Introduction
To date, breast cancer is the most common female
malignancy diagnosed, and the most frequent cause of cancer
incidence rate among women worldwide (1). Despite advances in diagnosis
oncology, the early detection and treatment of breast cancer
remains a significant problem due to its metastatic nature
(2). Preventing breast cancer
prior to its development may be the most effective way to reduce
mortality resulting from this disease (3). Previous research has emphasized that
breast cancer is a highly heterogeneous group of diseases that
differ in their prognosis and response to treatment. The
development of breast cancer is thought to occur via a multi-stage
process, and a large number of molecules are considered to be
involved in the tumorigenesis and progression (4). Therefore, it is critical to
investigate the mechanisms underlying breast tumorigenesis and to
develop therapeutic strategies that are effective for the early
diagnosis and management of breast cancer.
The tumor necrosis factor receptor associated factor
(TRAF) protein family were initially identified as adaptor proteins
that couple tumor necrosis factor receptor family members to
signaling pathways, and are signal transducers of
toll/interleukin-1 receptor family members (5). All TRAF proteins share a C-terminal
homology region, termed the TRAF domain, that is able to bind to
the cytoplasmic domain of receptors, and to other TRAF proteins.
Previous studies have confirmed that TRAF proteins are important
regulators of cell death and cellular responses to stress (6). To date, seven members of the TRAF
family have been identified, which are termed TRAF1-7 (7). Notably, TRAF6 is a unique adaptor
protein of the TRAF family that consists of 530 amino acids,
including a highly conserved TRAF6 domain at the C terminus, and an
activated coiled-coil domain at the N terminus. The interaction of
TRAF6 and other ubiquitin conjugating enzymes is required for IκB
kinase activation and downstream signaling of the nuclear factor
(NF)-κB transcription factor. TRAF6 also interacts with the
transforming growth factor β (TGF-β) receptor complex and is
required for mitogen activated protein kinase (MAPK) activation.
Since TRAF6 may activate several signaling pathways simultaneously,
it serves a crucial role in the growth, proliferation,
differentiation and death of cells (8). Furthermore, TRAF6 has been identified
in humans and mice (9).
Upregulation of TRAF6 expression has been detected in glioma,
osteosarcoma, esophageal squamous cell carcinoma, and colon and
lung cancer tissues (10).
However, there have been no further mechanistic studies regarding
the contribution of TRAF6 expression to the development of breast
cancer. Therefore, the present study aimed to investigate the
pathophysiological contribution of TRAF6 expression to breast
tumorigenesis in breast cancer patients.
Materials and methods
Tissue specimens
Human breast tissues (n=32) were obtained from
Jiangsu Cancer Hospital (Nanjing, China). The patients had not
previously received chemotherapy or radiotherapy. Adjacent healthy
tissues were used as a control (n=25). All cases were diagnosed by
two experienced pathologists. This study was approved by the
institutional review board at Jiangsu Cancer Hospital, and signed
informed consent was obtained from all patients and their
relatives.
Antibodies and reagents
Antibodies against the following proteins were
purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA):
TRAF6 (cat. no. 8028; 1:1,000), phosphorylated (p)-transforming
growth factor β-activated kinase 1 (TAK1); cat. no. 4508; 1:1,000),
TAK1 (cat. no. 5206; 1:1,000), p-protein kinase B (AKT)Ser473 (cat.
no. 4060; 1:1,000), AKT (cat. no. 4691; 1:1,000), p-glycogen
synthase kinase (GSK)3β (cat. no. 9322; 1:1,000), GSK3β (cat. no.
9315; 1:1,000), and GAPDH (cat. no. 2118; 1:1,000). The secondary
antibody was horseradish peroxidase-conjugated goat anti-rabbit IgG
(cat. no. 7074; 1:2,000; Cell Signaling Technology, Inc.). The
Bicinchoninic Acid (BCA) protein assay kit was purchased from
Pierce (Rockford, IL, USA). A FluorChem E imager (ProteinSimple;
Bio-Techne, Minneapolis, MN, USA) was used for visualization. The
TAK1 kinase inhibitor, 5Z-7-Oxozeaenol (5Z-O; cat. no. 3604), was
purchased from Tocris Bioscience (Bristol, UK).
Cell culture
The MCF-7 breast cancer cell line was preserved and
maintained at 37°C in high glucose complete Dulbecco's modified
Eagle's medium (DMEM; Hyclone; GE Healthcare Life Sciences, Logan,
UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare Life Sciences) in a humidified 5% CO2
incubator. The cells were treated with vehicle [0.1%
dimethylsulfoxide (DMSO)] or 5Z-O (1,000 nM) combined with 10% FBS
in high glucose complete DMEM medium for 24 h, as described
previously (11).
Cell viability assay
An MTT assay was conducted to analyze cell
viability. Briefly, cells were seeded into 96-well plates at a
density of 8×103/well and cultured with 100 µl complete medium
(DMEM high glucose containing 10% FBS). After 24 h, 20 µl 5 mg/ml
MTT solution (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was
added into each well. Cells were incubated for 4 h at 37°C before
the culture medium was removed. Following this, 150 µl DMSO
(Amresco, LLC, Solon, OH, USA) per well was added and mixed in
order to guarantee cytolysis and dissolution of the formazan
crystal. The absorbance was measured at a wavelength of 490 nM
using a microplate reader (CliniBio 128; ASYS Hitech GmbH,
Eugendorf, Austria). Cells treated with 0.1% DMSO instead of the
TAK1 inhibitor were used as blank control. Three independent
experiments were performed.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was performed as previously described
(12). Briefly, total RNA was
extracted from frozen human tissue using a TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
reverse-transcribed into cDNA using oligo (dT) primers with a
Transcriptor First Strand cDNA Synthesis kit (Roche Applied
Science, Penzberg, Germany). PCR amplifications were quantified
using SYBR®-Green PCR Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and normalized to GAPDH gene expression by the
2-∆∆Cq method (13). The primer
sequences used were as follows: Forward, GCA GCC CGT GTA ACT GGA
ATG A and reverse, GTC TTC TAG AGC CTG GGC CTT for Ki-67; and
forward, TGC GGC CGG GTT CAG GAG TCA and reverse, CAG GCA GGC GGG
AAG GAG GAA AGT for proliferating cell nuclear antigen (PCNA).
Cycling parameters consisted of 95°C 10 min, followed by 40 cycles
at 95°C for15 sec (denaturing) and 60°C for 30 sec (annealing and
extension). All samples were analyzed in triplicate for each
specific gene.
Western blotting
Western blotting was performed as previously
described (14). Total proteins
extracted from frozen human tissue were first lysed in
radioimmunoprecipitation assay lysis buffer (Biouniquer Technology,
Beijing, China) on ice for 30 min, followed by centrifugation at
14,000 × g for 15 min at 4°C to remove cell pellet. Protein
concentrations were measured using a BCA Protein Assay kit.
Proteins (50 µg) were separated by 8–12% SDS-PAGE (Invitrogen;
Thermo Fisher Scientific, Inc.) and transferred onto polyvinylidene
difluoridemembranes (Merck KGaA). The membranes were blocked in TBS
with Tween-20 containing 5% skimmed milk powder for 1 h at room
temperature, and incubated with various primary antibodies
overnight at 4°C. Following this, membranes were incubated with
secondary antibodies for 1 h at room temperature, and were
visualized with Enhanced Chemiluminescent reagents (cat. no.
170–5061; Bio-Rad Laboratories, Inc., Hercules, CA, USA) using a
FluorChem E imager according to the manufacturer's protocol.
Protein expression levels were normalized against GAPDH values. All
experiments were conducted at least in triplicate.
Statistical analysis
Data are presented as the mean ± standard deviation.
For two-group comparisons, Gaussian samples were compared using the
two-tailed Student's t-test, while non-Gaussian samples were
compared employing the non-parametric Mann-Whitney U test.
Statistical analyses were performed with SPSS software version 21.0
(IBM SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
TRAF6 expression is upregulated in
human breast cancer
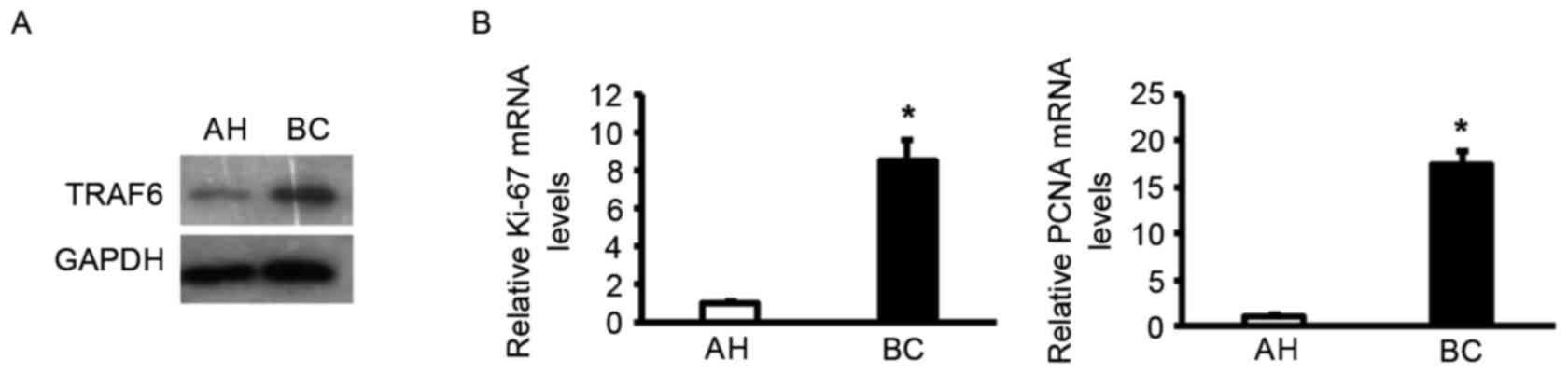
Protein expression levels of TRAF6 were determined
in breast cancer samples to determine if there was an association.
Western blotting revealed that the expression of TRAF6 was higher
in breast carcinoma specimens compared with adjacent healthy
tissues (Fig. 1A), which implies a
role of TRAF6 in tumor progression. RT-qPCR analysis was
subsequently performed to determine the association between TRAF6
expression and pathological features. Notably, TRAF6 expression was
positively associated with the induction of a series of cellular
markers for proliferation at the mRNA level, including Ki-67 and
PCNA (Fig. 1B). Collectively, the
altered pattern of TRAF6 expression suggested that TRAF6 may be
associated with cell proliferation in the development of human
breast cancer.
TRAF6 promotes breast carcinoma
proliferation in an TAK1-dependent manner
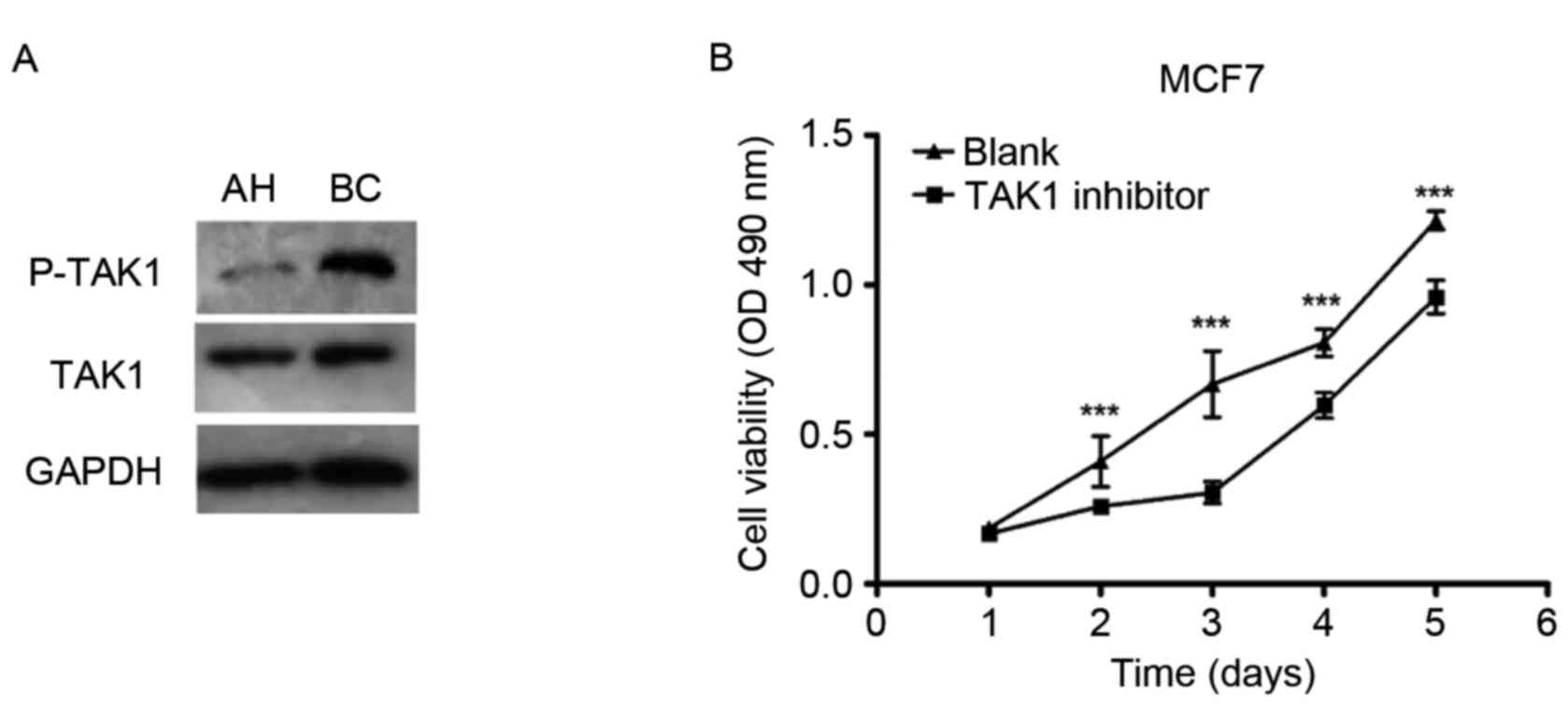
To elucidate the potential molecular mechanisms
underlying the effects of TRAF6 on cell proliferation in human
breast cancer, the expression levels/activities of TRAF6 signaling
molecules (the accessory molecules TAK1) were examined. Protein
expression levels of phosphorylated-TAK1 were demonstrated to be
upregulated in human breast carcinoma tissues compared with
adjacent healthy tissues, as indicated by western blot analysis
(Fig. 2A). However, total TAK1
protein expression levels did not differ between the groups
(Fig. 2A). The specific TAK1
inhibitor 5Z-O was used to clarify whether the TAK1 signaling
pathway is the main molecular event involved in the cell
proliferative role of TRAF6 in human breast cancer. 5Z-O
significantly impaired the effects of TRAF6 on tumor cell
proliferation in MCF-7 cells, as demonstrated by the MTT analysis
(Fig. 2B). These results indicated
that TAK1 is critical for the TRAF6-induced cell proliferative
effect that occurs during breast tumorigenesis.
TRAF6 regulates breast tumorigenesis
via activation of the AKT/GSK3β signaling pathways
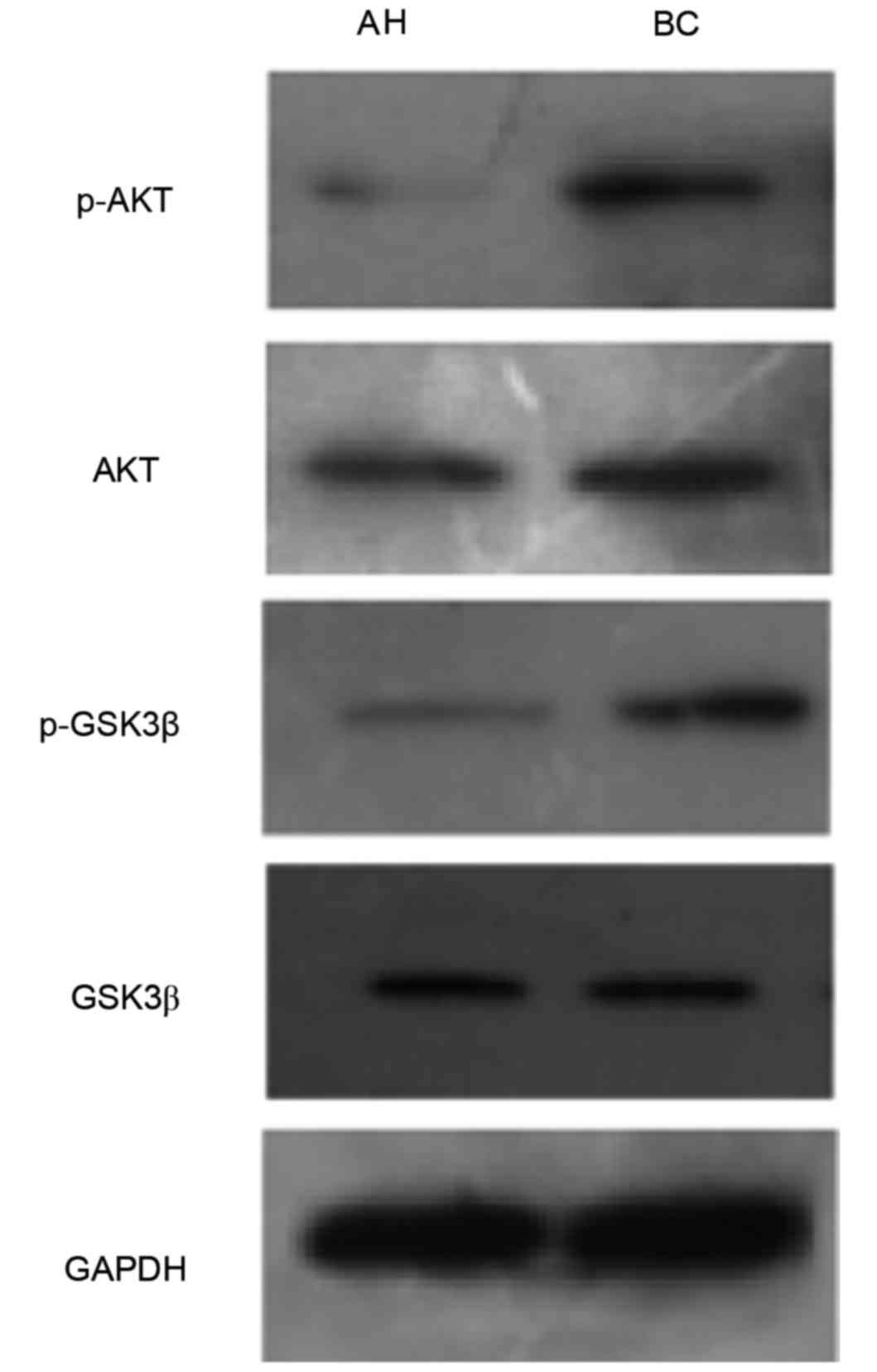
Increasing evidence has suggested that regulation of
the AKT/GSK3β signaling cascade may regulate cellular events
critical for tumorigenesis. Activation of the AKT/GSK3β signaling
pathway was therefore investigated. The expression levels p-AKT and
p-GSK3β were higher in the human breast carcinoma group compared
with the adjacent healthy tissues group. However, expression levels
of AKT and GSK3β did not differ between the two groups (Fig. 3). These results demonstrated that
the proliferative role of TRAF6 may be associated with activation
of AKT/GSK3β signaling during breast tumorigenesis.
Discussion
The present study provided evidence of the critical
role of the TRAF superfamily member TRAF6 in human breast cancer.
Notably, protein expression levels of TRAF6 were observed to be
upregulated in human breast carcinoma, compared with adjacent
healthy tissues. In addition, the cellular proliferative marker
Ki-67 and PCNA mRNA expression levels were significantly
upregulated in breast carcinoma tissues compared with adjacent
healthy tissues, indicating that TRAF6 expression may be associated
with these markers. Mechanistically, it was further demonstrated
that TRAF6 may bind to the accessory molecule TAK1, potentially via
the AKT/GSK3β signaling pathways, thereby initiating cell
proliferation. Therefore, TRAF6 may represent a promising
therapeutic target for the treatment of human breast cancer.
TRAF6 may regulate an adverse array of physiological
processes, including adaptive and innate immunity, bone metabolism,
and the development of several tissues, including lymph nodes,
mammary glands, skin and the central nervous system (5). Previous studies have demonstrated
that upregulation of TRAF6 occurs in many types of cancer, which
suggests that TRAF6 may serve an important role in tumorigenesis
and tumor progression (15,16).
The present study demonstrated that breast carcinoma specimens had
an increase in TRAF6 expression compared with the adjacent healthy
tissues, consistent with a study conducted by Bilir et al
(17) that revealed that serum
TRAF6 expression levels were higher in patients with non-metastatic
triple-negative breast cancer. The present study demonstrated that
mRNA expression levels of cellular markers for proliferation Ki-67
and PCNA were upregulated in breast cancer tissues compared with
adjacent healthy tissues, which indicated that TRAF6 may be
involved in cell proliferation in human breast tumorigenesis. Peng
et al (18) reported that
TRAF6 induced proliferation of U-87 MG human glioma cells.
Furthermore, the proteasome inhibitory effect of bortezomib on
TRAF6 was able to restrain multiple myeloma cell proliferation
(19). Therefore, further
investigation is required to elucidate the underlying mechanisms of
TRAF6 in breast tumorigenesis.
Previous studies have demonstrated that the
TRAF6-catalyzed Lys-63-linked polyubiquitination of TAK1 at Lys-34
may alter the conformation of TAK1 to function as a central, key
regulatory sensor for extra-cellular cues (20,21).
In the present study, it was observed that the accessory molecule
TAK1 was significantly augmented, suggesting that the
TRAF6-mediated regulation of tumor cell proliferation may be
directly associated with its downstream target, TAK1. This
hypothesis was supported by the findings that a blockade of TAK1
activation induces the TRAF6-elicited cell proliferative response.
Such findings are consistent with Xiao et al (22), who demonstrated that TRAF6 and its
intrinsic ubiquitin E3 ligase activity promotes myogenic
differentiation and muscle regeneration via TAK1. Therefore, the
proliferative role of TRAF6 in breast tumorigenesis may be
dependent, at least partially, on the activation of TAK1.
Furthermore, it is well documented that TRAF6-mediated TAK1
activity regulates cell survival, differentiation and inflammatory
responses via many diverse signals, including the NF-κB and MAPK
cascades (23). To further
understand the underlying mechanism responsible for TRAF6/TAK1
signaling, the activation of their downstream pathways was
investigated. The results of the present study indicated that
activation of AKT and its downstream activator GSK3β was greatly
enhanced. Consistent with previous studies, Yoon et al
(24) also demonstrated that TRAF6
positively regulates cell survival by regulating AKT/GSK3β
cascades. In terms of proliferation, previous studies have
confirmed that AKT/GSK3β signaling promotes cell cycle progression
by upregulating the positive regulators of cell growth including
PCNA and cyclin D1 (25–27). Indeed, in the MDA-MB-231 breast
cancer cell line, fangchinoline inhibited cell proliferation via
suppression of the AKT/GSK3β signaling pathway (28). Furthermore, dihydroartemisinin
inhibited cell proliferation via suppression of the AKT/GSK3β
signaling pathway in the A549 lung cancer cell line (29). Taken together, these results
indicated that AKT/GSK3β signaling pathway is involved in
TRAF6-mediated human breast cancer tumorigenesis, but is not the
primary molecular mechanism responsible. However, the potential
underlying mechanisms for its involvement in human breast cancer
progression remains to be fully elucidated; therefore, other
signaling pathways require investigating to further
understanding.
In conclusion, the current study provided evidence
that TRAF6, a member of the TRAF superfamily, functions as a
positive regulator of human breast tumorigenesis by applying the
accessory molecule TAK1 to activate the AKT/GSK3β signaling
pathway, thereby promoting cell proliferation at the mRNA level.
These results implicate TRAF6 as a potential therapeutic target for
the prevention and treatment of human breast cancer.
Acknowledgements
This study was funded by the National High
Technology Research and Development Program of China (grant no.
2014AA020604), the National Natural Science Foundation of China
(grant no. 81272470), the National Key Clinical Specialist
Construction Programs of China [grant no. 2013 (544)], the Major
Program of Natural Science Foundation of Jiangsu Province (grant
no. BL2014090) and the Natural Science Foundation of Jiangsu
Province (grant no. BK20151579).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
den Hollander P, Savage MI and Brown PH:
Targeted therapy for breast cancer prevention. Front Oncol.
3:2502013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Independent UK Panel on Breast Cancer
Screening: The benefits and harms of breast cancer screening: An
independent review. Lancet. 380:1778–1786. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li Y, Li L, Lin J, Hu X, Li B, Xue A, Shen
Y, Jiang J, Zhang M, Xie J and Zhao Z: Deregulation of RGS17
expression promotes breast cancer progression. J Cancer. 6:767–775.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bradley JR and Pober JS: Tumor necrosis
factor receptor-associated factors (TRAFs). Oncogene. 20:6482–6491.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee NK and Lee SY: Modulation of life and
death by the tumor necrosis factor receptor-associated factors
(TRAFs). J Biochem Mol Biol. 35:61–66. 2002.PubMed/NCBI
|
|
7
|
Chung JY, Park YC, Ye H and Wu H: All
TRAFs are not created equal: Common and distinct molecular
mechanisms of TRAF-mediated signal transduction. J Cell Sci.
115:679–688. 2002.PubMed/NCBI
|
|
8
|
Inoue J, Gohda J and Akiyama T:
Characteristics and biological functions of TRAF6. Adv Exp Med
Biol. 597:72–79. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ishida T, Mizushima S, Azuma S, Kobayashi
N, Tojo T, Suzuki K, Aizawa S, Watanabe T, Mosialos G, Kieff E, et
al: Identification of TRAF6, a novel tumor necrosis factor
receptor-associated factor protein that mediates signaling from an
amino-terminal domain of the CD40 cytoplasmic region. J Biol Chem.
271:28745–28748. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He Z, Huang C, Lin G and Ye Y:
siRNA-induced TRAF6 knockdown promotes the apoptosis and inhibits
the invasion of human lung cancer SPC-A1 cells. Oncol Rep.
35:1933–1940. 2016.PubMed/NCBI
|
|
11
|
Huang HL, Chiang CH, Hung WC and Hou MF:
Targeting of TGF-β-activated protein kinase 1 inhibits chemokine
(C-C motif) receptor 7 expression, tumor growth and metastasis in
breast cancer. Oncotarget. 6:995–1007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li L, Wang X, Chen W, Qi H, Jiang DS,
Huang L, Huang F, Wang L, Li H and Chen X: Regulatory role of CARD3
in left ventricular remodelling and dysfunction after myocardial
infarction. Basic Res Cardiol. 110:562015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li L, Chen W, Zhu Y, Wang X, Jiang DS,
Huang F, Wang L, Xiang F, Qin W, Wang Q, et al: Caspase recruitment
domain 6 protects against cardiac hypertrophy in response to
pressure overload. Hypertension. 64:94–102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhong L, Cao F and You Q: Effect of TRAF6
on the biological behavior of human lung adenocarcinoma cell.
Tumour Biol. 34:231–239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yao F, Han Q, Zhong C and Zhao H: TRAF6
promoted the tumorigenicity of esophageal squamous cell carcinoma.
Tumour Biol. 34:3201–3207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bilir C, Engin H, Can M, Likhan S,
Demirtas D, Kuzu F and Bayraktaroglu T: Increased serum tumor
necrosis factor receptor-associated factor-6 expression in patients
with non-metastatic triple-negative breast cancer. Oncol Lett.
9:2819–2824. 2015.PubMed/NCBI
|
|
18
|
Peng Z, Shuangzhu Y, Yongjie J, Xinjun Z
and Ying L: TNF receptor-associated factor 6 regulates
proliferation, apoptosis, and invasion of glioma cells. Mol Cell
Biochem. 377:87–96. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu H, Tamashiro S, Baritaki S, Penichet
M, Yu Y, Chen H, Berenson J and Bonavida B: TRAF6 activation in
multiple myeloma: A potential therapeutic target. Clin Lymphoma
Myeloma Leuk. 12:155–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Adhikari A, Xu M and Chen ZJ:
Ubiquitin-mediated activation of TAK1 and IKK. Oncogene.
26:3214–3226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sorrentino A, Thakur N, Grimsby S,
Marcusson A, von Bulow V, Schuster N, Zhang S, Heldin CH and
Landström M: The type I TGF-beta receptor engages TRAF6 to activate
TAK1 in a receptor kinase-independent manner. Nat Cell Biol.
10:1199–1207. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiao F, Wang H, Fu X, Li Y and Wu Z: TRAF6
promotes myogenic differentiation via the TAK1/p38
mitogen-activated protein kinase and Akt pathways. PLoS One.
7:e340812012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chung JY, Lu M, Yin Q, Lin SC and Wu H:
Molecular basis for the unique specificity of TRAF6. Adv Exp Med
Biol. 597:122–130. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoon K, Jung EJ and Lee SY: TRAF6-mediated
regulation of the PI3 kinase (PI3K)-Akt-GSK3beta cascade is
required for TNF-induced cell survival. Biochem Biophys Res Commun.
371:118–121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wikman H and Kettunen E: Regulation of the
G1/S phase of the cell cycle and alterations in the RB pathway in
human lung cancer. Expert Rev Anticancer Ther. 6:515–530. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vincenzi B, Schiavon G, Silletta M,
Santini D, Perrone G, Di Marino M, Angeletti S, Baldi A and Tonini
G: Cell cycle alterations and lung cancer. Histol Histopathol.
21:423–435. 2006.PubMed/NCBI
|
|
27
|
Gugger M, Kappeler A, Vonlanthen S,
Altermatt HJ, Ris HB, Lardinois D, Borner MM, Heighway J and
Betticher DC: Alterations of cell cycle regulators are less
frequent in advanced non-small cell lung cancer than in resectable
tumours. Lung Cancer. 33:229–239. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang CD, Yuan CF, Bu YQ, Wu XM, Wan JY,
Zhang L, Hu N, Liu XJ, Zu Y, Liu GL and Song FZ: Fangchinoline
inhibits cell proliferation via Akt/GSK-3beta/ cyclin D1 signaling
and induces apoptosis in MDA-MB-231 breast cancer cells. Asian Pac
J Cancer Prev. 15:769–773. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liao K, Li J and Wang Z:
Dihydroartemisinin inhibits cell proliferation via
AKT/GSK3β/cyclinD1 pathway and induces apoptosis in A549 lung
cancer cells. Int J Clin Exp Pathol. 7:8684–8691. 2014.PubMed/NCBI
|