Introduction
Biofilms are multicellular communities of bacteria
encased in an extracellular matrix of exopolysaccharides (EPS),
proteins and, occasionally, DNA (1,2). EPS
are thought to maintain the architecture of the biofilm and to
protect the bacteria by harboring them within the biofilm. The
composition and structure of the EPS component may vary according
to the microbial constituents, which are affected by local
environmental conditions; for example, by shear forces in fluid
environments, and can change over time (3–5). EPS
are a key component of the matrix of cariogenic oral biofilms and
are known virulence factors in the pathogenesis of dental caries
(6). EPS create complex
three-dimensional structures and compartmentalized acidic
microenvironments within a biofilm, which trigger the dominance of
pathogenic Streptococcus mutans within a mixed-species
system (7). Glucans constitute the
majority of the EPS in S. mutans biofilms (8–10).
S. mutans synthesizes glucans from dietary sucrose via
glucosyltransferases (GTFs). These enzymes are virulence factors in
the etiology and pathogenesis of dental caries (9). Glucans promote the adherence and
accumulation of S. mutans on the tooth surface and serve an
essential role in the development of caries-forming activity
(6). The majority of previous
research has focused on the bacterial aspects of these biofilms,
whereas the assembly of the S. mutans EPS and the functional
roles that these EPS serve in the microcosmic basic structures of
these biofilms have received limited attention.
S. mutans lives on the tooth surface at a
high cell density in dental plaque. In a process termed quorum
sensing (QS), which was first discovered in the marine bacterium
Vibrio fischeri, bacterial populations coordinate the
behavior of the community. It is now apparent that QS systems exist
in both Gram-positive and Gram-negative bacteria, and that they
have evolved to improve access to complex nutrients or
environmental niches or to collectively enhance the defense against
other microorganisms or eukaryotic-host defense mechanisms
(11–14). QS bacteria convey their presence to
one another by releasing and responding to the accumulation of
low-molecular-weight chemical signaling molecules called
autoinducers. Gram-positive bacteria generally accomplish
interspecies communication via processed oligopeptides and these
signals are detected via two-component signal transduction systems.
S. mutans produces both AI-2 and competence-stimulating
peptides (CSPs), which belong to the AI-1 family, via the
luxS and comC genes, respectively. The relationship
between QS and S. mutans biofilm structure has been
investigated and discussed previously. The ability of bacterial
cells to communicate and behave collectively as a group most likely
provides significant benefits for the colonization of hosts,
defense against competitors, adaptation to varying physical
conditions, cellular differentiation and species evolution
(15).
Polyamines are linear, organic polycations that are
fully protonated at physiological pH and are observed in most
bacterial species. Due to their cationic nature, polyamines are
able to interact with negatively charged molecules within the cell,
including DNA, RNA, proteins and phospholipids, thereby affecting
their structure and function (16). Polyamines are known to serve
important roles in biofilm formation, although the exact mechanisms
remains to be elucidated. Polyamines have been observed to affect
proteins that are important for biofilm formation in Yersinia
pestis and Escherichia coli (17–19),
and may serve as intercellular signaling molecules in Proteus
mirabilis (19). Norspermidine
(Nspd), with one methyl less than spermidine, serves a negative
role in numerous bacteria, with the exception of Vibrio. In
addition, exogenous Nspd may directly dissolve EPS, leading to the
detachment of Bacillus subtilis biofilms, although this hypothesis
remains controversial (20).
However, information is limited regarding the association between
polyamines and biofilm structure, particularly the EPS, which
comprise major structures in biofilms.
In the present study, the impact of Nspd on S.
mutans biofilm formation and biofilm three-dimensional
structure were investigated in vitro. By analyzing the
differentially expressed genes affected by Nspd, the present study
proposed that the basic structure of the S. mutans biofilm
appeared to be controlled by a QS system.
Materials and methods
Bacterial growth, agents and biofilm
formation
Streptococcus mutans UA159 was obtained from
Provincial Key Laboratory of Stomatology, Sun Yat-sen University
(Guangzhou, China). The medium used included brain heart infusion
broth (BHI; Becton Dickinson, Sparks, MD, USA). Nspd
(Sigma-Aldrich, St. Louis, MO, USA) was used in the present study.
The polyamine solutions were freshly prepared prior to each
use.
Effective Nspd concentration on the
biofilm formation of S. mutans and growth curve detection
Nspd at concentrations of 5, 2.5 and 1.25 mM, and
625, 320, 160, 80 and 5 µM were added to the S. mutans
suspensions in 96-well plates. The plates were subsequently
incubated anaerobically at 37°C for 24 h without agitation. The
biofilms that had formed in the BHI were washed twice and crystal
violet (CV) staining was performed to determine the biofilm biomass
as the blank control.
Nspd was added to the S. mutans suspensions
in tubes containing fresh liquid BHI to a final concentration of 5
mM prior to incubation. The tubes were subsequently incubated
anaerobically at 37°C for 12, 24, 36, 48, 60, 72, 84 and 96 h
without agitation. The number of viable bacteria in each of the
cultures was determined in 10-fold serial dilutions with sterile
water using the plate count methods. All experiments were performed
at least in triplicate and cultures lacking polyamines were used as
the blank control.
Effect of polyamines on biofilm
disassembly and dynamic light scattering of EPS
Biofilms that had formed over 24 h without
polyamines were washed twice to remove the media. Fresh BHI
containing 1% sucrose was added to the wells, and Nspd was added to
a final concentration of 5 mM. Fresh BHI-sucrose was added to the
control group. The plates were incubated at 37°C for 24 h under
anaerobic conditions without agitation. Following this, the
biofilms were washed twice and CV staining was performed to
evaluate the biofilm biomass.
The dynamic light scattering measurements were
performed according to methods reported previously, with some minor
changes (20). Biofilms formed in
BHI containing 1% (w/v) sucrose for 24 h were harvested and washed
twice to remove the supernatants. The biofilms were subsequently
collected and resuspended in a 0.4 M NaOH solution. The cells were
removed by centrifugation at 18,900 × g at 4°C for 10 min,
and the supernatants were mixed with cold isopropanol at a 5:1
ratio and incubated at 4°C overnight. The samples were centrifuged
at 7,000 × g at 4°C for 10 min, and the pellets were
harvested and subsequently lyophilized. The temperature was
maintained at 25°C. The dry, water-insoluble EPS samples were
diluted in water and were gently sonicated at 200 W for 5 min. The
final EPS concentrations were adjusted to 30 mg/ml. The light
scattering value of each sample was measured prior to and after the
addition of Nspd to a final concentration of 5 mM or 10 mM. The
dynamic light scattering measurements were obtained by focusing a
vertically polarized light (532 nm) onto the sample and collecting
the scattered light using a detector arranged at 90°. The data were
collected at 30 sec intervals for 10–30 min.
Stereomicroscopy
Biofilms that had formed in 24-well microtiter
plates for 96 h in the presence of 5 mM Nspd or control group were
washed twice to remove the supernatant and were naturally air-dried
at room temperature for 20 min. The biofilms were observed and
images were captured using a stereomicroscope at a magnification of
×5 (Leica M205A; Leica Microsystems, Wetzlar, Germany).
Confocal laser-scanning microscopy
(CLSM)
The biofilms of each group were grown on
glass-bottomed chamber slides for 12, 24, 36, 48, 60, 72, 84 and 96
h. The biofilms were washed twice, stained using 200 µl
polysaccharide label Fluorescent Brightener 28 at 300 mg/l
(Sigma-Aldrich) for 30 min and subsequently washed twice using PBS
to remove the excess dye.
In the cell- and EPS-staining experiment, the
samples were stained using the L-7012 LIVE/DEAD
BacLight™ Bacterial Viability kit (Molecular Probes
Inc., Eugene, OR, USA) according to the manufacturer's protocol
after staining with Fluorescent Brightener 28. The live and dead
cells in the biofilms were differentiated by staining using SYTO9
(green fluorescence) and propidium iodide (PI, red fluorescence).
The samples were subsequently washed twice using PBS to remove the
excess dyes.
Images of the stained specimens were captured using
a Carl Zeiss LSM 780 confocal laser-scanning microscope (Carl Zeiss
Microscopy, Jena, Germany) and analyzed using ZEN software (Zen
2012 light edition; Carl Zeiss MicroImaging, Inc., Thornwood, NY,
USA). Three randomly selected areas of each biofilm were
scanned.
Microarray analysis of the gene
expression
Samples
The biofilms in the control and Nspd groups were
allowed to form for 48 h prior to harvesting by centrifugation and
re-suspension in TRIzol reagent (Takara Bio Inc., Dalian, China).
Each suspension was subsequently transferred to an RNase-free 1.5
ml microcentrifuge tube. The quantity and quality of the RNA were
evaluated using a NanoDrop ND-1000 instrument. The RNA integrity
was assessed using standard denaturing agarose-gel
electrophoresis.
DNA microarray
The Streptococcus mutans UA159 microarray is
a broad array that represents well-known and predicted genes and
transcripts. Coupled with the gene prediction capability of NCBI
and the probe selection program of Agilent, this platform delivers
data of better quality and has less redundant gene coverage.
RNA labeling and array hybridization
Sample labeling and array hybridization were
performed according to the Agilent One-Color Microarray-Based Gene
Expression Analysis protocol (Agilent Technology, Inc., Santa
Clara, CA, USA). Briefly, the total RNA from each sample was
linearly amplified and labeled using Cy3-UTP. The labeled cRNAs
were purified using an RNeasy Mini kit (Qiagen GmbH, Hilden,
Germany). The concentration and specific activity of the labeled
cRNAs (pmol Cy3/µg cRNA) were measured using a NanoDrop ND-1000
instrument. One microgram of each labeled cRNA was fragmented by
adding 11 µl of 10X blocking agent and 2.2 µl of 25X fragmentation
buffer, and heating the mixture to 60°C for 30 min. Finally, 55 µl
of 2X GE hybridization buffer was added to dilute the labeled cRNA
(GE Healthcare Life Sciences, Chalfont, UK). A total of 100 µl
hybridization solution was dispensed into the gasket slide, which
assembled on the gene expression microarray slide. The slides were
incubated for 17 h at 65°C in an Agilent hybridization oven
(Agilent Technology, Inc.). The hybridized arrays were washed,
fixed and scanned using the Agilent DNA Microarray Scanner (part
number, G2505C; Agilent Technology, Inc.).
Data analysis
Agilent Feature Extraction software (version
11.0.1.1; Agilent Technology, Inc.) was used to analyze the
acquired array images. Quantile normalization and subsequent data
processing were performed using the GeneSpring GX v11.5.1 software
package (Agilent Technologies, Inc.). Following quantile
normalization of the raw data, the genes expressed in at least two
of the six samples were flagged as detected and were selected for
further analysis. Genes that were significantly differentially
expressed were identified through volcano plot filtering.
Hierarchical clustering was performed using Agilent GeneSpring GX
software (version 11.5.1). Pathway analysis was performed using the
standard enrichment computation method.
Statistical analysis
All experiments were performed in triplicate and
were repeated at least three times. The data were analyzed using
SPSS (version 17.0 for Windows) software. Two-group comparisons
were performed using Student's t-test. Significance was set as
P<0.05. The COMSTAT program was used to calculate the biomass,
number and size (volume, diameter and thickness) of the
microcolonies.
Results
Nspd effective concentration and
growth curve in BHI
Reductions of 75.9% biofilm volume was observed in
the Nspd group of 5 mM concentration (Fig. 1A) (P<0.05). The growth curve
revealed that S.mutans growth was inhibited during the early
stages, then rebounded and reaching their growth peaks at 36 h
(Fig. 1B).
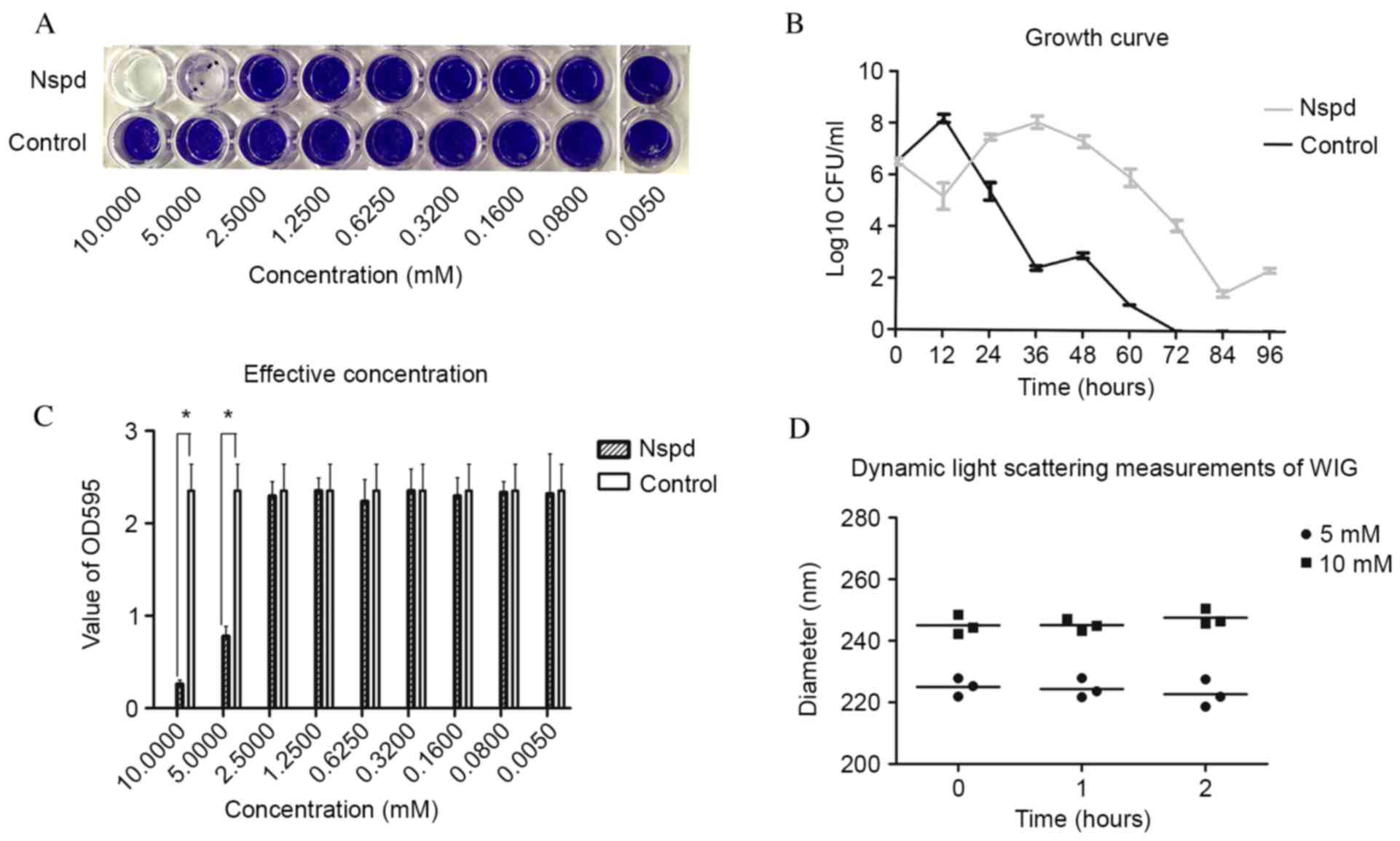 | Figure 1.(A) Biofilms that formed following
the addition of Nspd at 5 mM and incubated for 24 h, as visualized
using crystal violet staining. (B) Growth curve of Streptococcus
mutans. Nspd (5 mM) was added to the suspension, which was
incubated anaerobically at 37°C for 12, 24, 36, 48, 60, 72, 84 and
96 h without agitation. The results were expressed as the mean ±
standard deviation of triplicate assays. (C) Biofilm production was
evaluated using the OD595 values. The results are expressed as the
mean ± standard deviation of triplicate assays (*P<0.05, n=9
paired Student's t-test). (D) Dynamic light scattering
measurements. The diameters of the water-insoluble
exopolysaccharide were determined following treatment with 5 or 10
mM Nspd (n=3). Each measurement was repeated three times.
Correlation analysis indicated no linear relationship between the
diameter of the water-insoluble glucan and Nspd treatment
(P>0.05). Nspd, norspermidine; CFU, colony forming units; OD,
optical density; WIG, water-insoluble glucan. |
Nspd had no direct effect on EPS nor
caused any disassembly effect of S. mutans biofilm
In the disassembly experiment, Nspd was added to the
cultures for 24 h. Following this period, no significant
differences in the optical density values of each group was
observed (P>0.05). No collapse or vacuoles were observed in
these groups using a Leica M205A stereomicroscope (Leica
Microsystems, Wetzlar, Germany). As an independent approach to
detect an interaction between Nspd and EPS, dynamic
light-scattering experiments were performed. No direct effect of
Nspd on the EPS was detected in this experiment.
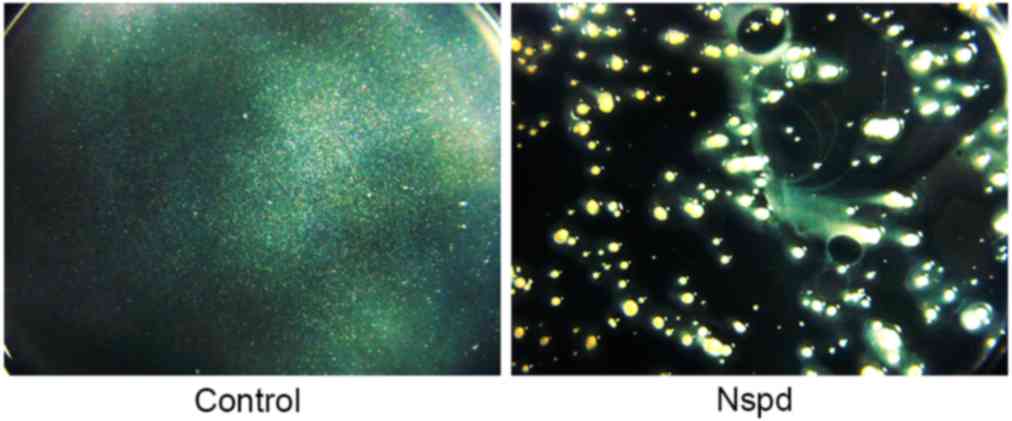
Non-homogeneous general appearance of
the biofilms in the experiment group was observed using
stereomicroscopy
The biofilms produced in the control group were
thick and firm, and formed intact blocks that were tightly attached
to the surface of the plate. These biofilms had a smooth surface
and uniform structure. The biofilms appeared to have white, smooth,
confluent and translucent membranes (Fig. 2).
The biofilms of the Nspd group were very different
compared with the control group. Their aggregates had a stellate
shape and had no connection to each other. Rough, non-homogeneous
and irregular structures were observed in these biofilms.
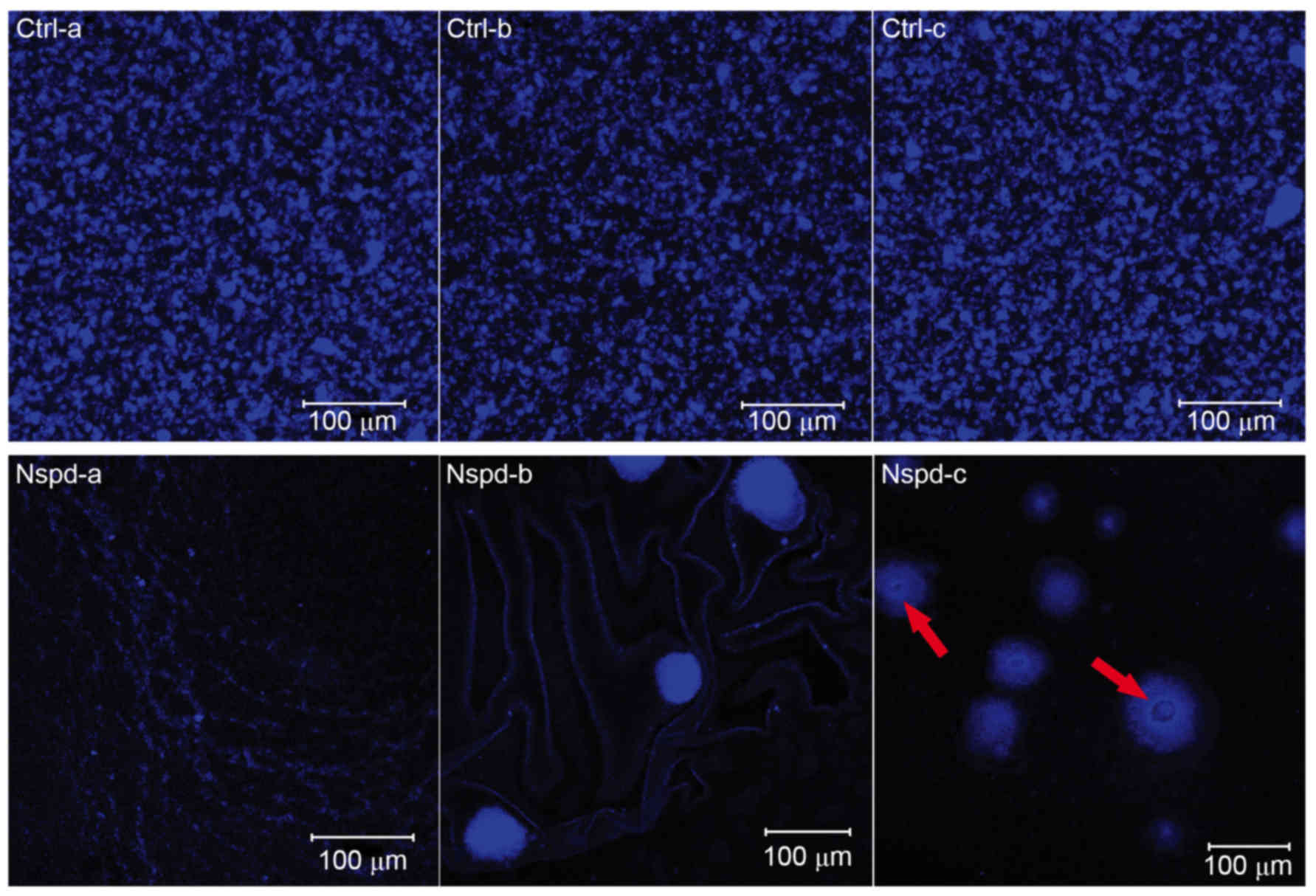
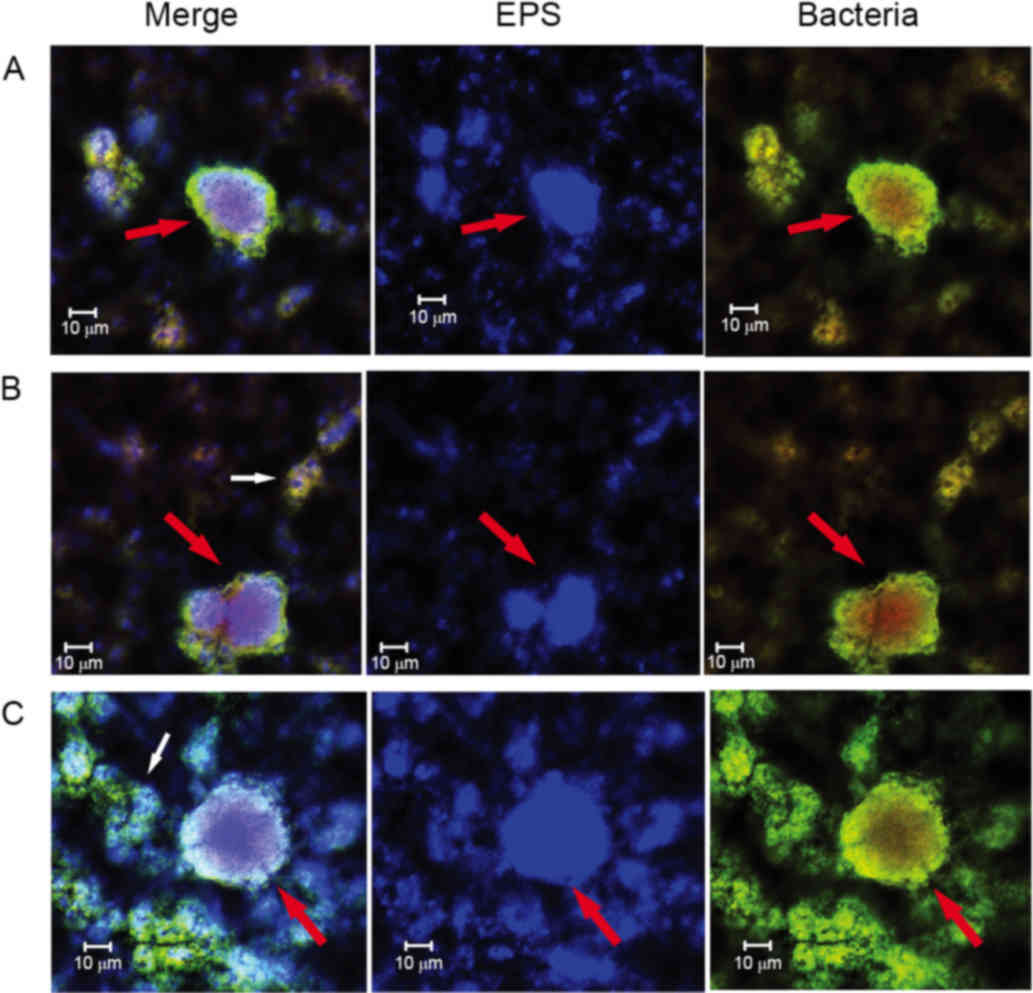
Diverse EPS architectures lead by Nspd
observed using CLSM
The EPS granules in the control group appeared as
globular structures and had similar sizes ranging from ~5–25 µm
(Fig. 3). The uniform and regular
EPS granules had a thickly dotted structure. The spaces within
these EPS granules were uniform and ranged from ~1–25 µm in
diameter. While in the experiment group, S. mutans formed
heterogeneous EPS structures. Some of these EPS structures
resembled fishnets and were wrinkled (Fig. 3, Nspd-a). Other structures were
small and were connected to one another via chain-like structures
(Fig. 3, Nspd-b). Larger
structures that resembled pearls with a central hole or tube were
also observed (Fig. 3, Nspd-c).
This diverse EPS structure resulted in an irregular biofilm with
varying thicknesses that appeared highly different from the
biofilms produced by the other groups.
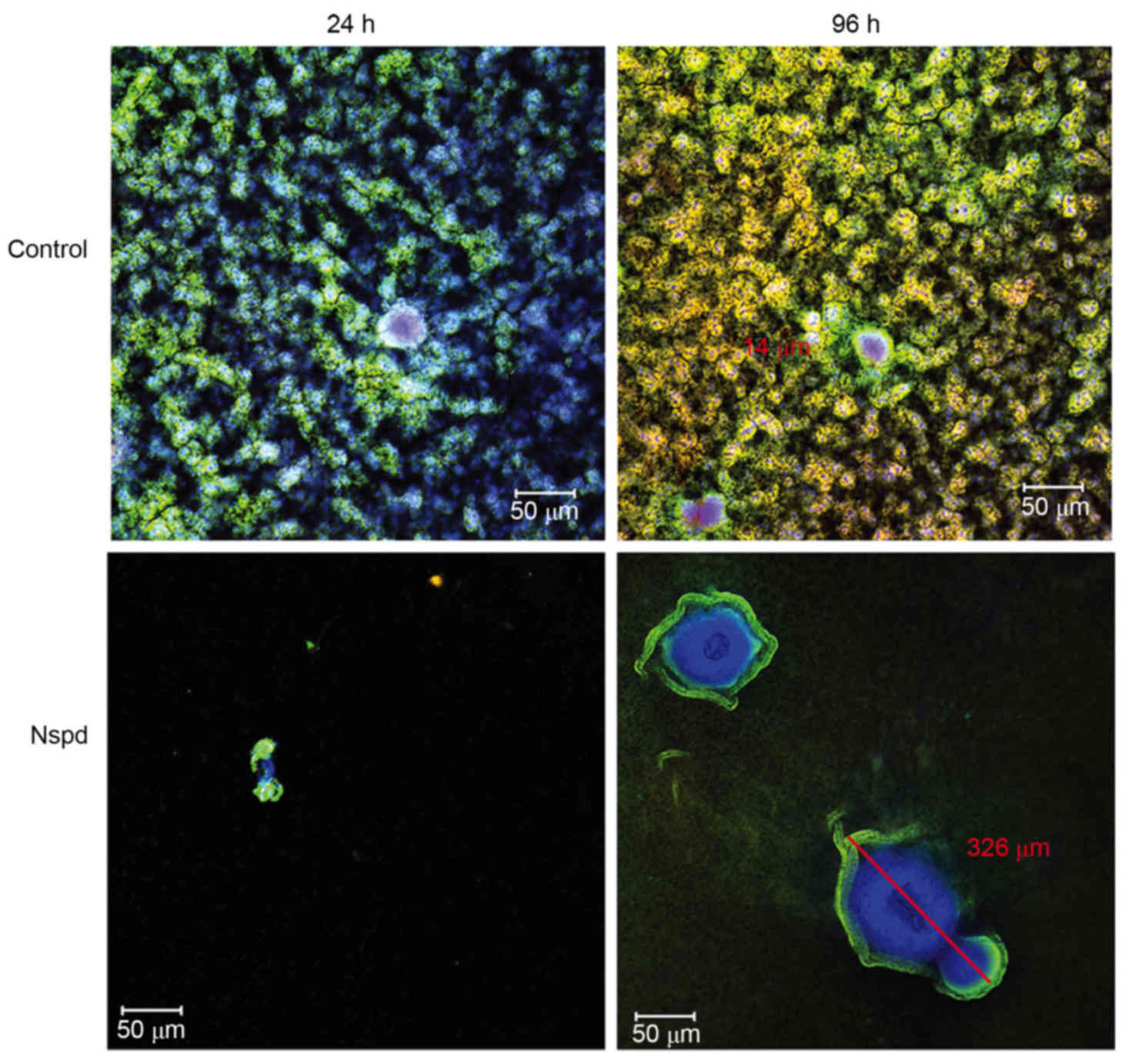
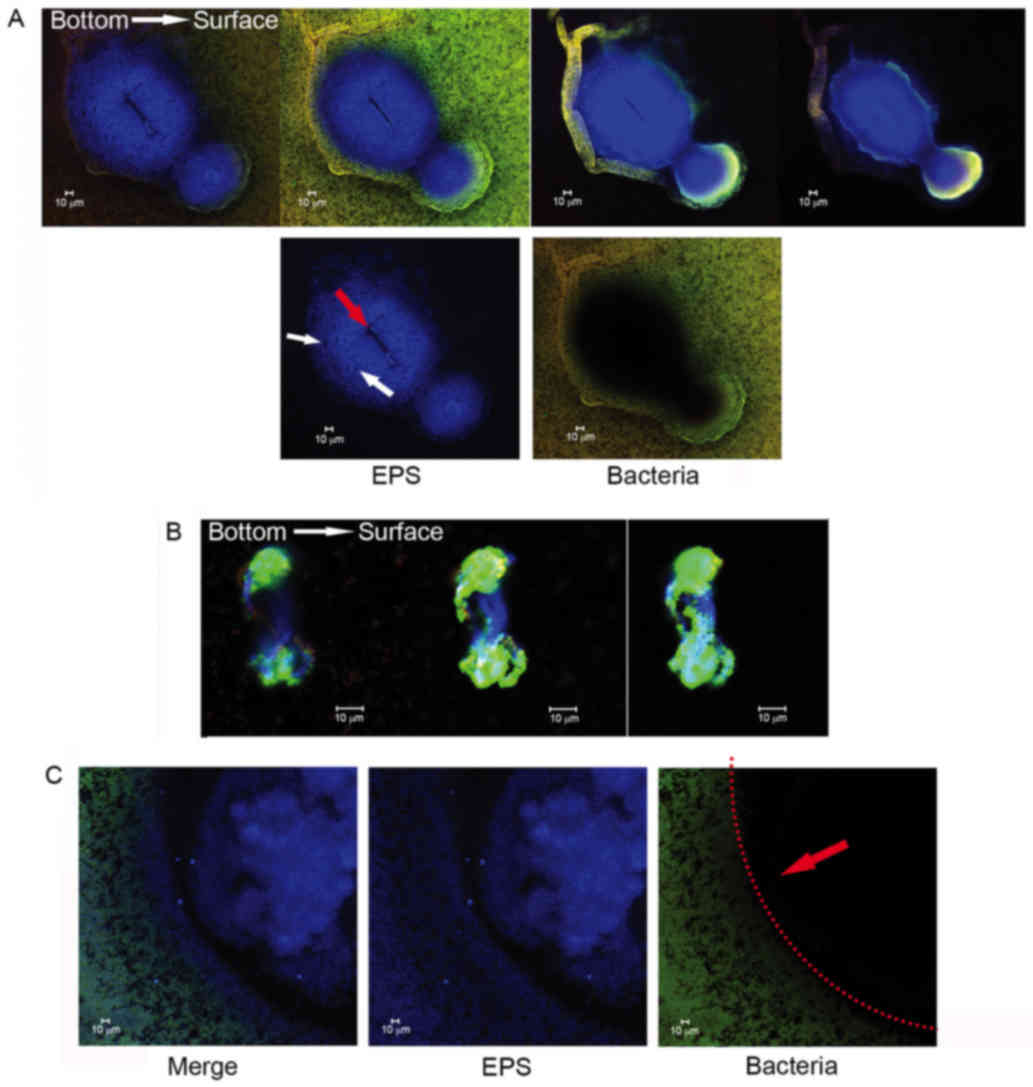
Three-dimensional distribution detail
for basic structure units in S. mutans biofilms observed using
CLSM
More detailed architectural differences were
revealed in cells along with EPS stained observations (Fig. 4). Little structural differences
were observed, however the number of dead cells increased between
24 and 96 h in the control group, while small aggregates expanded
to a huge architecture in the experiment group. This Similarly
sized and regularly shaped EPS granules were scattered equally
throughout the biofilms in the control group. Within these
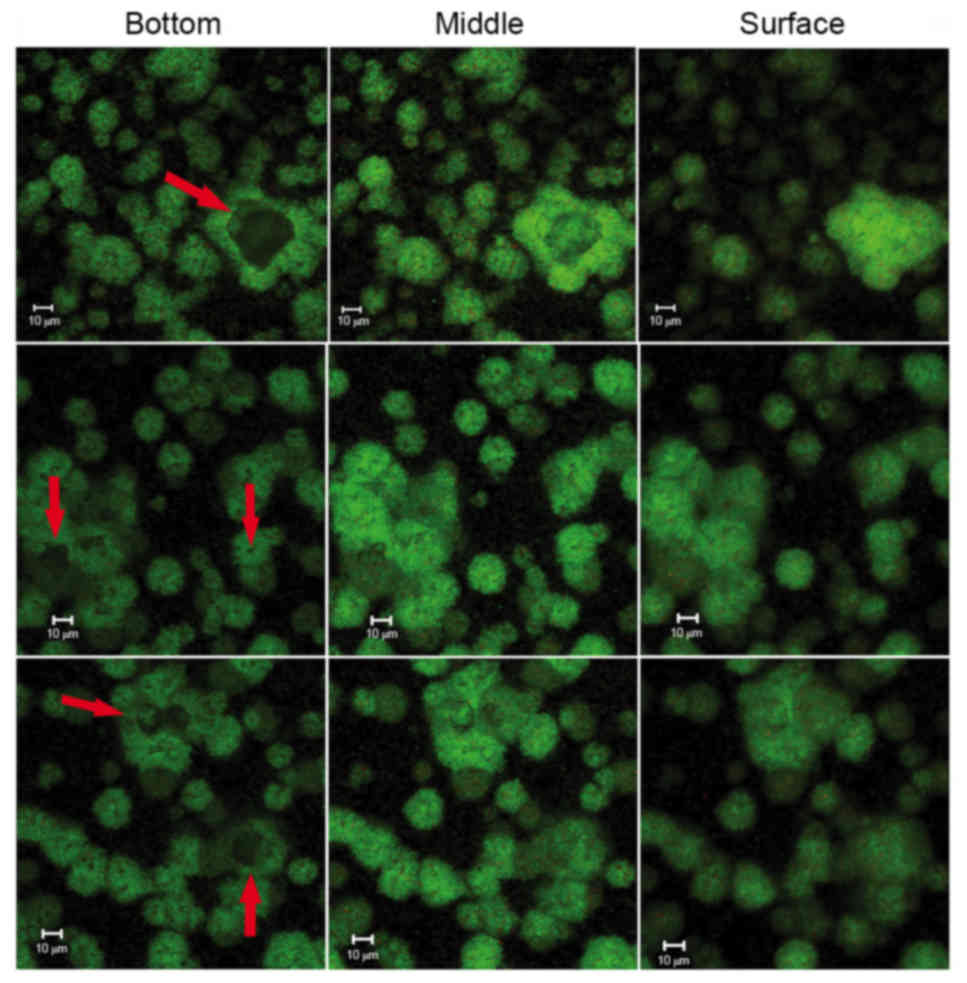
structures was a ‘space’ lacking stained bacteria, which made the
ball-like structures appear to be hollow spheres (Fig. 5). The ‘space’ gradually narrowed
from the adherent interface (bottom) to the biofilm surface. These
types of ‘hollow space’ appeared to be completely filled with
blue-stained EPS (Fig. 6). The
cells surrounded the EPS formed three-dimensional ball- or
stick-like structures. EPS was at the center of each ball-like
bacterial cluster and exhibited an even gel-like brightness. This
confirmed the exact basic structure assembly pattern of S.
mutans biofilms. The basic structures of the biofilm included
live and dead cells, which were organized into spheres or
irregular-shaped balls (Fig. 5).
These basic structures were uniform and well-distributed due to the
size of each unit being ~5–40 µm in diameter (Figs. 4 and 6). The clusters were distributed in a
regular pattern. The images demonstrated that the spherical
structural units consisting of bacteria surrounding EPS were the
basic units in the S. mutans biofilms (Fig. 6). The basic units were notably
larger in the Nspd group (Figs. 4
and 7). The large EPS structures
were irregular ball-like units with a thin bacterial layer on their
surfaces, and there were holes in the center of the huge EPS
clusters (Figs. 4 and 7). The course of the assembly of the EPS
structures and the formation of the basic units in the Nspd group
is shown (Figs. 4 and 7). During the first stage of ~24 h, a
small number of bacterial clusters aggregated and successfully
adhered to the surface of the chamber slide and began to secrete
EPS toward the underside of the center of the clusters. With the
cells continuing to secrete, the EPS with the units became more
compact and abundant over time and formed a central hole.
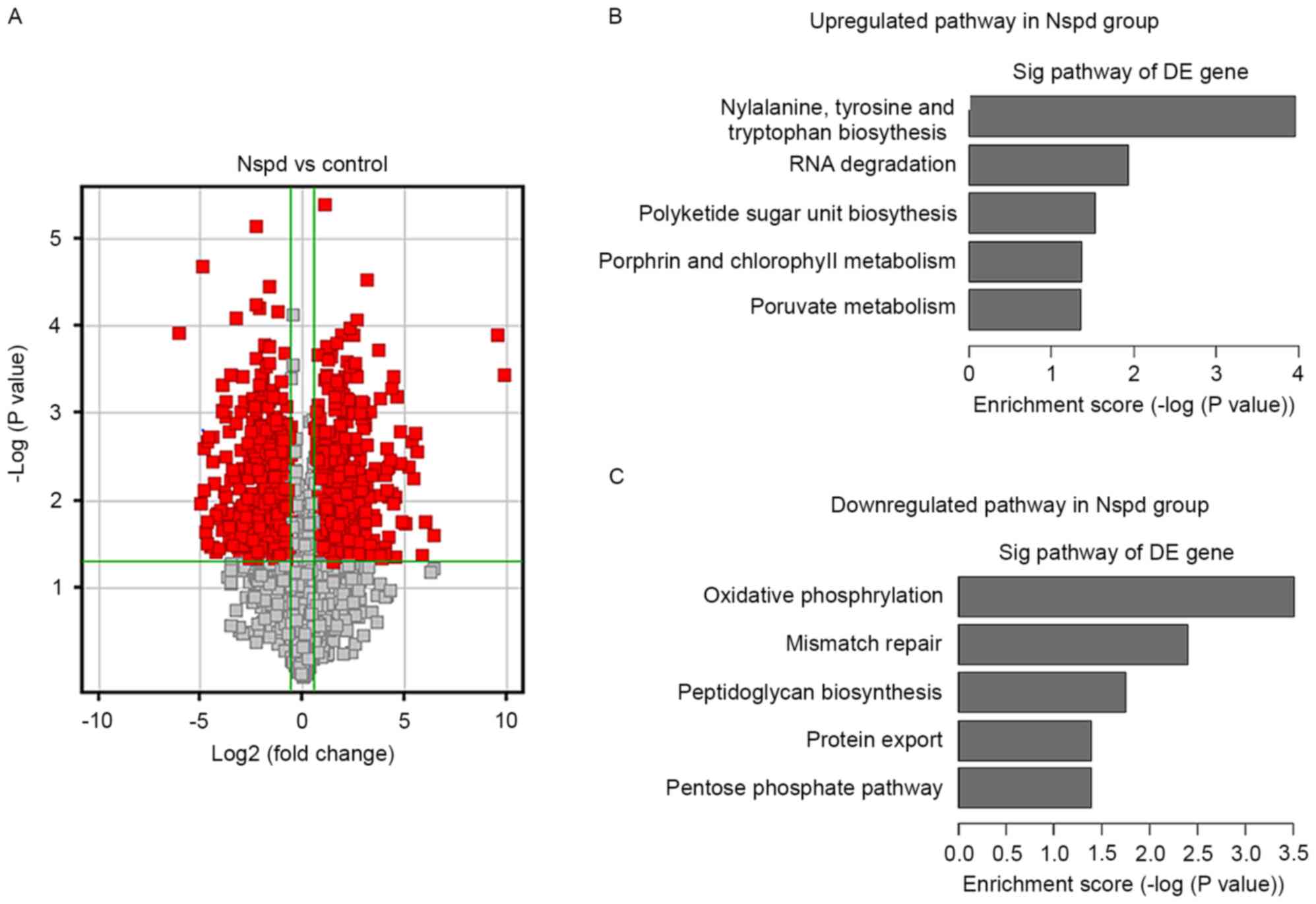
Effect of Nspd on gene expression
To determine how Nspd leads to the formation of huge
EPS clusters and basic structure, the present study examined the
gene expression of cells in the biofilm compared with that of the
control group. An overview of the S. mutans pathways
regulated by Nspd is presented in a volcano graph (Fig. 8). Volcano graphs demonstrated a
large difference in the levels of gene expression in the Nspd group
compared with those of the control group (Fig. 8A), and several up or downregulated
pathways were observed in the Nspd group (Fig. 8B and C).
>400 genes were differentially expressed in the
Nspd group compared with the control group (Table I). Since the present study focused
on biofilm-associated changes, information induced by Nspd, the
present study selected 24 genes that have been fully reported to be
closely associated with S. mutans biofilm formation and
polyamine transport. Among these 24 genes, the expression levels of
12 genes were significantly different (P<0.05), including
luxS, comC, comD, comE, gtfB, spa, scrR, brpA,
fruB, gbpD and potB. Although the expression levels of the
other genes were not significantly different at the 1.5-fold change
limit (P>0.05), the changes in their expression revealed certain
discrete and important tendencies associated with the differences
in the biofilm structures in the Nspd group. It was noted that the
expression of the QS-related genes lusX, comC, comD
and comE were remarkably downregulated by 7.9, 2.7, 8 and
15-fold, respectively (P<0.05).
 | Table I.Structure of biofilm differentially
expressed genes regulated by Nspd relative to the negative control
group. |
Table I.
Structure of biofilm differentially
expressed genes regulated by Nspd relative to the negative control
group.
| Gene name | Predicted
function | Regulation | Fold-change |
|---|
| gtfA | Sucrose
phosphorylase GtfA | Down | 1.5509206 |
| gtfBa |
Glucosyltransferase-I | Down | 1.4373798 |
| gtfC |
Glucosyltransferase-SI | Down | 2.0291426 |
| gtfD |
Glucosyltransferase-S | Down | 1.9489628 |
| scrRa | Sucrose operon
repressor | Up | 1.5087006 |
| spaPa | Cell surface
antigen SpaP | Down | 19.62776 |
| brpAa | Transcriptional
regulator | Up | 2.3073087 |
| fruBa |
Exo-beta-D-fructosidase | Down | 1.9644824 |
| relA | Stringent response
protein, ppGpp synthetase | Up | 2.0203063 |
| dexA | Dextranase | Up | 1.6448688 |
| dexBa | Dextran glucosidase
DexB | Up | 2.9510176 |
| luxSa |
S-ribosylhomocysteinase | Down | 7.883619 |
| comCa | Competence
stimulating peptide | Down | 2.6926205 |
| comDa | Histidine kinase of
the competence regulon, ComD | Down | 7.9834288 |
| comEa | Response regulator
of the competence regulon ComE | Down | 15.048688 |
| gbpA | Glucan-binding
protein GbpA | Up | 1.5979749 |
| gbpB | Secreted antigen
GbpB/SagA | Up | 1.5592909 |
| gbpC | Glucan-binding
protein GbpC | Up | 1.2180864 |
| gbpDa | Glucan-binding
protein D | Up | 3.98988328 |
| potA |
Spermidine/putrescine ABC transporter
ATP-binding protein | Up | 1.3464806 |
| potBa |
Spermidine/putrescine ABC transporter
permease | Down | 1.7071223 |
| potC |
Spermidine/putrescine ABC transporter
permease | Down | 1.1213877 |
| potD | ABC transporter
periplasmic spermidine/putrescine-binding protein | Down | 1.2626697 |
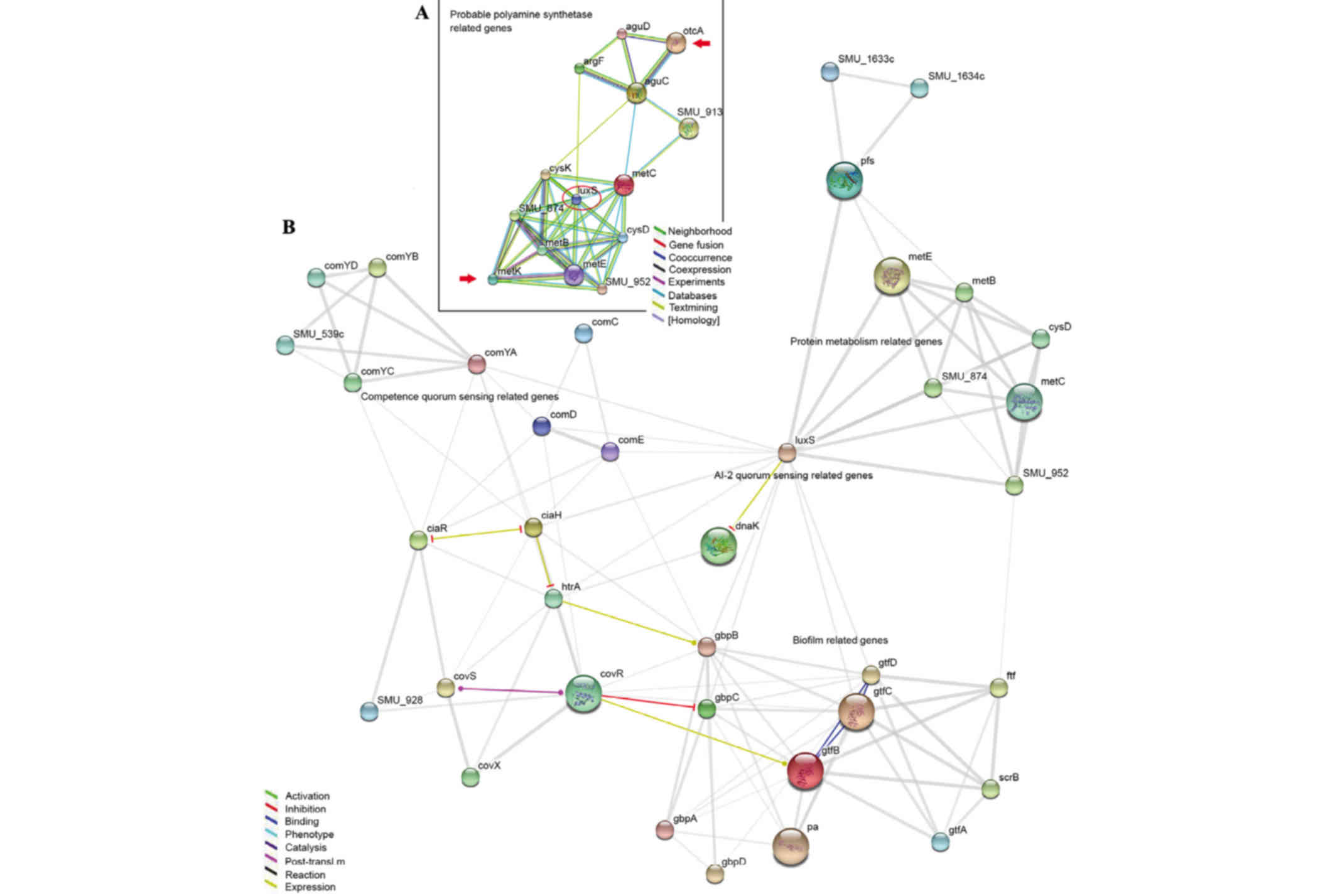
Gene connection map
To discover the associations between the genes that
were differently expressed led by Nspd and biofilm structure, the
present study used an online program that demonstrated the
connections among genes (http://string-db.org/newstring.) and created a gene
net map (Fig. 9) according to a
previous study (21). GTF encoded
genes (gtfA, B, C and D) and glucan-binding proteins encoded genes
(gbpA, B, C and D) directly affected biofilm architecture and
structure. The QS-associated genes included luxS and
competence-stimulating peptide comC, comD and
comE genes. Glucan production and binding assocaited genes
(gtfA, B, C and D and gbpA, B, C and D) were directly or indirectly
impacted by luxS, comC, comD and comE.
Certain clues were observed between polyamine and biofilms using
this map. otcA encodes putrescine carbamoyltransferase and
metK encodes argcarbomyltransferase. This gene-connection
map revealed that these polyamine-synthetic genes were most likely
associated with gtf and gbp through the luxS gene (Fig. 9B).
Discussion
Although information associated with the S.
mutans Spd/Put transporter can be observed in NCBI, no report
is available concerning these polyamines in S. mutans; the
spermidine synthetase remains unknown. Exogenous Nspd, the
allosteric form of spermidine, served an adverse role in S.
mutans by inhibiting its growth and leading to the formation of
irregular biofilm structures. No direct effect of Nspd on S.
mutans EPS was observed.
Notably, through observing the different
three-dimensional structures of the biofilm that were formed in the
presence of Nspd, the present research revealed that the basic
structure in S. mutans biofilms is composed of EPS
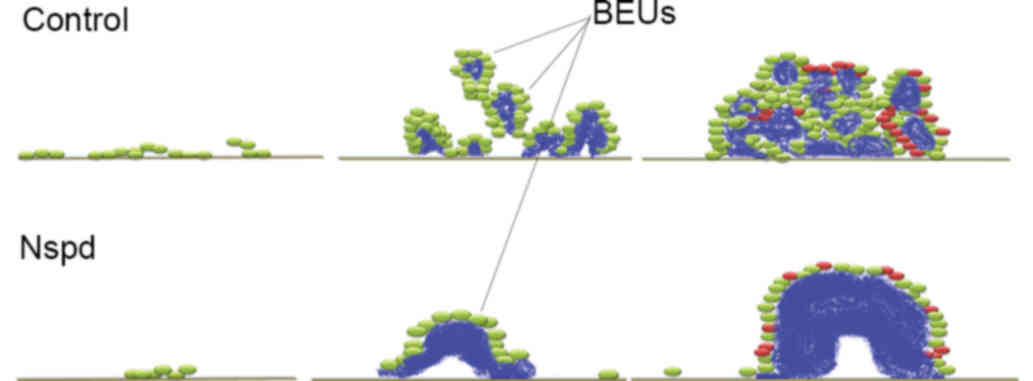
surrounded by bacterial units. This clear and distinct unit has not
been reported previously and it was termed it as ‘bacteria-EPS
units’ (BEUs; Figs. 6 and 7), as it is more visual in structural
detail than colony forming units. This finding may indicate the
mechanism by which EPS and bacteria assemble the EPS-rich S.
mutans biofilms formed in the presence of sucrose and somewhat
explain the QS mechanism.
In the present research, spherical or globular BEUs
were formed by bacterial clusters secreting EPS into the center of
cell clusters. The EPS were surrounded by bacteria rather than the
EPS, enmeshing the bacteria in this type of basic structure. This
result is in disagreement with certain theories. Xiao et al
(7) has reported that EPS were
detected surrounding and covering microcolonies, and suggested that
individual microcolonies encased in polysaccharides may serve as
the architectural units in mixed-species biofilms, including those
of S. mutans (7). These
structures were termed EPS-microcolony complexes. As noted in the
present study, the diameter of the EPS-microcolony complexes was
>40 µm compared with the 5–40 µm diameter of the BEUs observed
in the present study; therefore, it was speculated that the BEU may
be the most basic structure formed by S. mutans. Time-lapse
observations of organized BEUs of a larger size that occurred in
the Nspd group provided further evidence to support this
speculation. This type of organized BEU formed from bacterial
clusters that secreted EPS toward the adhesion interface and the
center of the cluster, and developed from a hat-like structure into
a globular structure. The present study speculated that the
bacteria gradually moved toward the exterior, as the EPS granule
grew larger. This type of EPS secretion pattern is somewhat similar
to the pattern of enamel secretion by ameloblasts (22). However, how the internal EPS
protects the bacteria in a basic structure such as a BEU remains to
be determined. It was suggested that the development of the biofilm
may answer this question; as the biofilm grew, more BEUs formed and
they became larger and more densely packed, until they finally
contacted one another and the bacteria were crowded within adjacent
BEUs (Fig. 10). A biofilm
contains a large numbers of BEUs, which would result in the
majority of the bacteria being enmeshed within the EPS.
Nspd treatment led to the formation of oversized and
undersized BEUs. The present study also proposed that EPS secretion
and accumulation at too rapid a rate can lead to some of the
defects observed in the oversized BEU-EPS aggregates in the Nspd
group. These defects included central holes and some depressions
(Fig. 7). It was suggested that
the central holes in the oversized BEUs in the Nspd group can be
the early gathering sites of the bacteria and the original
EPS-secretion points; clear evidence of this hypothesis would be to
trace the tracks of the bacteria while they are secreting EPS.
Hydrodynamics, nutrient concentrations, carbon
sources, motility, genetics, QS (23–25)
and glucan-binding proteins (26)
have been identified as factors that affect biofilm architecture.
To understand how S. mutans regulates or controls the size
of the BEUs and forms a regular, smooth, homogeneous biofilm, whole
genome expression microarray analysis of S. mutans cells in
the biofilms formed in the Nspd and control groups was performed
with the goal of determining the cause of the differences in
biofilm formation. Using the same conditions without agitation, the
present study was able to exclude hydrodynamics, nutrient
concentration and genetic factors as responsible for the
differences between the two groups. Therefore, our attention was
focused on alterations in the expression of the cell-surface
antigen SpaP, glucosyltransferase, glucan-binding proteins and
members of regulatory systems, including the QS system and
covR.
For the Nspd group, the present study focused on 20
genes with severely compromised expression that served a role in
biofilms. It was also revealed that the expression of a gene
encoding a surface antigen I (SpaP), which is important for the
initial attachment stage of biofilm formation (27–31),
was markedly decreased. This phenomenon can explain why most areas
consisted of a rather thin layer of bacteria or contained no
bacteria. A sharp reduction in the ability of bacteria to adhere to
the surface led to dramatic deficiencies in the biofilm basement.
EPS are an important component of the microbial biofilm
extracellular matrix as it contributes to the overall biofilm
architecture and to the resistance phenotype of the bacteria in
biofilms (5–33). An irregular architecture results in
the loss of protection of the bacteria and leads to decreased
virulence. In S. mutans biofilms, the EPS are composed
predominantly of glucans. The glucosyltransferases (Gtfs) of S.
mutans are constituents of the pellicle and can synthesize
glucans in situ using sucrose (34,35).
The polymers that form on the surface provide bacterial binding
sites for subsequent colonization and local accumulation of S.
mutans and other organisms (36,37).
Gtfs also bind numerous oral bacteria, even those that do not
synthesize Gtfs (8,35,38),
thereby converting them into original glucan producers (35). In the Nspd group, the expression
levels of four glucosyltransferase-encoding genes, gtfA, B,
C and D (39–42) were all reduced compared with the
levels of the control group. Although a number of oversized EPS
structures formed in the biofilms of the Nspd group, the total
quantity of EPS was less than that of the control group. The
glucan-binding protein family includes the glucan-protein receptor,
the function of which is necessary to sustain the architecture of
biofilms (26,43–45).
The effect of such proteins was demonstrated by the large
structures containing EPS in the Nspd group. The expression levels
of four glucan-binding protein-encoding genes, gbpA, B, C
and D, were all upregulated in the Nspd group. The overexpression
of these genes may have led to too much EPS binding and induced the
formation of the huge aggregates of EPS mentioned above.
SpaP, glucosyltransferase and glucan-binding
proteins are considered direct factors that affect the architecture
of biofilms, whereas the QS system indirectly regulates biofilm
architecture by affecting the above-mentioned proteins. In the
present study, the expression levels of comC, comD,
comE and luxS were markedly reduced in the Nspd
group. As mentioned in above, S. mutans produces both AI-2
and competence-stimulating peptides (CSP belongs to the AI-1 group)
via the luxS and comC genes, respectively. The
relationship between QS and S. mutans biofilm structure has
been studied and discussed by a certain number of researchers
(26,46,47).
Inactivation of any of the individual genes under investigation
results in the formation of an abnormal biofilm. The comC
deletion mutant, which is unable to produce or secrete CSP, forms
biofilms with an altered architecture, whereas the comD and
comE S. mutans mutants, which are defective in sensing and
responding to CSP, form biofilms with a reduced biomass (46). Previous reports regarding the
effect of an interspecies QS system on the architecture of S.
mutans biofilms have attracted our attention. Biofilms formed
by the luxS mutant have a more granular appearance compared
with the relatively smooth, confluent layer normally produced by
wild-type cells (47,48). This granular appearance was similar
to the appearance of the biofilm formed by the Nspd group in the
present study. Huang et al (49) reported that biofilm formation by
the luxS mutant was accelerated during the mid-exponential
phase and that the differences between the mutant and wild-type
biofilms were markedly greater when carbohydrates were added to the
medium. These findings suggested that AI-2 may affect the early
stages of biofilm formation by inhibiting exopolysaccharide matrix
production (49). Consistent with
these previous reports (47–49),
luxS expression was notably downregulated in the Nspd group
in the present study, which may have led to the acceleration of EPS
aggregation, and subsequently led to the formation of oversized
BEUs.
In conclusion, the present study proposed a
mechanism for the assembly of three-dimensional structures in S.
mutans biofilms. The limited size and regularity of the BEUs,
due to regulation by the QS system, guaranteed that the majority of
the bacteria dispersed themselves among and covered the EPS, which
protected the S. mutans bacteria in the biofilm from
unfavorable factors. The following appeared to explain certain
aspects of the formation of normal BEUs: i) The size of the EPS
granule in the BEU was predetermined and limited; ii) The bacteria
were crowded by more than surrounded by the EPS in the biofilm;
iii) The bacteria secreted EPS into the center of the aggregates
while moving toward the exterior; iv) The interface between the
bacteria and EPS was a globular interface; and v) The BEUs always
combined to form clusters.
Acknowledgements
The authors would like to thank Professor Christine
Wu and Dr Wei Li (College of Dentistry, University of Illinois) for
their assistance and advice in repeating the experiments. Professor
Yutao Jian, Mr. Jingtao Wang and Mr. Tao He (Provincial Key
Laboratory of Stomatology, Sun Yat-sen University) for their
assistance in the confocal laser-scanning microscopic analysis. The
present study was supported by the National Natural Science
Foundation of China (no. 81371132). The funders had no role in the
study design, data collection and analysis, decision to publish, or
preparation of the manuscript.
References
|
1
|
Wallace HM, Fraser AV and Hughes A: A
perspective of polyamine metabolism. Biochem J. 376:1–14. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Romero D and Kolter R: Will biofilm
disassembly agents make it to market? Trends Microbiol. 19:304–306.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Flemming HC, Neu TR and Wozniak DJ: The
EPS matrix: The ‘house of biofilm cells’. J Bacteriol.
189:7945–7947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Flemming HC and Wingender J: The biofilm
matrix. Nat Rev Microbiol. 8:623–633. 2010.PubMed/NCBI
|
|
5
|
Branda SS, Vik S, Friedman L and Kolter R:
Biofilms: The matrix revisited. Trends Microbiol. 13:20–26. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bowen WH and Koo H: Biology of
Streptococcus mutans-derived glucosyltransferases: Role in
extracellular matrix formation of cariogenic biofilms. Caries Res.
45:69–86. 2011. View Article : Google Scholar
|
|
7
|
Xiao J, Klein MI, Falsetta ML, Lu B,
Delahunty CM, Yates JR III, Heydorn A and Koo H: The
exopolysaccharide matrix modulates the interaction between 3D
architecture and virulence of a mixed-species oral biofilm. PLoS
Pathog. 8:e10026232012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hamada S, Tai S and Slade HD: Binding of
glucosyltransferase and glucan synthesis by Streptococcus mutans
and other bacteria. Infect Immun. 21:213–220. 1978.PubMed/NCBI
|
|
9
|
Koo H, Xiao J, Klein MI and Jeon JG:
Exopolysaccharides produced by Streptococcus mutans
glucosyltransferases modulate the establishment of microcolonies
within multispecies biofilms. J Bacteriol. 192:3024–3032. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Loesche WJ: Role of Streptococcus mutans
in human dental decay. Microbiol Rev. 50:353–380. 1986.PubMed/NCBI
|
|
11
|
Fuqua C, Parsek MR and Greenberg EP:
Regulation of gene expression by cell-to-cell communication:
Acyl-homoserine lactone quorum sensing. Annu Rev Genet. 35:439–468.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kleerebezem M and Quadri LE: Peptide
pheromone-dependent regulation of antimicrobial peptide production
in Gram-positive bacteria: A case of multicellular behavior.
Peptides. 22:1579–1596. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miller MB and Bassler BL: Quorum sensing
in bacteria. Annu Rev Microbiol. 55:165–199. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miller MB, Skorupski K, Lenz DH, Taylor RK
and Bassler BL: Parallel quorum sensing systems converge to
regulate virulence in Vibrio cholerae. Cell. 110:303–314. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shapiro JA: Thinking about bacterial
populations as multicellular organisms. Annu Rev Microbiol.
52:81–104. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karatan E and Watnick P: Signals,
regulatory networks, and materials that build and break bacterial
biofilms. Microbiol Mol Biol Rev. 73:310–347. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wortham BW, Oliveira MA, Fetherston JD and
Perry RD: Polyamines are required for the expression of key Hms
proteins important for Yersinia pestis biofilm formation. Environ
Microbiol. 12:2034–2047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sakamoto A, Terui Y, Yamamoto T, Kasahara
T, Nakamura M, Tomitori H, Yamamoto K, Ishihama A, Michael AJ,
Igarashi K and Kashiwagi K: Enhanced biofilm formation and/or cell
viability by polyamines through stimulation of response regulators
UvrY and CpxR in the two-component signal transducing systems and
ribosome recycling factor. Int J Biochem Cell Biol. 44:1877–1886.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sturgill G and Rather PN: Evidence that
putrescine acts as an extracellular signal required for swarming in
Proteus mirabilis. Mol Microbiol. 51:437–446. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kolodkin-Gal I, Cao S, Chai L, Böttcher T,
Kolter R, Clardy J and Losick R: A self-produced trigger for
biofilm disassembly that targets exopolysaccharide. Cell.
149:684–692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue). D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nishikawa S: Correlation of the
arrangement pattern of enamel rods and secretory ameloblasts in pig
and monkey teeth: A possible role of the terminal webs in
ameloblast movement during secretion. Anat Rec. 232:466–478. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Harmsen M, Yang L, Pamp SJ and
Tolker-Nielsen T: An update on Pseudomonas aeruginosa biofilm
formation, tolerance and dispersal. FEMS Immunol Med Microbiol.
59:253–268. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stoodley P, Dodds I, Boyle JD and
Lappin-Scott HM: Influence of hydrodynamics and nutrients on
biofilm structure. J Appl Microbiol. 85:(Suppl 1). 19S–28S. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parsek MR and Tolker-Nielsen T: Pattern
formation in Pseudomonas aeruginosa biofilms. Curr Opin Microbiol.
11:560–566. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lynch DJ, Fountain TL, Mazurkiewicz JE and
Banas JA: Glucan-binding proteins are essential for shaping
Streptococcus mutans biofilm architecture. FEMS Microbiol Lett.
268:158–165. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Crowley PJ, Brady LJ, Michalek SM and
Bleiweis AS: Virulence of a spaP mutant of Streptococcus mutans in
a gnotobiotic rat model. Infect Immun. 67:1201–1206.
1999.PubMed/NCBI
|
|
28
|
Qu YP, Liu JG, Yang P and Yang DQ:
Construction and expression of spap/A eukaryotic expression plasmid
of Streptococcus mutans in mammalian cells. Shanghai Kou Qiang Yi
Xue. 18:61–65. 2009.(In Chinese). PubMed/NCBI
|
|
29
|
Durán-Contreras GL, Torre-Martínez HH, de
la Rosa EI, Hernández RM and de la Garza Ramos M: spaP gene of
Streptococcus mutans in dental plaque and its relationship with
early childhood caries. Eur J Paediatr Dent. 12:220–224.
2011.PubMed/NCBI
|
|
30
|
Davis E, Kennedy D, Halperin SA and Lee
SF: Role of the cell wall microenvironment in expression of a
heterologous SpaP-S1 fusion protein by Streptococcus gordonii. Appl
Environ Microbiol. 77:1660–1666. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sato Y, Okamoto-Shibayama K and Azuma T: A
mechanism for extremely weak SpaP-expression in Streptococcus
mutans strain Z1. J Oral Microbiol. 3:2011.
|
|
32
|
Itoh Y, Wang X, Hinnebusch BJ, Preston JR
and Romeo T: Depolymerization of beta-1,6-N-acetyl-D-glucosamine
disrupts the integrity of diverse bacterial biofilms. J Bacteriol.
187:382–387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sutherland IW: The biofilm matrix-an
immobilized but dynamic microbial environment. Trends Microbiol.
9:222–227. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rolla G, Ciardi JE and Schultz SA:
Adsorption of glucosyltransferase to saliva coated hydroxyapatite.
Possible mechanism for sucrose dependent bacterial colonization of
teeth. Scand J Dent Res. 91:112–117. 1983.PubMed/NCBI
|
|
35
|
Vacca-Smith AM and Bowen WH: Binding
properties of streptococcal glucosyltransferases for
hydroxyapatite, saliva-coated hydroxyapatite, and bacterial
surfaces. Arch Oral Biol. 43:103–110. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schilling KM and Bowen WH: Glucans
synthesized in situ in experimental salivary pellicle function as
specific binding sites for Streptococcus mutans. Infect Immun.
60:284–295. 1992.PubMed/NCBI
|
|
37
|
Venkitaraman AR, Vacca-Smith AM, Kopec LK
and Bowen WH: Characterization of glucosyltransferaseB, GtfC, and
GtfD in solution and on the surface of hydroxyapatite. J Dent Res.
74:1695–1701. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
McCabe RM and Donkersloot JA: Adherence of
Veillonella species mediated by extracellular glucosyltransferase
from Streptococcus salivarius. Infect Immun. 18:726–734.
1977.PubMed/NCBI
|
|
39
|
Yamashita Y, Bowen WH and Kuramitsu HK:
Molecular analysis of a Streptococcus mutans strain exhibiting
polymorphism in the tandem gtfB and gtfC genes. Infect Immun.
60:1618–1624. 1992.PubMed/NCBI
|
|
40
|
Chia JS, Hsieh CC, Yang CS and Chen JY:
Purification of glucosyltransferases (GtfB/C and GtfD) from mutant
strains of Streptococcus mutans. Zhonghua Min Guo Wei Sheng Wu Ji
Mian Yi Xue Za Zhi. 28:1–12. 1995.PubMed/NCBI
|
|
41
|
Tsai YW, Chia JS, Shiau YY, Chou HC, Liaw
YC and Lou KL: Three-dimensional modelling of the catalytic domain
of Streptococcus mutans glucosyltransferase GtfB. FEMS Microbiol
Lett. 188:75–79. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yoshida A and Kuramitsu HK: Streptococcus
mutans biofilm formation: Utilization of a gtfB promoter-green
fluorescent protein (PgtfB:gfp) construct to monitor development.
Microbiology. 148:3385–3394. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lynch DJ, Michalek SM, Zhu M, Drake D,
Qian F and Banas JA: Cariogenicity of Streptococcus mutans
glucan-binding protein deletion mutants. Oral Health Dent Manag.
12:191–199. 2013.PubMed/NCBI
|
|
44
|
Banas JA, Fountain TL, Mazurkiewicz JE,
Sun K and Vickerman MM: Streptococcus mutans glucan-binding
protein-A affects Streptococcus gordonii biofilm architecture. FEMS
Microbiol Lett. 267:80–88. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Banas JA, Hazlett KR and Mazurkiewicz JE:
An in vitro model for studying the contributions of the
Streptococcus mutans glucan-binding protein A to biofilm structure.
Methods Enzymol. 337:425–433. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li YH, Tang N, Aspiras MB, Lau PC, Lee JH,
Ellen RP and Cvitkovitch DG: A quorum-sensing signaling system
essential for genetic competence in Streptococcus mutans is
involved in biofilm formation. J Bacteriol. 184:2699–2708. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Merritt J, Qi F, Goodman SD, Anderson MH
and Shi W: Mutation of luxS affects biofilm formation in
Streptococcus mutans. Infect Immun. 71:1972–1979. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yoshida A, Ansai T, Takehara T and
Kuramitsu HK: LuxS-based signaling affects Streptococcus mutans
biofilm formation. Appl Environ Microbiol. 71:2372–2380. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang Z, Meric G, Liu Z, Ma R, Tang Z and
Lejeune P: luxS-based quorum-sensing signaling affects Biofilm
formation in Streptococcus mutans. J Mol Microbiol Biotechnol.
17:12–19. 2009. View Article : Google Scholar : PubMed/NCBI
|