITC antibody in the solution prior to and
following coupling using the Lowry protein concentration assay,
with BSA as a protein standard (23).
Preparation of LFA device
Streptavidin and goat anti-mouse IgG were printed on
a porous nitrocellulose membrane to form the test line (T line) and
control line (C line), respectively, using a HM3010 BioJet
dispenser (BioDot, Inc., Irvine, CA, USA). To detect the genotype
of each SNP, two complementary strips were run using half the
volume of the same PCR product separately. One strip detects
wild-type alleles (WT channel) and the other detects mutant alleles
(M channel). The probe solution containing PGMNs with an
anti-digoxin antibody or anti-FITC antibody was dispensed on the
conjugate pad of WT channel and M channel of the LFA, respectively.
These strips were placed in a card box and stored in a sealed
aluminum foil bag with desiccant silica gel at room temperature.
The strips remain stable for 12 months.
PCR-GoldMag based-LFA for the visual
detection of ApoE genotypes
According to the principle of ARMS-PCR (24), the reverse primers were designed as
biotin-labeled common primers, and the forward primers are allele
specific primers with the nucleotide at their 3′ terminus
corresponding to the SNP site. To detect the genotype of each
sample, two complementary reactions (ApoE 158 tube and ApoE 112
tube) were run separately. The primers were combined in two
reaction mixtures to yield predicted amplification products of 451
bp (Reaction ApoE 158: ApoE 158C, ApoE 158T) and 588 bp (Reaction
ApoE 112: ApoE 112T, ApoE 112C). Each PCR reaction was performed in
a total volume of 50 µl containing 10X PCR buffer (10 mM Tris HCl,
1.5 mM MgCl2), 2.5 µl DMSO, 0.2 mM dNTP mixture (dATP, dCTP, dGTP
and dUTP), 0.5 U HotMaster Taq DNA Polymerase, 0.5 U of UDG
polymerase, 0.1 µM common reverse primer and 4 µl DNA template (4
µl TE buffer without DNA served as a control in all experiments).
The ApoE 158 reaction mixture also contained 0.05 µM ApoE 158C and
ApoE 158T forward primers. Similarly, the ApoE 112 reaction mixture
contained 0.05 µM ApoE 112T and ApoE 112C forward primers. An
Applied Biosystems 2720 PCR Thermal Cycler (Thermo Fisher
Scientific, Inc.) was used for the amplification. The amplification
was commenced with a UDG incubation step (50°C for 2 min), initial
denaturation/UDG inactivation step (95°C for 5 min), followed by 30
cycles of 95°C for 30 sec and 65°C for 1 min, and a final extension
at 65°C for 5 min. Subsequent to PCR amplification, the whole
reaction volume (50 µl) was loaded on the sample pad of the WT
channel and M channel of LFA strips and the results were
analyzed.
The reference DNA samples with different ApoE
genotypes, confirmed by direct sequencing (Sangon Biotech Co.,
Ltd., Shanghai, China), were used to validate the method. The 3′
penultimate or antepenultimate nucleotide (underlined in Table I) was mismatched to enhance
specificity of the assay (25).
The sensitivity was evaluated by varying the concentrations of DNA
samples.
Clinical application
Human whole blood samples (n=305) were obtained from
the Shaanxi Provincial People's Hospital (Xi'an, China). All
subjects were self-reported to be from the Chinese Han populations
and unrelated to each other. Informed written consent was obtained
for participation in the present study, which was approved by the
Human Subjects Ethical Committee of Northwest University. Genomic
DNA was extracted from 200 µl whole blood sample by Whole Blood
Genomic DNA Isolation Kit from Xi'an GoldMag Nanobiotech Co., Ltd.
(Xi'an, China) according to manufacturer's instructions. All DNA
samples were tested to determine the genotypes of ApoE in a
double-blind trial. Each sample was analyzed using PGMNs-based LFA
strips and DNA sequencing (Sangon Biotech Co., Ltd.) as a
comparison.
Statistical analysis
Statistical analyses were performed using Microsoft
Excel version 14.0 (Microsoft Corporation, Redmond, WA, USA) and
SPSS software version 16.0 (SPSS, Inc., Chicago, IL, USA).
Statistical differences were determined using a Chi-squared test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Preparation of GoldMag probes for
LFA
GoldMag nanoparticles were synthesized and
characterized as described previously (21). The general scheme to functionalize
nanoflower GMNs with CTAB and PSS coating, is represented in
Fig. 1A. The addition of cationic
surfactant CTAB to the Fe3O4/Au/Fe3O4 particles inhibited the
absorption of additional Fe3O4 petals to the Fe3O4/Au core shell
structure. CTAB stabilized the core/shell structure by neutralizing
layers of surface positive charge to prevent aggregation. However,
CTAB-coated particle dispersions are frequently destabilized in
salt and buffer solution, resulting in partial aggregation and low
recovery yields (26). In the
present study, CTAB-capped coated Fe3O4/Au particles were
stabilized by wrapping the CTAB layer with PSS (Fig. 1A).
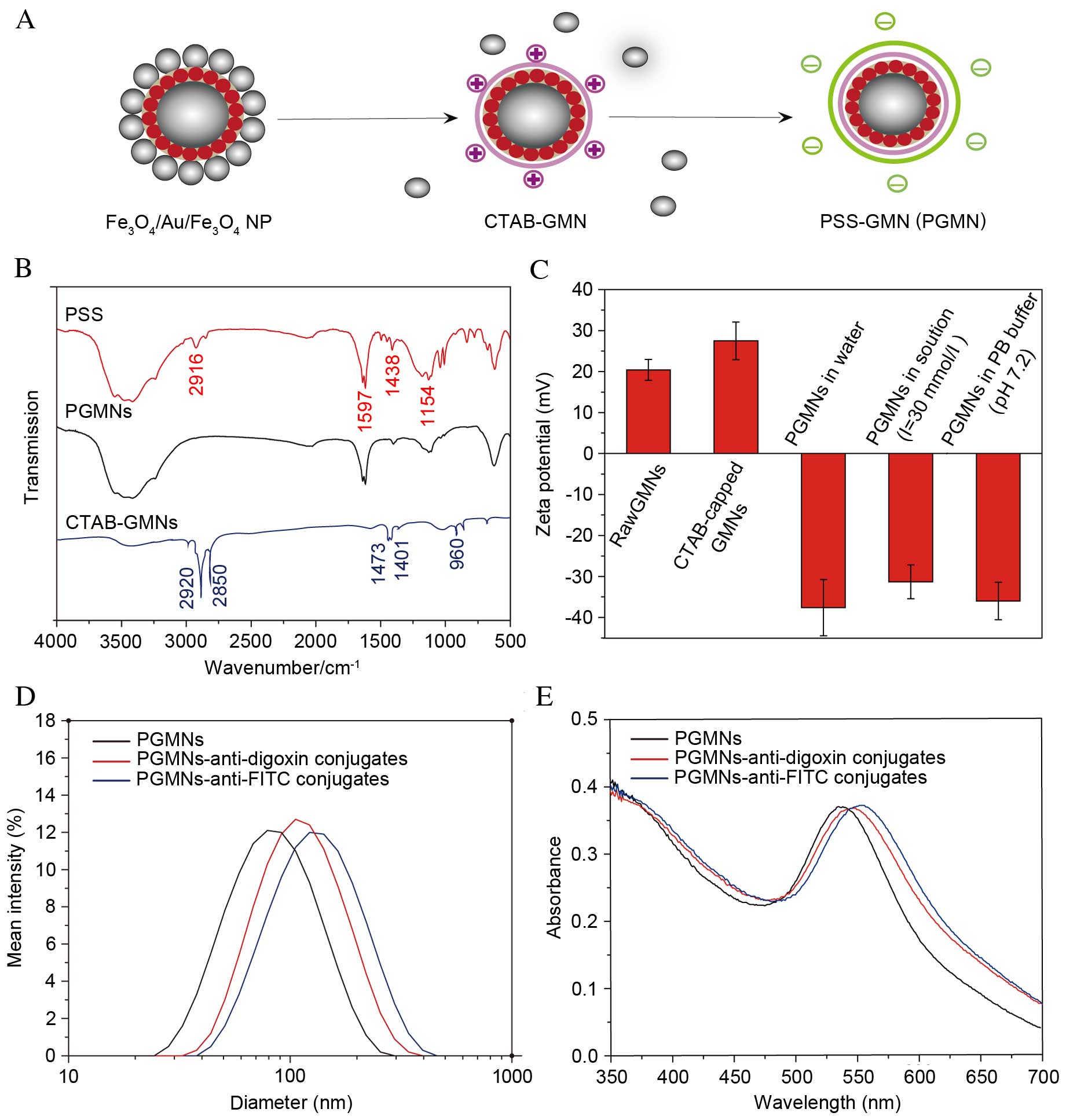 | Figure 1.Generating PGMNs. (A) Schematic
illustration of the process of surface modification, capping of the
particles with surfactant CTAB and a follow-up coating of PSS. (B)
FT-IR spectroscopy of CTAB-capped GMNs, pure PSS and PGMNs. (C)
Zeta potential of GMNs, CTAB-capped GMNs and PGMNs suspended in
water, PGMNs suspended in high electrolyte solution (I=30 mmol/l),
and PGMNs suspended in PB. (D) Size distribution of PGMNs and
PGMNs-antibody conjugates monitored by the dynamic light scattering
analyzer. Reasonable increase of hydrodynamic size indicates the
successful conjugation. (E) UV-vis spectrum of PGMNs-antibody
conjugates revealed a corresponding red shift of surface plasmon
resonance after PGMNs were conjugated with antibodies. NP,
nanoparticle; CTAB, cetyltrimethylammonium bromide; GMN, GoldMag
nanoparticle; PSS, polystyrenesulfonate; PGMN,
polyelectrolyte-coated GMN; FITC, fluorescein isothiocyanate. |
The successful attachment of PSS was demonstrated in
FT-IR spectra (Fig. 1B). The PSS
spectrum has typical absorption features of neat polystyrene and
sulfonate group (SO3−) (27,28),
including a primary absorption of the stretching vibrations that
correspond to asymmetric stretching of -CH2- at 2,916
cm−1, C-C stretching of sp2 hybridized carbon
atoms at 1,597 cm−1, para-disubstituted benzenes
(C=C) at 1,438 cm−1, symmetric vibrations of
-SO3 groups in PSS chains at 1,158 cm−1 and
stretching of hydroxyl groups of -SO2-OH over the region
3,700–3,000 cm−1. The spectra for CTAB-coated GMNs
exhibited the distinctive absorption peaks centered at 2,920,
2,850, 1,401, and 960 cm−1, which represented symmetric
(2,920 and 2,850 cm−1) and asymmetric (1,401
cm−1) stretching of the C-H bond in CTAB and quaternary
amine (960 cm−1) stretching of CTAB (19), respectively. For PGMNs, the
characteristic absorption bands of PSS were observed, whereas the
distinctive peaks of CTAB were not detected. This result
demonstrated the presence of the polymer on the particle,
indicating that the PSS functionalized GMNs had been successfully
prepared.
The stepwise conjugation of functional groups on the
GMNs was then monitored by measuring the surface charges at
different stages of synthesis. As presented in Fig. 1C, a positive zeta potential (+27.5
mV) in neutral pH (water) of CTAB-capped GMNs, is attributed to the
positive charge of the trimethyl ammonium group [-N
(CH3)3+] of CTAB. Following the
PSS coating of the nanoparticle surfaces, the zeta potential
shifted to negative values (−37.6 mV). This reversal of zeta
potential upon the PSS coating indicates the presence of PSS on the
nanoparticle surfaces, the PSS exhibits a constant negative charge
above pH 2 (18,29). Negative zeta potential formulations
help repel each particle in the suspension, ensuring long-term
stability and avoiding particle aggregation (30). When PGMNs were added into sodium
chloride solution with an ionic strength of 30 mmol/l or 1X PB
buffer at pH 7.2, a slight decrease in zeta potential of PGMNs was
observed due to lowering of ion screen effects of PSS in the
electrolyte solution.
For assay development, PGMNs were conjugated to
anti-digoxin antibodies or anti-FITC antibodies to construct
GoldMag probes. Bioconjugation of particles with targeted
antibodies was confirmed by dynamic light scattering measurement,
SPR band analysis and the protein concentration assay. As
demonstrated in Fig. 1D, the mean
hydrodynamic diameter of particles increased from 68 to 95 nm
(PGMNs-anti-digoxin) or 107 nm (PGMNs-anti-FITC) following
conjugation. The increased hydrodynamic size following the
conjugation suggested that antibodies were effectively coupled to
the particles (31). UV-vis data
(Fig. 1E) indicated that
antibody-labeled PGMNs exhibited a marked red shift compared with
the unlabeled ones, from 538 to 547 nm (PGMNs-anti-digoxin) or 552
nm (PGMNs-anti-FITC) where the plasmon resonance band appears
(32). This shift is likely to be
caused by the surface chemistry change of the nanoparticles from
PSS to antibody (33). The
optimization of PGMNs-antibody conjugates was defined by the
protein concentration assay as described in a previous report
(23), and the appropriate amount
of anti-digoxin or anti-FITC antibodies immobilized on the PGMNs
was about 60 and 48 µg/mg, respectively.
Principles of PCR-GoldMag LFA
The principle of visual detection of ApoE genotypes
using PCR-GoldMag LFA system is illustrated in Fig. 2. The method consists of the
following two steps (Fig. 2A): i)
Amplification of one sample in two reactions, ApoE 158 and ApoE 112
tubes; and ii) PCR products of each tube are loaded onto the sample
pads of two complementary LFA strips following amplification. One
strip detects wild-type alleles (termed ‘WT channel’) and the other
detects mutant alleles (termed ‘M channel’). The LFA strip is
composed of five parts (Fig. 2B):
A sample pad, a conjugate pad, a strip of nitrocellulose membrane,
an absorbent pad and a plastic backing. Detection of each PCR
product is performed using two complementary strips (WT and M
channel) with conjugate pads pre-dispensed with PGMNs-anti-digoxin
antibody conjugates and PGMNs-anti-FITC antibody conjugates,
respectively.
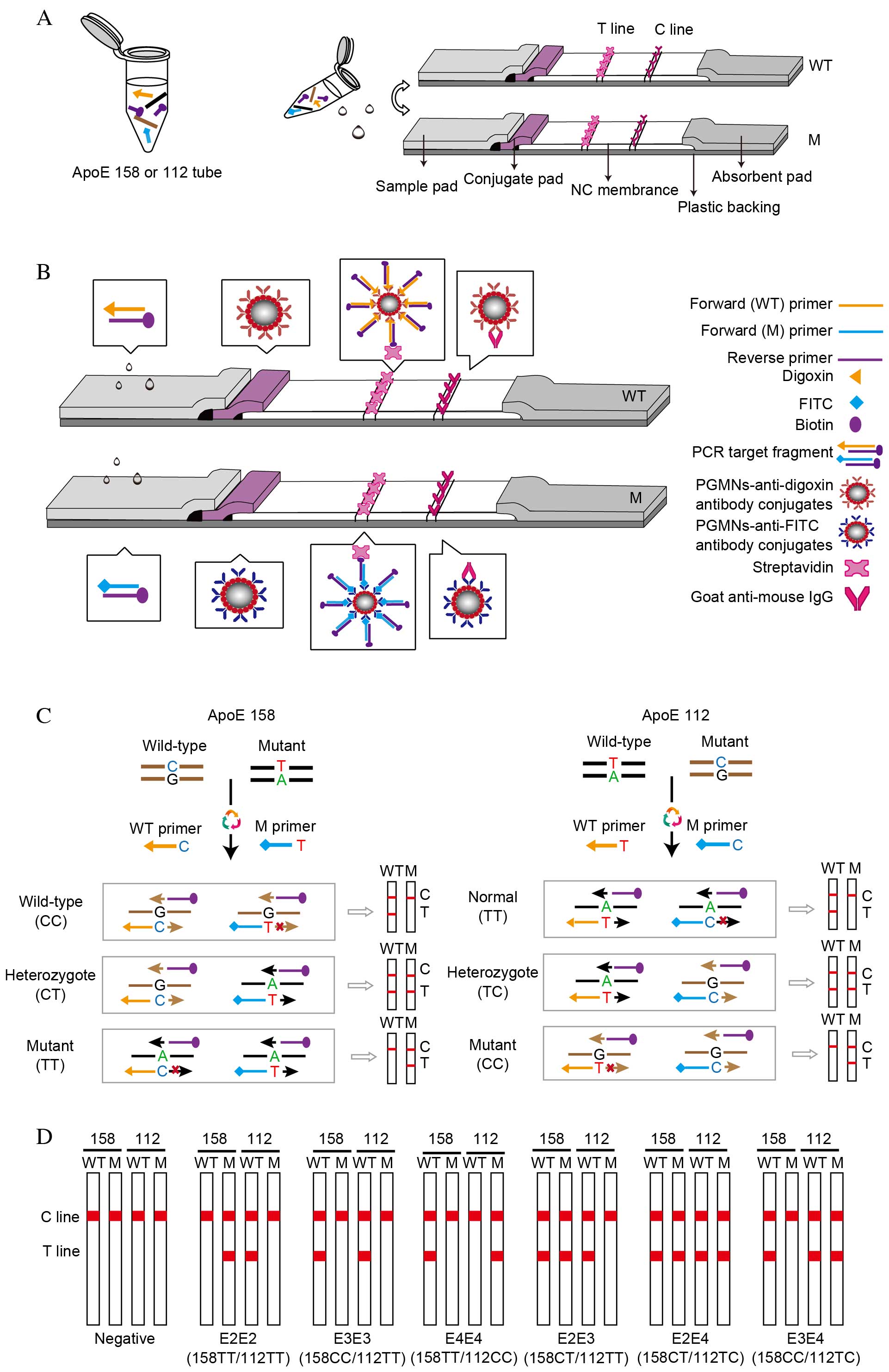 | Figure 2.Schematic of the PCR-GoldMag LFA
system. (A) Detecting one sample requires two reactions: ApoE 158
and ApoE 112. Detection of each PCR product using two complementary
strips (WT channel and M channel). (B) Schematic diagram of the LFA
strips based on the antibody-functionalized PGMN probes for visual
detection of the PCR products. The conjugate pads of WT and M
channel contain dispensed digoxin-conjugated PGMNs and
FITC-conjugated PGMNs, respectively. The target fragments carrying
the labels of digoxin/FITC and biotin are able to conjugate with
PGMNs-anti-digoxin/FITC antibody complexes, which are captured by
streptavidin on the test line with an appearance of a red band on
the WT/M channel. The excessive PGMNs-antibody complex is
precipitated by goat anti-mouse IgG on the control line. (C)
Schematic illustration of the formation of digoxin/FITC- and
biotin-conjugated duplex DNA complexes based on multiple
amplification refractory mutation system-PCR. (D) Representative
diagrams of LFAs in the presence of different ApoE phenotypes and
negative control. LFA, lateral flow assay; ApoE, apolipoprotein E;
T line, test line; C line, control line; NC, nitrocellulose; WT,
wild-type channel; M, mutant channel; FITC, fluorescein
isothiocyanate; PCR, polymerase chain reaction; PGMN,
polyelectrolyte-coated GoldMag nanoparticle. |
To reduce the labor and cost, the multi-ARMS-PCR
analysis was used to amplify wilt-type and mutant target SNP
alleles in a single reaction tube, by simultaneously using
allele-specific forward primers and a common, biotin-labeled
reverse primer (Fig. 2C).
Wild-type primers were labeled with digoxin and mutant primers were
labeled with FITC. Genotyping of ApoE 158 and ApoE 112 was
performed in two separate PCR tubes, with ApoE 158-specific primers
to produce 158-specific amplicons (451 bp) and the other with ApoE
112-specific primers to produce 112-specific amplicons (588 bp).
The PCR amplicons were synthesized with the 3′-end of the primer
complementary to the template. Thus, amplicons from the wild-type
DNA were biotin and digoxin labeled, whereas the mutant amplicons
were biotin and FITC labeled (Fig.
2C).
The visual detection of the amplicons was performed
on the LFA strips. No purification of the PCR products was required
prior to detection by LFA. In the test procedure, the PCR products
migrate through the membrane strip. PCR target fragments, if
labeled with digoxin, form a complex with the pre-fabricated
PGMNs-anti-digoxin antibody conjugates on the adjacent conjugate
pad of the WT channel strip. Similarly, PCR target fragments
labeled with FITC form a complex with the pre-fabricated
PGMNs-anti-FITC antibody conjugates of the M channel strip. The
subsequent DNA-PGMNs-antibody conjugates migrate across the
membrane strip until captured by pre-immobilized streptavidin on
the T line forming the positive result of a red band. The red band
at the T line of the WT channel strip indicates the presence of
wild-type fragments, and the red band at T line of the M channel
strip indicates presence of mutant fragments. The excess of
PGMNs-antibody conjugates is captured at the C line by goat
anti-mouse IgG that suggests the successful performance of the LFA
system.
The final genotyping result of a sample for one
locus requires visual inspection of the color development on the T
lines of both the WT and M channel strips (Fig. 2C). For wild-type sample, a distinct
red band is only visible on the T line of the WT channel, whereas
the T line of the M channel does not have a color band. By
contrast, for homozygous mutant samples, the red band appears
exclusively on the M channel strip and not on the WT channel strip.
However, when red bands with similar intensities are present on the
T lines of WT and M strips, it indicates a heterozygous mutant
sample. The detection of different ApoE phenotypes and negative
control (no DNA) is illustrated in Fig. 2D.
Performance of the LFA for ApoE
genotyping
To obtain the optimal performance, potential
variables, including ARMS, LFA preparation and detection
conditions, were considered. All primers were designed to avoid
cross-hybridization. In order to ensure the specificity of the
primers, an additional mismatch at the penultimate or
antepenultimate nucleotide of the 3′ terminus of allele specific
forward primers was introduced based on principle of ARMS-PCR
(24). To increase the specificity
and to reduce the PCR running-time, a combination of primer
annealing and extension steps in the PCR was performed. By using a
two-step PCR, the running-time was greatly reduced (30 cycles
within 60 min).
Specificity of the LFAs
The specificity of ApoE genotyping was an important
consideration for the present study. For specificity analysis, six
ApoE genotypes (E2/E2, E3/E3, E4/E4, E2/E3, E2/E4, and E3/E4) were
analyzed using the LFA system (Fig.
3A). In addition, ApoE genotyping results detected by DNA
sequencing were performed as a comparison (Fig. 3B). The GoldMag-based LFA can
clearly differentiate one base mutations between wild-type target
DNA and mutant target DNA, allowing successfully distinction
between the six ApoE genotypes.
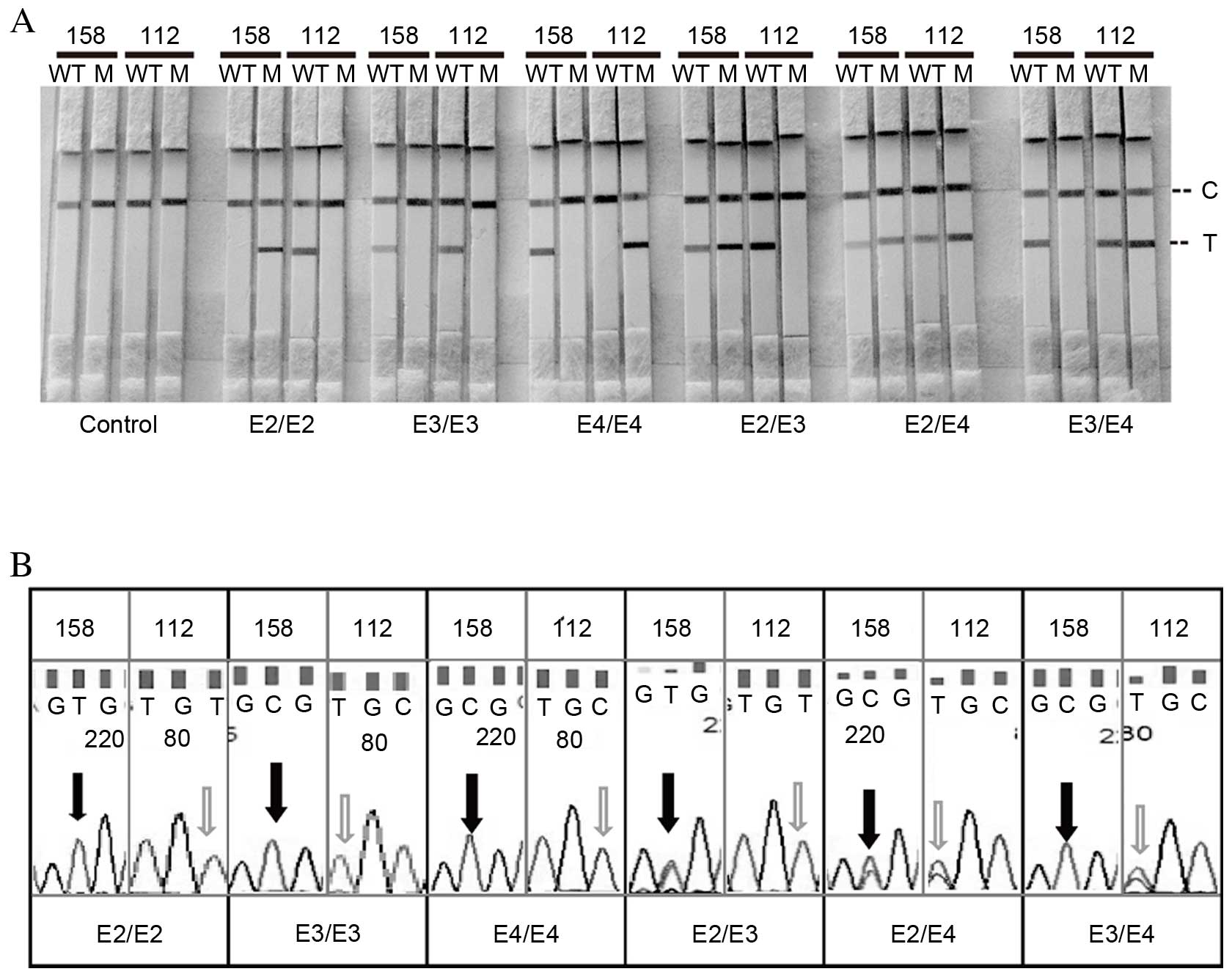 | Figure 3.Specificity. Specificity test results
of the amplification refractory mutation system method using 4 µl
each of DNA templates by (A) LFA and (B) direct DNA sequencing.
LFA, lateral flow assay; 158, apolipoprotein E 158 polymorphism;
112, apolipoprotein E 112 polymorphism; WT, wild-type channel; M,
mutant channel; C, control line; T, test line; E2, Cys158/Cys112
allele; E3, Cys158/Arg112 allele; E4, Arg158/Arg112 allele. |
Sensitivity of the LFAs
Under the optimum conditions, the sensitivity of LFA
was evaluated by using various quantities of DNA samples with known
ApoE genotypes. Different quantities of target DNA were used for
sensitivity analysis (5, 10, 20, 50, 100, 200, 400, 800 and 1,000
ng, and 0 ng as a negative control). Fig. 4 presents the representative images
of LFAs using different concentrations of ApoE 158 (Fig. 4A) and ApoE 112 (Fig. 4B) reference DNA templates. No red
band was observed on the T line of the LFA in the absence of DNA
(control). The signal intensity correspondingly increased as the
target DNA concentration increased. The red bands on the T line
were observed with ≥10 ng of target DNA, which was considered to be
the limit of detection of this assay.
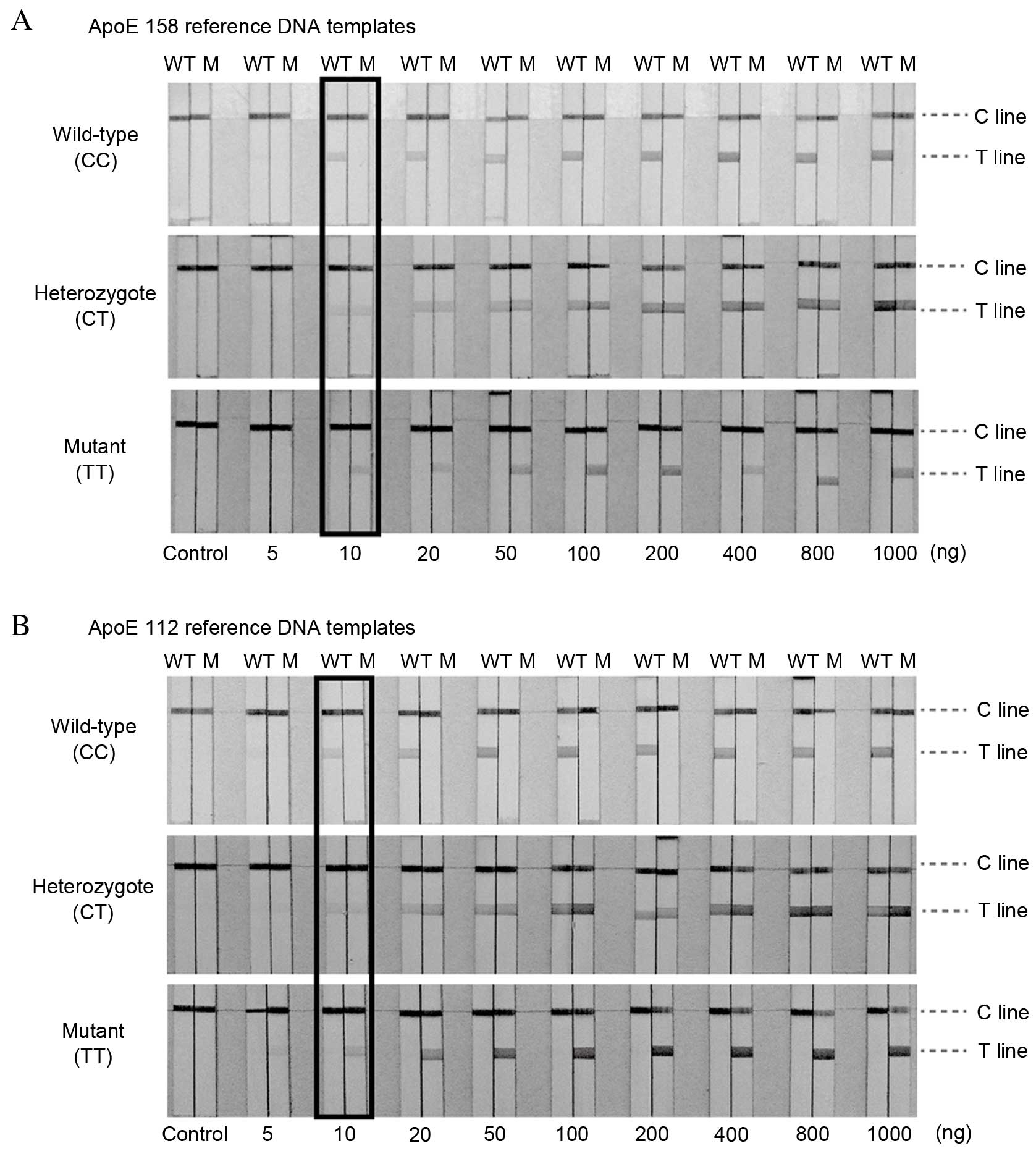 | Figure 4.Sensitivity. Typical responses of
LFAs with increasing quantities of (A) ApoE 158 and (B) ApoE 112
reference DNA templates. From left to right, the quantities of
target DNA were 0, 5, 10, 20, 50, 100, 200, 400, 800 and 1,000 ng.
The minimum amount of target DNA for PCR that reacted with the
particle conjugates causing coloration in the T-line is considered
to be the detection limit of the assay, and the limit of detection
of PCR-GoldMag LFA was as low as 10 ng. The limits of detection
obtained with LFA are highlighted. LFA, lateral flow assay; ApoE,
apolipoprotein E; 158, ApoE 158 polymorphism; 112, ApoE 112
polymorphism; WT, wild-type channel; M, mutant channel. |
Detection of clinical samples
To test the robustness of the proposed method for
the ApoE genotypes, 305 clinical samples from a Han Chinese cohort
were analyzed using the PCR-GoldMag LFA. In addition to the LFA,
direct DNA sequencing was performed. All analyzed samples produced
an unambiguous ApoE genotype. Of the 305 samples analyzed in the
current study, the allele frequency of E2, E3 and E4 was 6.98,
80.48 and 12.54%, respectively. The frequency of each of the ApoE
genotypes is presented in Table
II. All genotyping results (100%) were in accordance with the
results of direct DNA sequencing. In the subjects (n=305), the
overall allele distribution was similar to a previous study
(34), with no significant
difference in allele distribution (Chi-squared test, P=0.598).
These results indicate the reliability of using the PCR-GoldMag LFA
method for ApoE genotyping.
 | Table II.Genotypes and frequency of ApoE
variants in the Han Chinese cohort. |
Table II.
Genotypes and frequency of ApoE
variants in the Han Chinese cohort.
| Genotype | ApoE 158 variable
nucleotides | ApoE 112 variable
nucleotides | Frequency (%) |
|---|
| E2/E2 | TT | TT | 0.95 |
| E3/E3 | CC | TT | 67.62 |
| E4/E4 | CC | CC | 1.90 |
| E2/E3 | CT | TT | 8.24 |
| E2/E4 | CT | TC | 3.81 |
| E3/E4 | CC | TC | 17.48 |
Discussion
The present study demonstrated a PCR-GoldMag LFA for
visual detection of ApoE genotypes using PSS-functionalized GoldMag
nanoparticles as a carrier. To the best of our knowledge, this
represents the first attempt to perform ApoE genotyping by
integrating the multi-ARMS-PCR with LFA. GoldMag nanoparticles
allow visual detection of the PCR product by observing the red
color on an LFA strip. A prerequisite for immunoassay development
of GMNs is sufficient functionalization, which maintains GMNs in a
stable colloidal and monodispersed state with the ability to
conjugate to targeted moieties. This was achieved in the present
study with a fast and simple coating procedure using PSS. The
detection capabilities of the PCR-GoldMag LFA system were examined
using various quantities of DNA samples, with known ApoE genotypes.
The system accurately assesses a broad detection range of initial
starting genomic DNA quantity from 10 ng to 1,000 ng, with the
limit of detection reaching 10 ng. The specificity of the method
was also confirmed using known ApoE genotypes, and no false
positive results were observed. The performance of the LFA was
further confirmed using 305 clinical samples and demonstrated to be
a reliable method for determining the ApoE genotype. The entire
protocol of the established method, including PCR and the LFA, can
be performed in 1.5 h. No purification of the PCR products or
expensive detection instruments is required. The assay is also easy
to use and does not require highly qualified trained personnel to
be performed.
It has been previously proposed that the ApoE
polymorphism may be associated with a high of developing certain
diseases. The exposure of ApoE4 to contemporary environmental
conditions (Western diet and longer lifespan) may have rendered it
a susceptibility allele for coronary artery disease and Alzheimer's
disease (35). The present study
determined the frequency of the ApoE alleles, including the ApoE4
allele, in a cohort of northern Chinese Han population
subjects.
The allelic frequencies of ApoE vary substantially
around the world. Recently, a large study analyzed the ApoE allele
distribution in China (34),
including 19 separate cohorts, reporting distributions of 8.5, 83.0
and 8.5% for the E2, E3 and E4 alleles, respectively (n=3,679).
This particular previous study also found a conspicuous
south-to-north gradient of ApoE4 frequencies in China, with the
proportion of ApoE4 carriers at 4.9% in subjects from Kunming and
increased to 17.5% subjects from Harbin (34). In the subjects of the current study
(n=305), the overall allele distribution was similar to the
previous study, with no significant difference in allele
distribution (Chi-squared test, P=0.598).
In conclusion, the current study demonstrated that
the PCR-GoldMag lateral flow assay is a simple, sensitive, rapid
and cost-effective method for ApoE genotyping. This novel approach
may be adapted for the detection of other important SNPs and be
readily utilized for wide applications in molecular diagnosis
laboratories and for point-of-care genotype analysis.
Acknowledgements
This study was supported by the Project of National
Great New Drug Research and Development China (grant no.
2012ZX09506001-001, YC), the National Natural Science Foundation of
China (grant no. 31200749) and the National Institute of Health
(grant no. P20RR016457 from the INBRE Program of the National
Center for Research Resources, YW).
References
|
1
|
Hirschhorn JN and Daly MJ: Genome-wide
association studies for common diseases and complex traits. Nat Rev
Genet. 6:95–108. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shabo A: Integrating genomics into
clinical practice: Standards and regulatory challenges. Curr Opin
Mol Ther. 10:267–272. 2008.PubMed/NCBI
|
|
3
|
Bertram L and Tanzi RE: Thirty years of
Alzheimer's disease genetics: The implications of systematic
meta-analyses. Nat Rev Neurosci. 9:768–778. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu CC, Kanekiyo T, Xu H and Bu G:
Apolipoprotein E and Alzheimer disease: Risk, mechanisms and
therapy. Nat Rev Neurol. 9:106–118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lahiri DK, Sambamurti K and Bennett DA:
Apolipoprotein gene and its interaction with the environmentally
driven risk factors: Molecular, genetic and epidemiological studies
of Alzheimer's disease. Neurobiol Aging. 25:651–660. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hackler R, Schäfer JR, Motzny S, Brand S,
Kleine TO, Kaffarnik H and Steinmetz A: Rapid determination of
apolipoprotein E phenotypes from whole plasma by automated
isoelectric focusing using PhastSystem and immunofixation. J Lipid
Res. 35:153–158. 1994.PubMed/NCBI
|
|
7
|
Nishimura M, Satoh M, Nishimura S,
Kakinuma S, Sato K, Sawai S, Tsuchida S, Kazama T, Matsushita K,
Kado S, et al: Human apolipoprotein e resequencing by proteomic
analysis and its application to serotyping. PloS One. 9:e853562014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu YY, Delgado R, Costello R, Sunderland
T, Dukoff R and Csako G: Quantitative assessment of apolipoprotein
E genotypes by image analysis of PCR-RFLP fragments. Clin Chim
Acta. 293:213–221. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yi L, Wu T, Luo W, Zhou W and Wu J: A
non-invasive, rapid method to genotype late-onset Alzheimer's
disease-related apolipoprotein E gene polymorphisms. Neural Regen
Res. 9:69–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghebranious N, Ivacic L, Mallum J and
Dokken C: Detection of ApoE E2, E3 and E4 alleles using MALDI-TOF
mass spectrometry and the homogeneous mass-extend technology.
Nucleic Acids Res. 33:e1492005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Donohoe GG, Salomäki A, Lehtimaki T,
Pulkki K and Kairisto V: Rapid identification of apolipoprotein E
genotypes by multiplex amplification refractory mutation system PCR
and capillary gel electrophoresis. Clinical Chem. 45:143–146.
1999.
|
|
12
|
Poli M, Gatta LB, Dominici R, Lovati C,
Mariani C, Albertini A and Finazzi D: Apolipoprotein E haplotyping
by denaturing high-performance liquid chromatography. Clin Chem Lab
Med. 43:512–518. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Koch W, Ehrenhaft A, Griesser K, Pfeufer
A, Müller J, Schömig A and Kastrati A: TaqMan systems for
genotyping of disease-related polymorphisms present in the gene
encoding apolipoprotein E. Clin Chem Lab Med. 40:1123–1131. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ben-Avi L, Durst R, Shpitzen S,
Leitersdorf E and Meiner V: Apolipoprotein E genotyping: Accurate,
simple, high throughput method using ABI Prism SNaPshot Multiplex
System. J Alzheimers Dis. 6:497–501. 2004.PubMed/NCBI
|
|
15
|
Lie P, Liu J, Fang Z, Dun B and Zeng L: A
lateral flow biosensor for detection of nucleic acids with high
sensitivity and selectivity. Chem Commun (Camb). 48:236–238. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei Q, Xiang Z, He J, Wang G, Li H, Qian Z
and Yang M: Dumbbell-like Au-Fe3O4 nanoparticles as label for the
preparation of electrochemical immunosensors. Biosens Bioelectron.
26:627–631. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang C, Shen G, Shen Y and Zhang X: The
development of an electrochemical immunosensor using a thiol
aromatic aldehyde and PAMAM-functionalized Fe3O4@Au nanoparticles.
Anal Biochem. 485:66–71. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mehtala JG, Zemlyanov DY, Max JP, Kadasala
N, Zhao S and Wei A: Citrate-stabilized gold nanorods. Langmuir.
30:13727–13730. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang D, Ma JZ, Zhang QL, Li NN, Yang J,
Raju PA, Peng ML, Luo YL, Hui WL, Chen C and Cui Y:
Polyelectrolyte-coated gold magnetic nanoparticles for immunoassay
development: Toward point of care diagnostics for syphilis
screening. Anal Chem. 85:6688–6695. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hui W, Zhang S, Zhang C, Wan Y, Zhu J,
Zhao G, Wu S, Xi D, Zhang Q, Li N and Cui Y: A novel lateral flow
assay based on GoldMag nanoparticles and its clinical applications
for genotyping of MTHFR C677T polymorphisms. Nanoscale.
8:3579–3587. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hui W, Shi F, Yan K, Peng M, Cheng X, Luo
Y, Chen X, Roy V, Cui Y and Wang Z: Fe3O4/Au/Fe3O4 nanoflowers
exhibiting tunable saturation magnetization and enhanced
bioconjugation. Nanoscale. 4:747–751. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi F, Hui W, Chen C and Cui Y: Surface
modification and characterization of Fe3O4/Au
composite nanoparticles. NANO. 6:145–151. 2011. View Article : Google Scholar
|
|
23
|
Yang D, Ma J, Peng M, Zhang Q, Luo Y, Hui
W, Jin T and Cui Y: Building nanoSPR biosensor systems based on
gold magnetic composite nanoparticles. J Nanosci Nanotechnol.
13:5485–5492. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Little S: Amplification-refractory
mutation system (ARMS) analysis of point mutations. Curr Protoc Hum
Genet: Chapter 9:Unit 9.8. 2001.doi: 10.1002/0471142905.hg0908s07.
View Article : Google Scholar
|
|
25
|
Liu J, Huang S, Sun M, Liu S, Liu Y, Wang
W, Zhang X, Wang H and Hua W: An improved allele-specific PCR
primer design method for SNP marker analysis and its application.
Plant Methods. 8:342012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rostro-Kohanloo BC, Bickford LR, Payne CM,
Day ES, Anderson LJ, Zhong M, Lee S, Mayer KM, Zal T, Adam L and
Dinney CP: The stabilization and targeting of
surfactant-synthesized gold nanorods. Nanotechnology.
20:4340052009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee HY, Rwei SP, Wang L and Chen PH:
Preparation and characterization of core-shell
polyaniline-polystyrene sulfonate@Fe3O4
nanoparticles. Materials Chemistry and Physics. 112:805–809. 2008.
View Article : Google Scholar
|
|
28
|
Zalakain I, Politakos N, Ramos JA,
Saralegi A, Etxeberria H, Mondragon I, Corcuera A and Eceiza A:
Chemical and morphological characterization of sulfonated
polystyrene brushes in different environments. Eur Polym J.
49:2120–2127. 2013. View Article : Google Scholar
|
|
29
|
Dorris A, Rucareanu S, Reven L, Barrett CJ
and Lennox RB: Preparation and characterization of
polyelectrolyte-coated gold nanoparticles. Langmuir. 24:2532–2538.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Billotey C, Wilhelm C, Devaud M, Bacri JC,
Bittoun J and Gazeau F: Cell internalization of anionic maghemite
nanoparticles: Quantitative effect on magnetic resonance imaging.
Magn Reson Med. 49:646–654. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ahmadi A, Shirazi H, Pourbagher N and
Omidfar K: Synthesis and characterization of core-shell Au Fe oxide
nanocomposites and their application for detecting immunological
interaction. Monoclon Antib Immunodiagn Immunother. 33:74–79. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
McFarland AD, Haynes CL, Mirkin CA, Van
Duyne RP and Godwin HA: Color my nanoworld. J Chem Educ.
81:544A2004. View Article : Google Scholar
|
|
33
|
Kaur K and Forrest JA: Influence of
particle size on the binding activity of proteins adsorbed onto
gold nanoparticles. Langmuir. 28:2736–2744. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu P, Qin YH, Jing CX, Lu L, Hu B and Du
PF: Does the geographical gradient of ApoE4 allele exist in China?
A systemic comparison among multiple Chinese populations. Mol Biol
Rep. 38:489–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Corbo RM and Scacchi R: Apolipoprotein E
(APOE) allele distribution in the world. Is APOE*4 a ‘thrifty'
allele? Ann Hum Genet. 63:301–310. 1999. View Article : Google Scholar
|


















