Introduction
Peptidyl-prolyl cis/trans isomerase, NIMA
interacting-1 (Pin1), a peptide-prolyl amide linkage isomerase
discovered in 1996, is a protein that regulates mitosis. Pin1 also
regulates the conformation of phosphorylatable substrates and
influences their physiological function by specifically catalyzing
the cis-trans isomerism of the phosphorylated (p)Ser/Thr-Pro amide
linkage (1). Previous biological
research has indicated that Pin 1 is overexpressed in multiple
tumor cells, including cervical cancer, prostate cancer, lung
cancer, breast cancer, liver cancer and melanoma (2–5).
Overexpression of Pin1 is a significant marker of the occurrence
and development of tumors (2–5).
Multiple known oncogenes and cancer suppressor proteins are
substrates of Pin1. Pin1 activates the protein activity of
oncogenes and promotes the occurrence and development of tumors by
regulating the conformation of phosphorylatable substrates
(6–7). Hence, Pin1 inhibition is a possible
novel mechanism to exploit for antitumor therapy.
Research on crystal structures of a known Pin1
small-molecule inhibitor and Pin1 compounds has demonstrated that
the pharmacophore characteristics of this inhibitor are two
hydrophobic centers and one H-bond acceptor (8–10).
Pin1 binds to the H-bond acceptor in the small-molecule inhibitor
by binding the side chains of basic amino acid residues, namely,
Lys63, Arg68, and Arg6, to the cavity. Thus, Pin1 contributes to
the orientation and binding of small molecules in the binding
cavity. His59, His157, Met130 and Phe134 constitute a hydrophobic
center, which is usually defined as the prolyl pocket. The other
hydrophobic center is a flat region that consists of His59, Ala118
and Leu122; this center binds to bulk-mass aromatic structural
segments. By investigating and screening this structure, the
imidazoline ketone herbicide, imazamethabenz was demonstrated to
exhibit the capacity to bind to Pin1 (11). The present study demonstrated that
this compound inhibits the activity of Pin1 in vitro. MTT
cytotoxicity tests, caspase-3 enzyme expression quantity tests and
cellular migration and invasion tests verified that imazamethabenz
effectively inhibited the proliferation, migration and invasion of
the human breast cancer cell line MCF-7. This result provides a
basis for developing novel types of Pin1 inhibitors.
Materials and methods
Materials
Imazamethabenz and
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
were purchased from Aladdin Shanghai Biochemical Technology Co.,
Ltd. (Shanghai, China). Stock solutions of compounds (10 mM) were
prepared in DMSO and stored at −80°C. The solution was diluted to
working concentrations using double-distilled, deionized water.
Pin1 in prokaryotic expression vector pMAL-c2X and the expression
in E. coli and purification of the Pin1 fusion protein as
previously been described by Zhang et al (12). The synthetic substrate peptide
[suc-Ala-Glu-Pro-Phe-peptide nucleic acid (pNA)] and bovine serum
albumin were purchased from Sangon Biotech Co., Ltd. (Shanghai,
China). The MCF-7 cell line was obtained from the Animal
Experimental Center of Sun Yat-Sen University (Guangzhou, China).
All antibodies were purchased from Sigma-Aldrich; Merck KGaA
(Darmstadt, Germany).
Molecular docking
The X-ray crystal structure of Pin1 complexed with
the carboxylate inhibitor was obtained from the RCSB Protein Data
Bank (PDB code: 3JYJ) (13). The
docking studies were predicted using AUTODOCK 4.0 software (The
Scripps Research Institute, La Jolla, CA, USA). The dimensions of
the active site box were selected to be large enough to contain the
entire protein molecule. All docking calculations were performed
using a Lamarckian genetic algorithm. The default FlexX parameters
were used. The most likely conformation of the ligand was selected
based on its binding affinity and optimal binding arrangement with
the hydrophobic centers and H-bond acceptors of Pin1.
Inhibitory activity of Pin1
enzyme
Pin1 inhibitory activity was evaluated through the
proteinase-coupled method (14). A
mixture of 250 µM synthetic substrate peptide
[suc-Ala-Glu-Pro-Phe-peptide nucleic acid (pNA)], 0.2 mM
dithiothreitol, 100 µg/ml bovine serum albumin, and 25 nM Pin1 in
150 µl 35 mM HEPES-KOH (pH 7.8) was added to the DMSO solution. The
indicated concentrations (0, 0.5, 2.5, 5, 10, 20, 40 80 µM) of
imazamethabenz were then added to the solution. The mixture was
preincubated for 10 min at 10°C. The hydrolysis of the substrate
peptide was initiated by adding an excess amount of α-chymotrypsin
(150 µl) of 0.8 mg/ml protease in 35 mM HEPES-KOH (pH 7.8). The
absorbance of the released p-nitroaniline at 390 nm was determined
every second for 10 min with an Agilent 8453 UV-vis
spectrophotometer (Agilent Technologies, Inc., Santa Clara, CA,
USA). Initially, α-chymotrypsin rapidly digested the substrate
peptides in the trans form, then subsequently hydrolyzed the
peptides as they were isomerized from the cis form to the trans
form by Pin1. The observed reaction rate of the isomerization phase
was designated as Pin1 activity. Inhibitory activity was calculated
and expressed as
(k(inh)-k0)/(k(noinh)-k0)
×100 (%); where k(inh) is the observed
pseudo-first-order rate constant in the presence of the inhibitor,
k(noinh) is the rate constant without the inhibitor and
k0 is the constant in the absence of Pin1.
MTT assay
MCF-7 cells were seeded in 96-well plates at a
concentration of 5×103 cells/well and exposed to various
concentrations of imazamethabenz (0.1, 0.5, 2.5, 5, 10, 20, 40 and
80 µM). Following treatment for 48 h, 10 µl MTT solution was added
to each well and then incubated for 4 h. Following this, 100 µl
DMSO was added to each well. Optical density was detected at 490
nm. Three independent experiments were performed. IC50
values were calculated using Origin 8.0 software (OriginLab
Corporation, Northampton, MA, USA).
Western blot
Following 4 d of treatment with or without
imazamethabenz (0, 0.5, 1 and 2.5 µM), the MCF-7 cells were
harvested and washed with PBS (pH 7.4) three times. The collected
cells were lysed in 150 µl of extraction buffer consisting of 100
µl solution A (50 mM glucose, 25 mM Tris-HCl, pH 8, 0 mM EDTA and 1
mM phenylmethylsulfonyl fluoride) and 50 µl solution B (50 mM
Tris-HCl, pH 6.8, 6 M urea, 6% 2-mercaptoethanol, 3% sodium dodecyl
sulfate and 0.003% bromophenol blue). The soliquid was then
centrifuged at 11,310 × g at 4°C for 5 min. The supernatant
(10 µl for each sample) was loaded onto 10% polyacrylamide gel and
transferred to a microporous polyvinylidene difluoride membrane.
After blocking non-specific binding with 5% BSA (room temperature
for 1 h), the membranes were subsequently incubated with primary
antibodies against vascular endothelial growth factor (VEGF; cat.
no. V4758; 1:500; Sigma-Aldrich, Merck KGaA), matrix
metalloproteinase 9 (MMP-9; cat. no. SAB4501896; 1:500;
Sigma-Aldrich, Merck KGaA) and β-actin (cat. no. A1978; 1:1,000;
Sigma-Aldrich, Merck KGaA), overnight at 4°C, followed by
incubation with horseradish peroxidase-conjugated anti-mouse (cat.
no. A9044; 1:1,000; Sigma-Aldrich, Merck KGaA) or anti-rabbit
secondary antibody (cat. no. A4914; 1:1,000; Sigma-Aldrich, Merck
KGaA) at room temperature for 30 min. The signals were detected
using an enhanced chemiluminescence substrate (Bioss Biotechnology,
Beijing, China).
Wound healing assay
This assay was performed as previously described
(15). The cells were grown to
100% confluence in 12-well plates for 24 h. The cell monolayer was
scratched with a 200 µl pipette tip to obtain constant widths. The
cells were then incubated with 0, 1 and 2.5 µl of imazamethabenz
for 1 h at 37°C. Cell migration was visualized at magnification
×200 and photographed after 24 h using a Nikon Eclipse 80i light
microscope (Nikon Corporation, Tokyo, Japan). The wound area was
quantified using the program ImageJ version 2.0 (NIH, Bethesda, MD,
USA).
Matrigel invasion assay
This assay was performed as previously described
(15). Matrigel invasion assays
were performed using a 12-well Transwell insert with 8 µm pore size
from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The upper
wells of the inserts were coated with 100 µl Matrigel (1 mg/ml) and
incubated at 37°C for 4 h. The coating was then washed once with
serum-free medium. MCF-7 cells (1×105 cells) with or
without imazamethabenz (0, 1 and 2.5 µM) and 0.2 ml Dulbecco's
modified Eagle's medium (Meilun Biology Technology, Dalian, China)
were added to the well. Following incubation for 48 h, the cells in
the top well were removed. The cells on the underside of the
membrane were dyed using 0.4 g/l crystal violet (Aladdin Shanghai
Biochemical Technology Co., Ltd.) and counted by light microscopy
based on 5 randomly selected fields at magnification ×200. Three
independent experiments were performed.
Statistical analysis
Each experiment was performed 3–4 times. All data
were analyzed by the SPSS 17.0 software (SPSS, Inc., Chicago, IL,
USA). The statistical significance of differences was determined by
Student's t-test for comparison between one-way analysis of
variance. P<0.05 was considered to indicate a statistically
significant difference.
Results and Discussion
Molecular docking
The present study adopted the AUTODOCK 4.0 docking
program and RCSB Protein Data Bank (PDB code: 3JYJ) (13), and investigated the effect of
binding between imazamethabenz and Pin1, providing a basis for
structural modification.
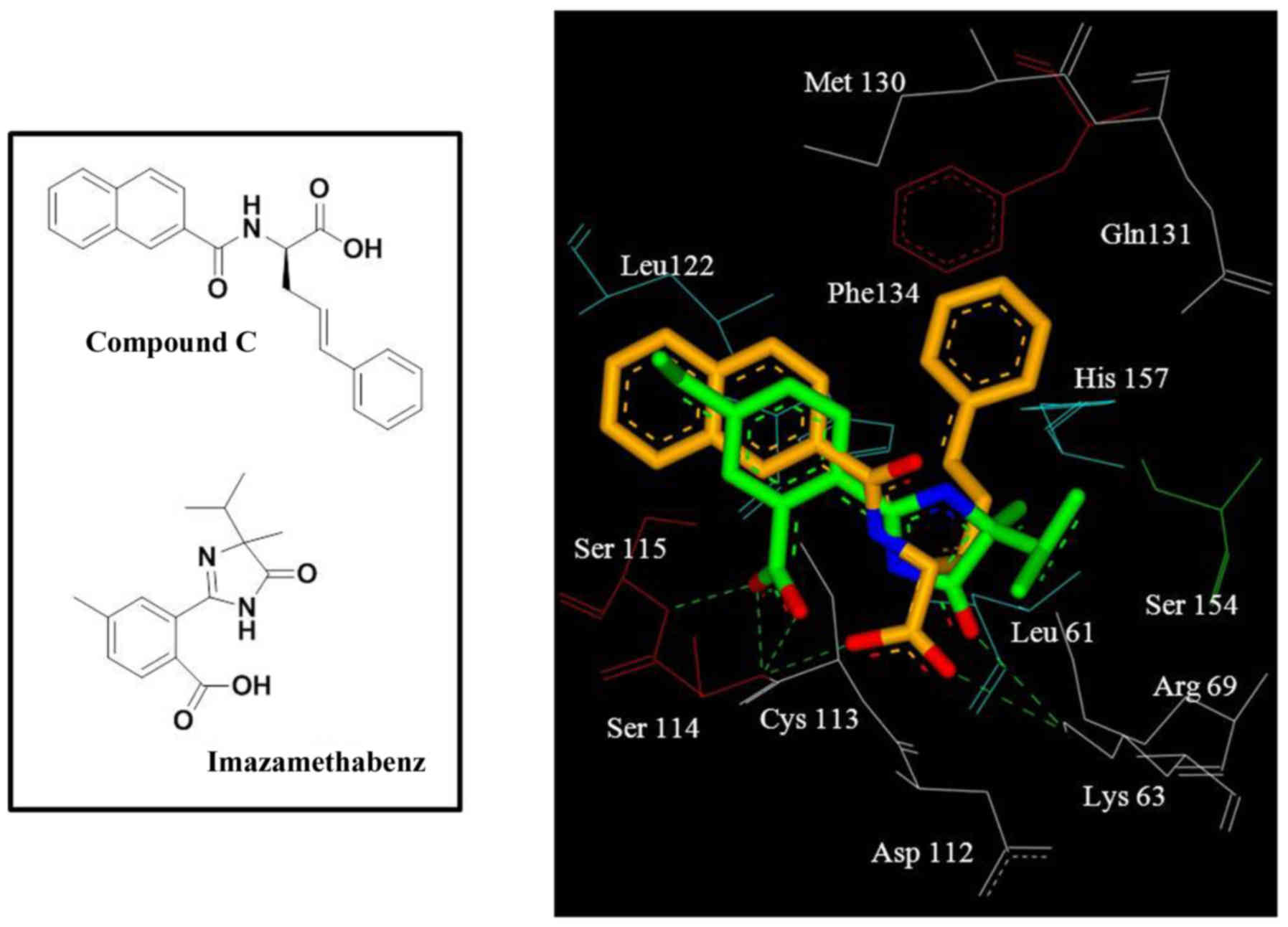
Docking results revealed that imazamethabenz and
Pin1 exhibited a good binding effect (Fig. 1). Hydroxyl in the oxalate base and
Pin1 bound to the characteristic basic aminoacid residue Ser114 to
form hydrogen-bond interactions in the cavity and 2-carbonyl in the
oxalate base. Furthermore, Ser114 and Ser115 formed a hydrogen
bond, and carbonyl in imidazoline ketone and Lys63 formed a
hydrogen bond. These findings indicated that the oxalate base is a
hydrogen-bond acceptor of the Pin1 inhibitor. The benzene ring
reached out to a relatively flat hydrophobic region, which
consisted of side chains Leu122 and Cys113 (Fig. 1). This result revealed that
imazamethabenz and Pin1 have multiple combination modes.
Further examination revealed that, compared with the
reference compound, compound C (13), in the crystal complex, the benzene
ring of imazamethabenz did not completely occupy the binding site
and had no binding group with the prolyl combination cavity, which
consists of the side chains Gln131, Phe134, and His157 (Fig. 1). Hence, introducing an aromatic
ring in the 1,2-position of the benzene ring would increase the
number of groups that bind to the prolyl combination cavity, and
this phenomenon would boost the binding force between this type of
compound and Pin1. This result may help to improve imazamethabenz
derivatives as selective Pin1 inhibitors.
Inhibitory activity of Pin1
The present study used a chymotrypsin coupling
experiment, designated Suc-Ala-Glu-Pro-Phe-pNA as the substrate,
and evaluated the inhibitory effect of the target compound on Pin1
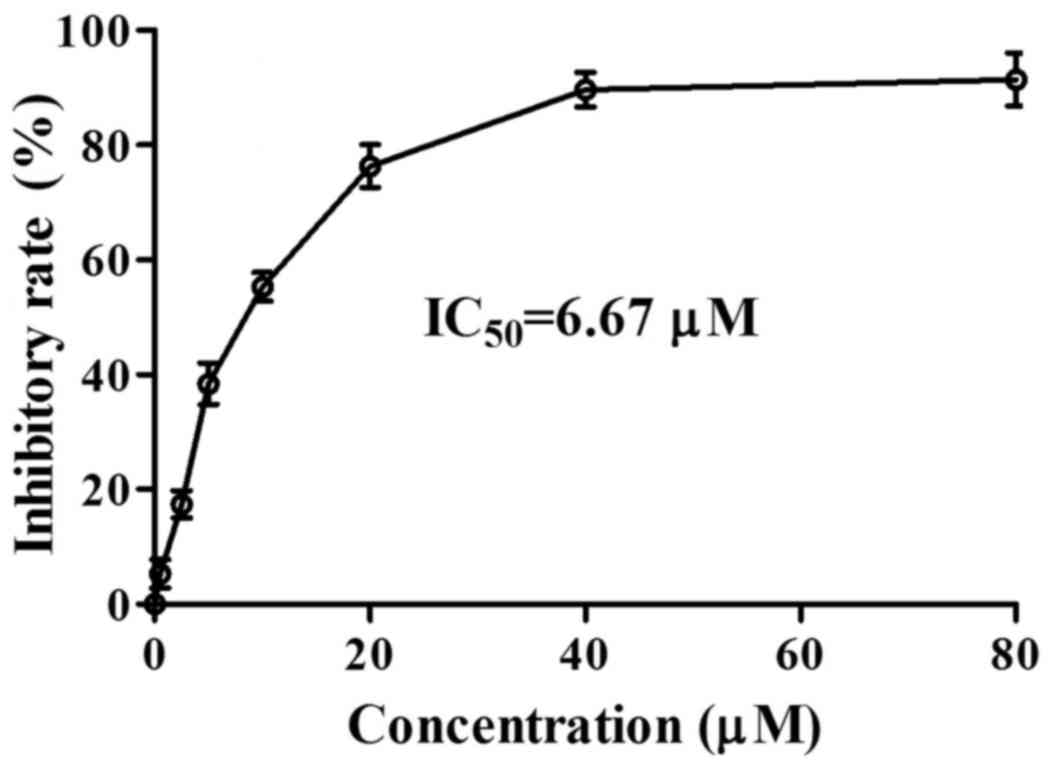
(16). Imazamethabenz inhibited
the activity of the Pin1 enzyme in vitro (Fig. 2), and its inhibitory activity
(IC50=6.67 µM) was equivalent with various Pin1 enzyme
inhibitors, as previously reported (17). Based on this result, the capacity
of imazamethabenz to induce apoptosis in tumor cells was further
surveyed.
Imazamethabenz inhibits survival,
migration and invasion of MCF-7
Pin1 overexpression induces cell proliferation and
inhibits apoptosis through multiple mechanisms (3–5).
Proteins associated with malignant tumors, including VEGF, MMP9, B
cell lymphoma-2, c-myc and p53, as well as nuclear factor-κB and
cyclin E, are active substrates of Pin1 (18–20).
Hence, it was hypothesized that Pin1 inhibition in tumor cells
overexpressing Pin1 would inhibit certain pathological features
that are important for tumor cell survival.
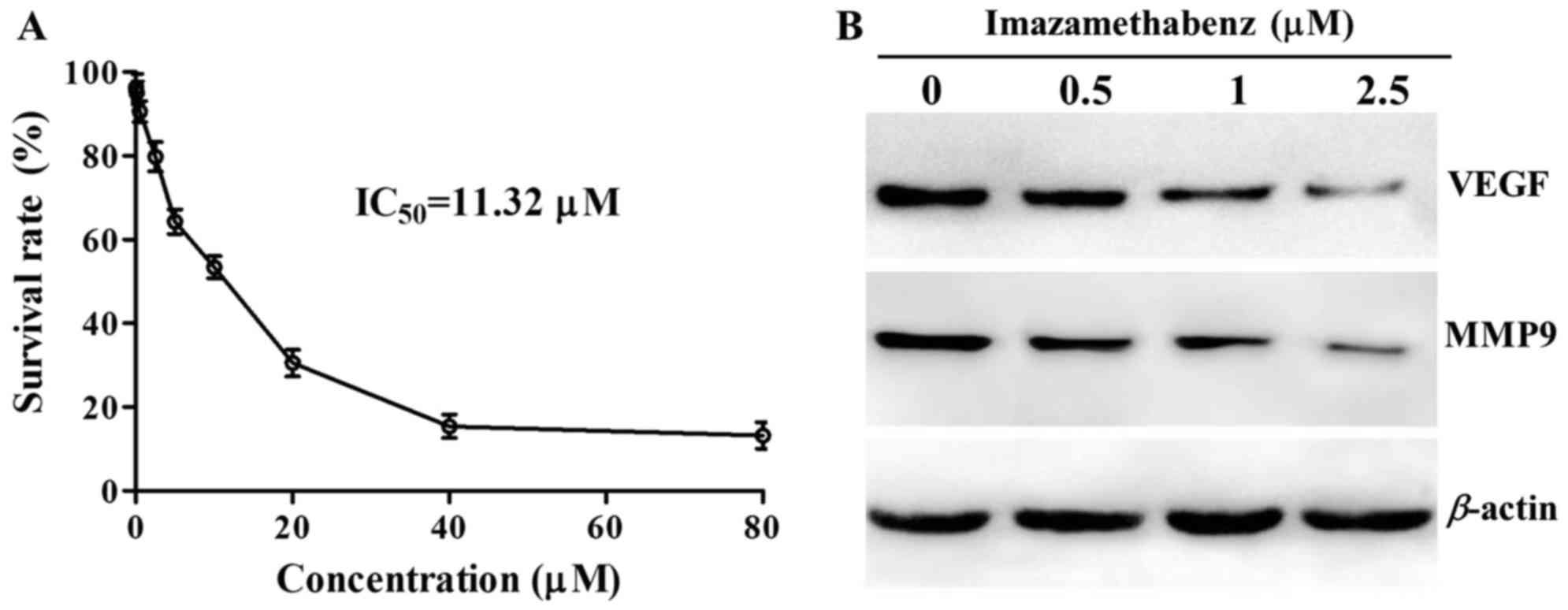
The results of the MTT assay are depicted in
Fig. 3A. At a concentration of
1–80 µM imazamethabenz significantly inhibited HepG-2 cell
proliferation and the IC50 value was 11.32 µM. Western
blot experiments were then performed following incubation of MCF-7
cells with imazamethabenz for 4 days. The protein expression levels
of VEGF and MMP9 were reduced in the MCF-7 cells following exposure
to imazamethabenz, in a concentration dependent manner (Fig. 3B). The capacity of imazamethabenz
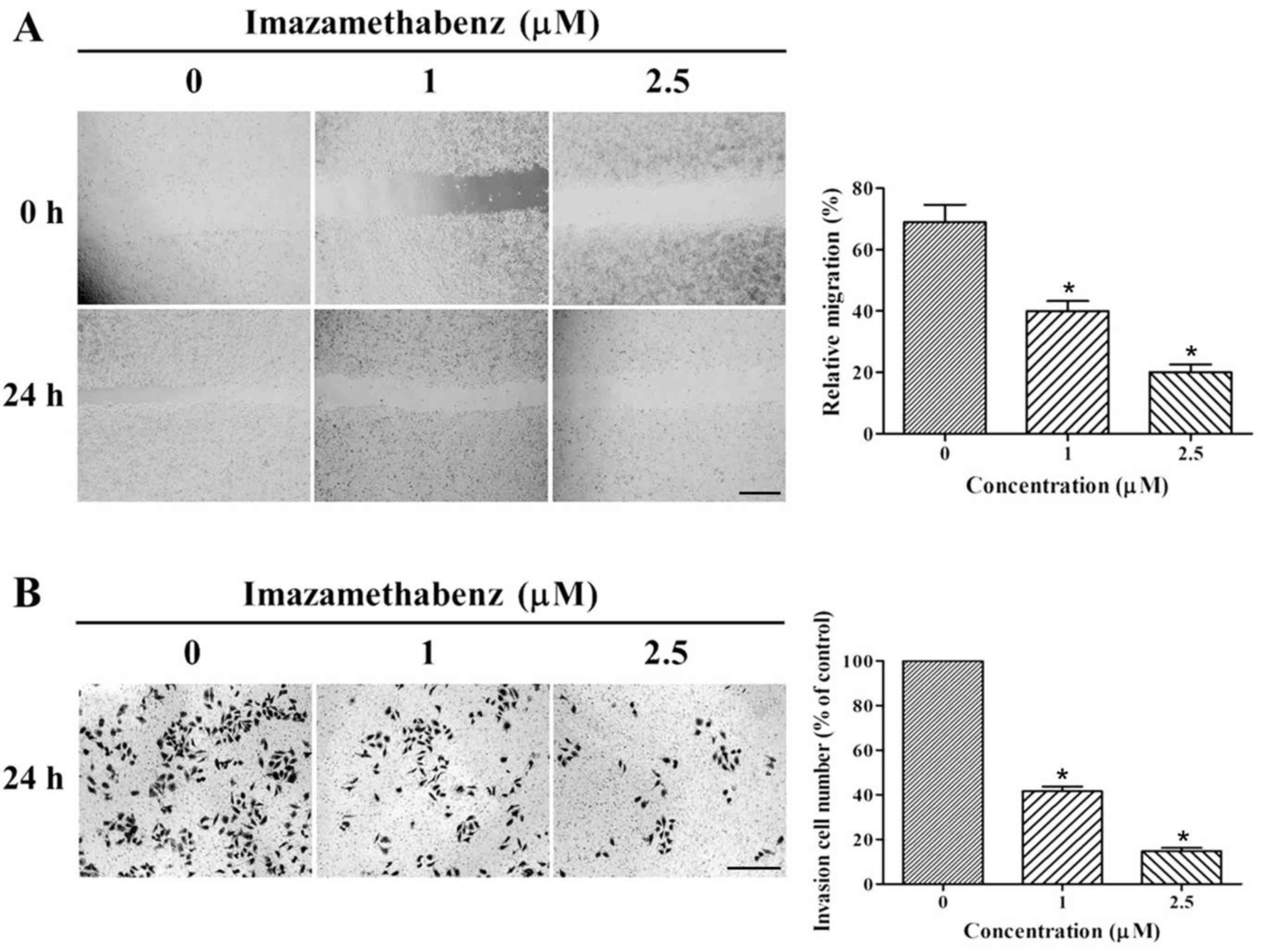
to inhibit MCF-7 cellular migration and invasion was then assessed
through further wound healing assays and transwell invasion assays,
respectively. The results of the wound healing assay are depicted
in Fig. 4A. Following 24 h
incubation of imazamethabenz with MCF-7 cells, the size of the
scratched region decreased less than in cells incubated without
imazamethabenz. The migration rates for 1 and 2.5 µM imazamethabenz
were 40.84 and 19.32%, respectively (P<0.05); therefore
imazamethabenz has an inhibitory effect on endothelial cell
migration. Based on the results of wound healing assay, Matrigel
invasion assays were used to observe the influence of
imazamethabenz on the in vitro invasion ability of MCF-7
cells, and the results are depicted in Fig. 4B. Following treatment of MCF-7
cells with 1 or 2.5 µM imazamethabenz for 24 h, the number of
transmembrane cells visibly decreased in a dose-dependent manner,
and the invasion ability of the cells was inhibited. Furthermore,
compared with the control group, the number of invading cells
decreased to 43.54 and 15.67%, respectively (P<0.05). This
finding indicated that Pin1 may be an important molecule involved
in the process of tumor cell migration and invasion.
To the best of our knowledge, the present study
demonstrated for the first time that the imidazoline ketone
herbicide, imazamethabenz, is a potential small-molecule inhibitor
of Pin1. The molecular docking experiment revealed the binding mode
of imazamethabenz and laid a foundation for further structural
modification. A previous study performed a Pin1 enzyme inhibition
experiment revealed that the enzyme inhibitory ability of
imazamethabenz is able to compete with that of other Pin1
inhibitors (17). Furthermore,
imazamethabenz inhibited the activity of Pin1 in vitro.
Furthermore, the mechanism of imazamethabenz in inducing cellular
apoptosis and a decrease in cell survival, migration and invasion
may be exerted by regulating phosphorylated protein activity in the
cell cycle. Under the effect of low concentrations of
chemotherapeutics, which do not influence the activity of cellular
survival, imazamethabenz effectively inhibited the migration and
invasion of breast cancer cells and decreased the levels of VEGF
and MMP9, which are involved in angiogenesis and metastasis,
respectively. In conclusion, based on these results, imazamethabenz
is able to inhibit tumor growth and migration by influencing
protein Pin1, which is associated with cell cycle control,
indicating imazamethabenz may be used as a potential compound for
future tumor treatments.
Acknowledgements
The authors would like to thank the National Nature
Science Foundation of China (grant no. 81302701) and the Grants for
Scientific Research of BSKY (grant. no. XJ201226) from Anhui
Medical University, that supported the present study.
References
|
1
|
Lu KP, Hanes SD and Hunter T: A human
peptidyl-prolyl isomerase essential for regulation of mitosis.
Nature. 380:544–547. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bao L, Kimzey A, Sauter G, Sowadski JM, Lu
KP and Wang DG: Prevalent overexpression of prolyl isomerase Pin1
in human cancers. Am J Pathol. 164:1727–1737. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yeh ES and Means AR: PIN1, the cell cycle
and cancer. Nat Rev Cancer. 7:381–388. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sorrentino G, Comel A, Mantovani F and Del
Sal G: Regulation of mitochondrial apoptosis by Pin1 in cancer and
neurodegeneration. Mitochondrion. 19:88–96. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin CH, Li HY, Lee YC, Calkins MJ, Lee KH,
Yang CN and Lu PJ: Landscape of Pin1 in the cell cycle. Exp Biol
Med (Maywood). 240:403–408. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Keune WJ, Jones DR and Divecha N: PtdIns5P
and Pin1 in oxidative stress signaling. Adv Biol Regul. 53:179–189.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marsolier J and Weitzman JB: Pin1: A
multi-talented peptidyl prolyl cis-trans isomerase and a promising
therapeutic target for human cancers. Med Sci (Paris). 30:772–778.
2014.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoon HE, Kim SA, Choi HS, Ahn MY, Yoon JH
and Ahn SG: Inhibition of Plk1 and Pin1 by 5′-nitro-indirubinoxime
suppresses human lung cancer cells. Cancer Lett. 316:97–104. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu L, Jin J, Liu C, Zhang C, Sun Y, Guo
Y, Fu D, Chen X and Xu B: Synthesis and biological evaluation of
novel quinazoline-derived human Pin1 inhibitors. Bioorg Med Chem.
19:2797–2807. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nakagawa H, Seike S, Sugimoto M, Ieda N,
Kawaguchi M, Suzuki T and Miyata N: Peptidyl prolyl isomerase
Pin1-inhibitory activity of D-glutamic and D-aspartic acid
derivatives bearing a cyclic aliphatic amine moiety. Bioorg Med
Chem Lett. 25:5619–5624. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Newsome WH and Collins PG: Determination
of imazamethabenz in cereal grain by enzyme-linked immunosorbent
assay. Bull Environ Contam Toxicol. 47:211–216. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Daum S, Wildemann D, Zhou XZ,
Verdecia MA, Bowman ME, Lücke C, Hunter T, Lu KP, Fischer G and
Noel JP: Structural basis for high-affinity peptide inhibition of
human Pin1. ACS Chem Biol. 2:320–328. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong L, Marakovits J, Hou X, Guo C,
Greasley S, Dagostino E, Ferre R, Johnson MC, Kraynov E, Thomson J,
et al: Structure-based design of novel human Pin1 inhibitors (II).
Bioorg Med Chem Lett. 20:2210–2214. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kofron JL, Kuzmic P, Kishore V,
Colón-Bonilla E and Rich DH: Determination of kinetic constants for
peptidyl prolyl cis-trans isomerases by an improved
spectrophotometric assay. Biochemistry. 30:6127–6134. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yuan SS, Hou MF, Hsieh YC, Huang CY, Lee
YC, Chen YJ and Lo S: Role of MRE11 in cell proliferation, tumor
invasion, and DNA repair in breast cancer. J Natl Cancer Inst.
104:1485–1502. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu C, Jin J, Chen L, Zhou J, Chen X, Fu
D, Song H and Xu B: Synthesis and biological evaluation of novel
human Pin1 inhibitors with benzophenone skeleton. Bioorg Med Chem.
20:2992–2999. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Min SH, Zhou XZ and Lu KP: The role of
Pin1 in the development and treatment of cancer. Arch Pharm Res.
39:1609–1620. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin CH, Li HY, Lee YC, Calkins MJ, Lee KH,
Yang CN and Lu PJ: Landscape of Pin1in the cell cycle. Exp Biol Med
(Maywood). 240:403–408. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Litchfield DW, Shilton BH, Brandl CJ and
Gyenis L: Pin1: Intimate involvement with the regulatory protein
kinase networks in the global phosphorylation landscape. Biochim
Biophys Acta. 1850:2077–2086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim MR, Choi HS, Heo TH, Hwang SW and Kang
KW: Induction of vascular endothelial growth factor by
peptidyl-prolyl isomerase Pin1 in breast cancer cells. Biochem
Biophys Res Commun. 369:547–553. 2008. View Article : Google Scholar : PubMed/NCBI
|