Introduction
Stroke is the fourth leading cause of mortality and
disability in the United States (1), and 10–20% of strokes are caused by
carotid artery disease (2,3). According to epidemiological data, ~7
million adults in the United States have suffered a stroke
(4), of which ischemic strokes
account for ~90% (5). In addition,
~10% of ischemic strokes are caused by carotid artery stenosis
(CAS) (6). CAS refers to a
narrowing or constriction of the lumen of the carotid artery,
usually attributed to atherosclerosis (7). The dynamic and complex process of
atherosclerosis remains to be fully understood; however, it is well
known that atherosclerosis is characterized by the accumulation of
lipid particles and fibrous elements, associated with migration and
proliferation of smooth muscle cells, in the large arteries
(8–10).
Over the past decade, inflammation and the immune
response in atherosclerosis have garnered attention. Previous
studies have indicated that low-density lipoprotein particles and
their content inside the vessel wall are susceptible to oxidation
by free radicals, which may initiate the accumulation and invasion
of macrophages, eventually lead to a narrowing of the major
arteries (9,11).
During the progression of atherosclerosis, an
imbalance between anti-inflammatory and proinflammatory cytokines
serves an important role. Previous studies have attempted to
identify the immune-associated genes that are involved in
atherosclerosis, and achievements have been made (12,13).
Superoxide dismutase, which is expressed at higher levels in
regions of laminar flow, may combat oxidative stress, and hence
limit vascular cell adhesion molecule-1 (VCAM-1) expression and the
expression of other inflammatory pathways (12). Nitric oxide arises from endothelial
nitric oxide synthase, which is known to be a shear
stress-regulated gene, and can inhibit VCAM gene expression through
a novel pathway involving inhibition of the activation of nuclear
factor-κB, the central transcription factor in vascular
inflammation (13). In addition,
previous studies have reported that interleukin (IL)-35 may
upregulate the expression of anti-inflammatory cytokines (14–16).
Huang et al (17)
investigated the effects of IL-35 on atherosclerosis and
hypothesized that IL-35 could be considered a novel target for the
treatment of atherosclerosis. However, the majority of genes
relevant to atherosclerosis remain unknown.
Mycophenolate mofetil (MMF) is an inhibitor of the
enzyme inosine monophosphate dehydroxygenase (IMPDH), and exerts a
powerful cytostatic effect on activated T cells by interfering with
their DNA synthesis (18). In the
present study, gene expression data were obtained from a Gene
Expression Omnibus (GEO) dataset uploaded by van Leuven et
al (19), which included 20
carotid endarterectomy samples from patients with CAS (>70%
diameter stenosis on angiography) that were randomly assigned to
the following treatment groups: Treatment with 1,000 mg MMF (n=9)
or placebo (n=11). Patients were treated with MMF or placebo for ≥2
weeks prior to undergoing carotid endarterectomy (CEA). van Leuven
et al (19) reported that
the inflammatory process in human atherosclerotic plaques could be
modified by short-term treatment with MMF, as determined using mRNA
expression profiling. However, this previous study did not analyze
the expression data in detail, nor did it determine how MMF
functioned in the treatment of symptomatic CAS (SCAS) or the
molecular mechanisms of SCAS.
In the present study, the gene expression data were
used to identify differentially-expressed genes (DEGs) between
MMF-treated and placebo-treated groups, with the aim of identifying
potential genes associated with atherosclerosis, which may be
considered targets for novel gene therapy. A total of 210 DEGs
between the MMF and placebo groups were identified with a threshold
of P<0.05. After analyzing the regulatory effects, a regulatory
network was constructed based on the DEGs. Subsequently, the data
were processed by bioinformatic analyses, including hierarchical
clustering, Gene Ontology (GO) terms (molecular function,
biological processes and cellular components) analysis and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Finally,
the 19 most significant DEGs were screened; the results of these
analyses indicated that apelin (APLN) and v-kit Hardy-Zuckerman 4
feline sarcoma viral oncogene homolog (KIT) may be valuable for
characterizing the mechanism underlying immunomodulatory therapy in
atherosclerosis.
Materials and methods
Datasets
The GSE13922 original mRNA expression profile used
in the present study was downloaded from the National Center of
Biotechnology Information GEO (http://www.ncbi.nlm.nih.gov/geo/). The platform used
to analyze these data was the GPL6255 Illumina humanRef-8 v2.0
expression beadchip (Illumina, San Diego, CA, USA).
Identification of DEGs
Background correction and quartile data
normalization of the downloaded data were performed using the
robust multi-array average (RMA) algorithm (20). Probes without a corresponding gene
symbol were filtered and the average value of gene symbols with
numerous probes was calculated. The expression profile dataset,
including 13,985 genes for the 20 samples, was subsequently
obtained. Student's t-test was used to identify DEGs between the
MMF and placebo groups using the R software LIMMA package (version
3.3.1; www.r-project.org) (21). Genes with P<0.05 were considered
DEGs and genes with P<0.01 were considered the most significant
DEGs between the two treatment groups. The most significant DEGs
were screened between the MMF and placebo groups using principal
components analysis (PCA). Cluster analysis of the most significant
DEGs was applied to generate a heat map, which allowed for
visualization of the differential gene expression between the two
groups.
Protein-protein interaction (PPI)
network construction and analysis
The PPI network was constructed from 210 DEGs using
the STRING online database (http://www.string-db.org/). PPI pairs with an
interaction score >0.4 were used to construct the PPI network.
Subsequently, the regulatory relationships between genes were
analyzed according to the topological properties of the network.
With a threshold of P<0.05 and |logFC|≥0.5, the key genes in the
network were further screened.
Functional analysis of various
DEGs
In order to identify biological functions associated
with the pathogenesis of atherosclerosis, bioinformatics analyses,
including hierarchical clustering, GO (22) terms (molecular function, biological
processes, cellular components) analysis and KEGG (23) pathway analysis, were conducted for
the most significant DEGs, using the online Database for
Annotation, Visualization and Integrated Discovery tool (24), based on the method of Expression
Analysis Systemic Explorer (EASE) test (25). The enrichment threshold was an EASE
score of 0.1.
Results
Data processing
A total of 13,985 genes in 20 samples were obtained
after preprocessing of the expression profile. The original
expression datasets were processed into expression estimates using
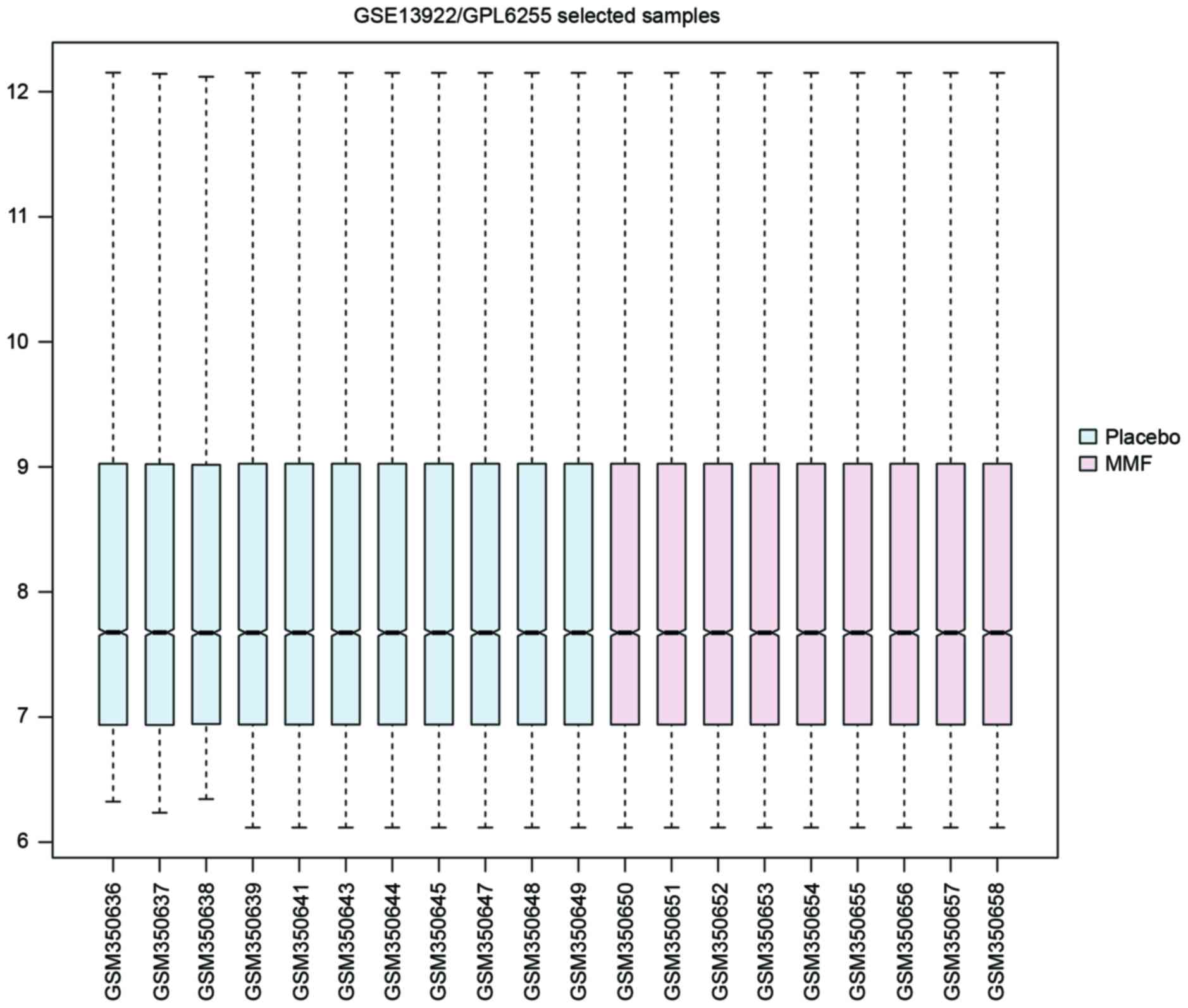
the RMA method. As presented in the box plot in Fig. 1, the median of different samples
was almost the same following normalization, which indicated a
great degree of standardization.
Identification of DEGs
The DEGs between the MMF and placebo groups were
identified using LIMMA package (21) in R software. A total of 210 DEGs
were identified with the threshold of P<0.05, including 19 most
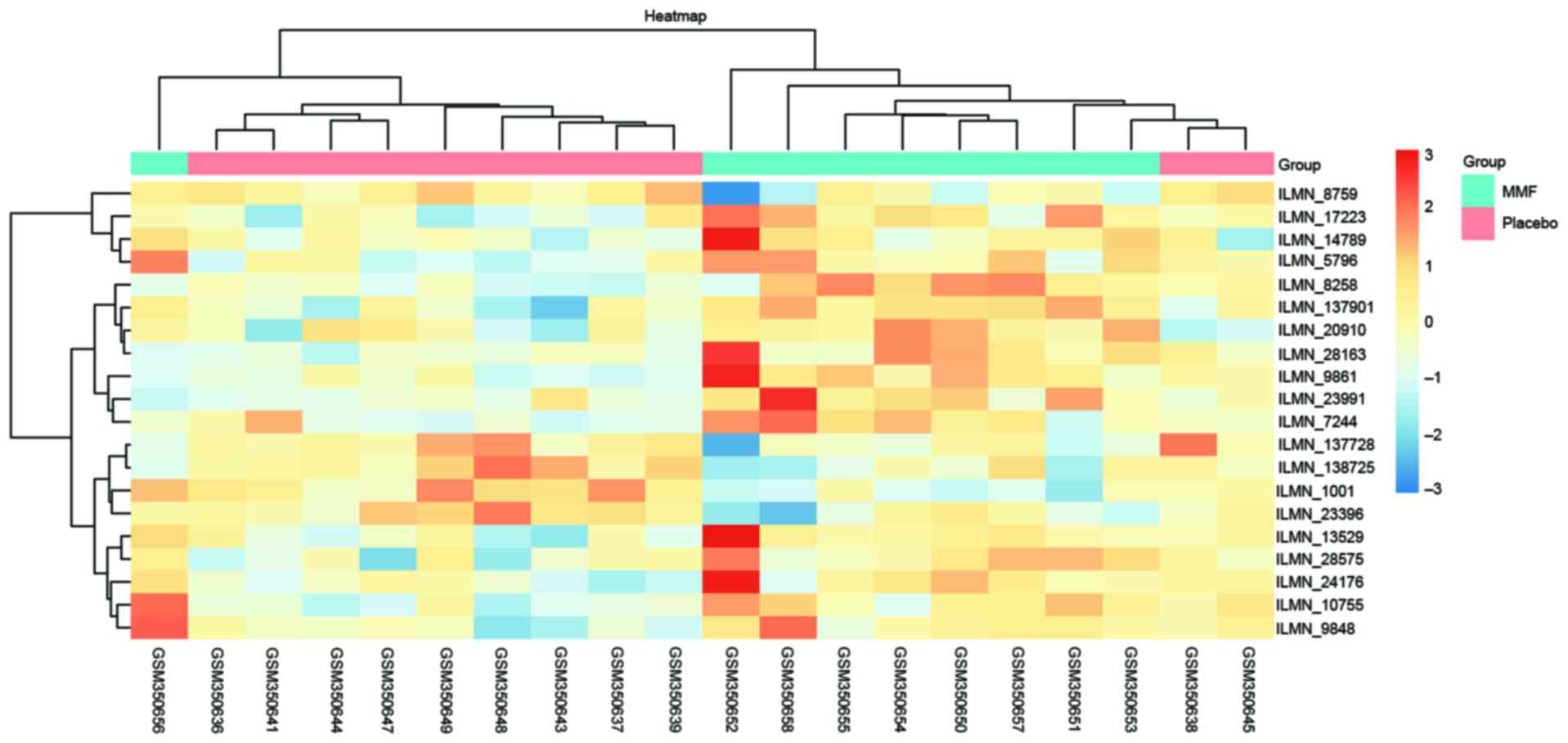
significant DEGs with the threshold of P<0.01 (Table I). Analysis of the most significant
DEGs revealed that there were 14 up- and 5 downregulated genes
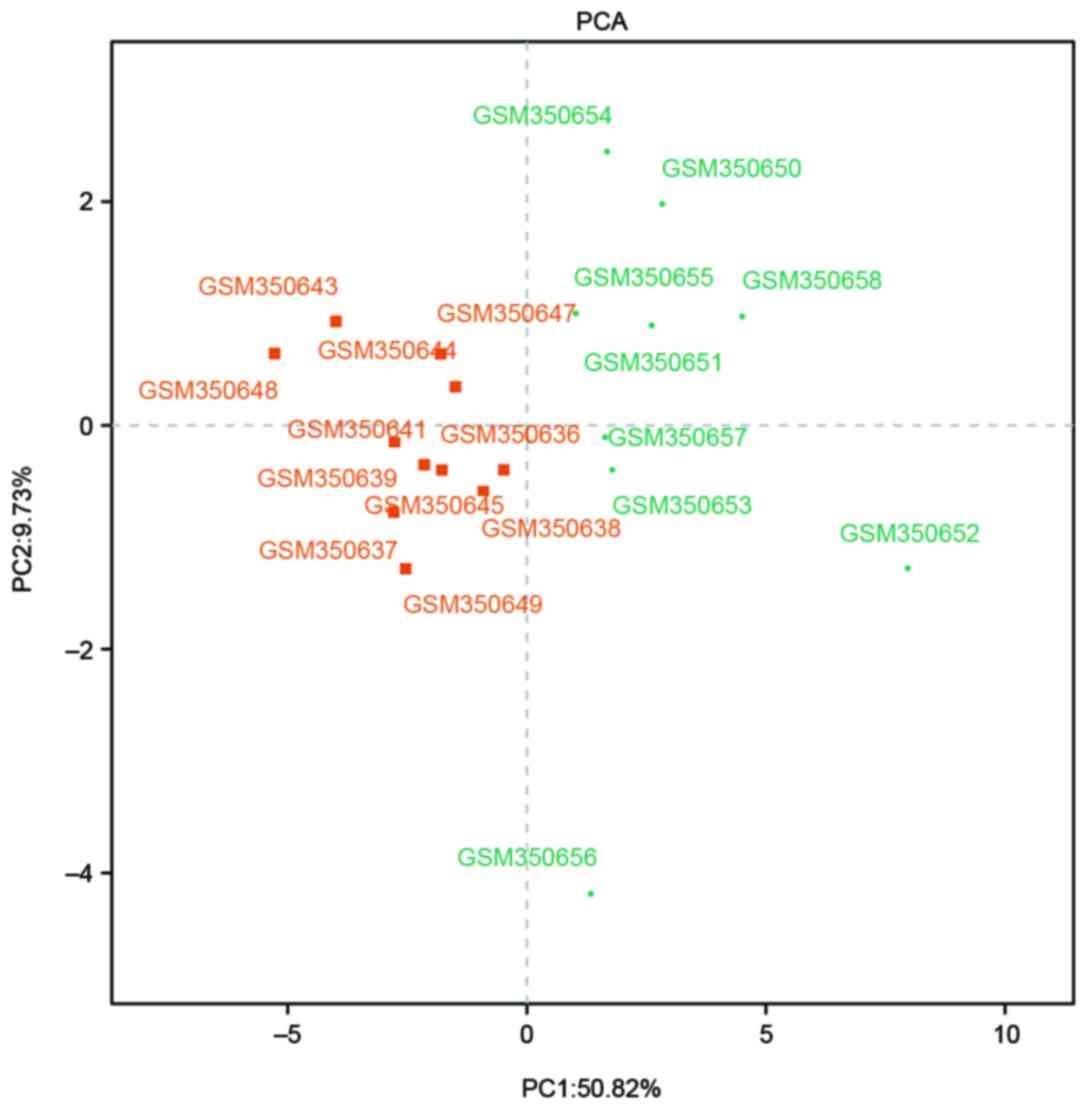
(Fig. 2). As presented in Fig. 3, all of the selected DEGs were
screened using PCA to distinguish between the MMF and placebo
groups. In the first principal components, 50.82% of variances were
explained, whereas in the second principal component, 9.73% of
variances were explained. In total, the resolution degree of
variances was 60.55%.
 | Table I.List of the 19 most significant DEGs
(P<0.01). |
Table I.
List of the 19 most significant DEGs
(P<0.01).
| Gene symbol | Gene name | P-value | logFC |
|---|
| AK4 | Adenylate kinase
4 | 0.0042 | 0.4736 |
| APLN | Apelin | 0.0092 | 0.5261 |
| CKB | Creatine kinase,
brain | 0.0011 | 0.9570 |
| CSF2 | Colony stimulating
factor 2 (granulocyte-macrophage) | 0.0097 | 0.2979 |
| EBI3 | Epstein-Barr virus
induced 3 | 0.0048 | −0.7416 |
| ECE1 | Endothelin
converting enzyme 1 | 0.0088 | 0.1873 |
| FAM102A | Family with
sequence similarity 102, member A | 0.0057 | −0.2496 |
| GFRA2 | GDNF family
receptor α2 | 0.0068 | 0.7860 |
| GPM6B | Glycoprotein
M6B | 0.0064 | 0.7289 |
| HDC | Histidine
decarboxylase | 0.0045 | 0.9700 |
| KIT | V-Kit
Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 0.0018 | 0.6283 |
| LMO4 | LIM domain only
4 | 0.0037 | 0.3623 |
| MGST2 | Microsomal
glutathione S-transferase 2 | 0.0049 | −0.3100 |
| MRPL30 | Mitochondrial
ribosomal protein L30 | 0.0031 | 0.1626 |
| NSDHL | NAD(P) dependent
steroid dehydrogenase-like | 0.0082 | 0.3269 |
| POLR2I | Polymerase (RNA) II
(DNA directed) polypeptide I, 14.5kDa | 0.0053 | 0.2292 |
| RRM1 | Ribonucleotide
reductase catalytic subunit M1 | 0.0094 | 0.2653 |
| SCIN | Scinderin | 0.0022 | −0.9358 |
| WDR41 | WD repeat domain
41 | 0.0042 | 0.4693 |
PPI network construction and
analysis
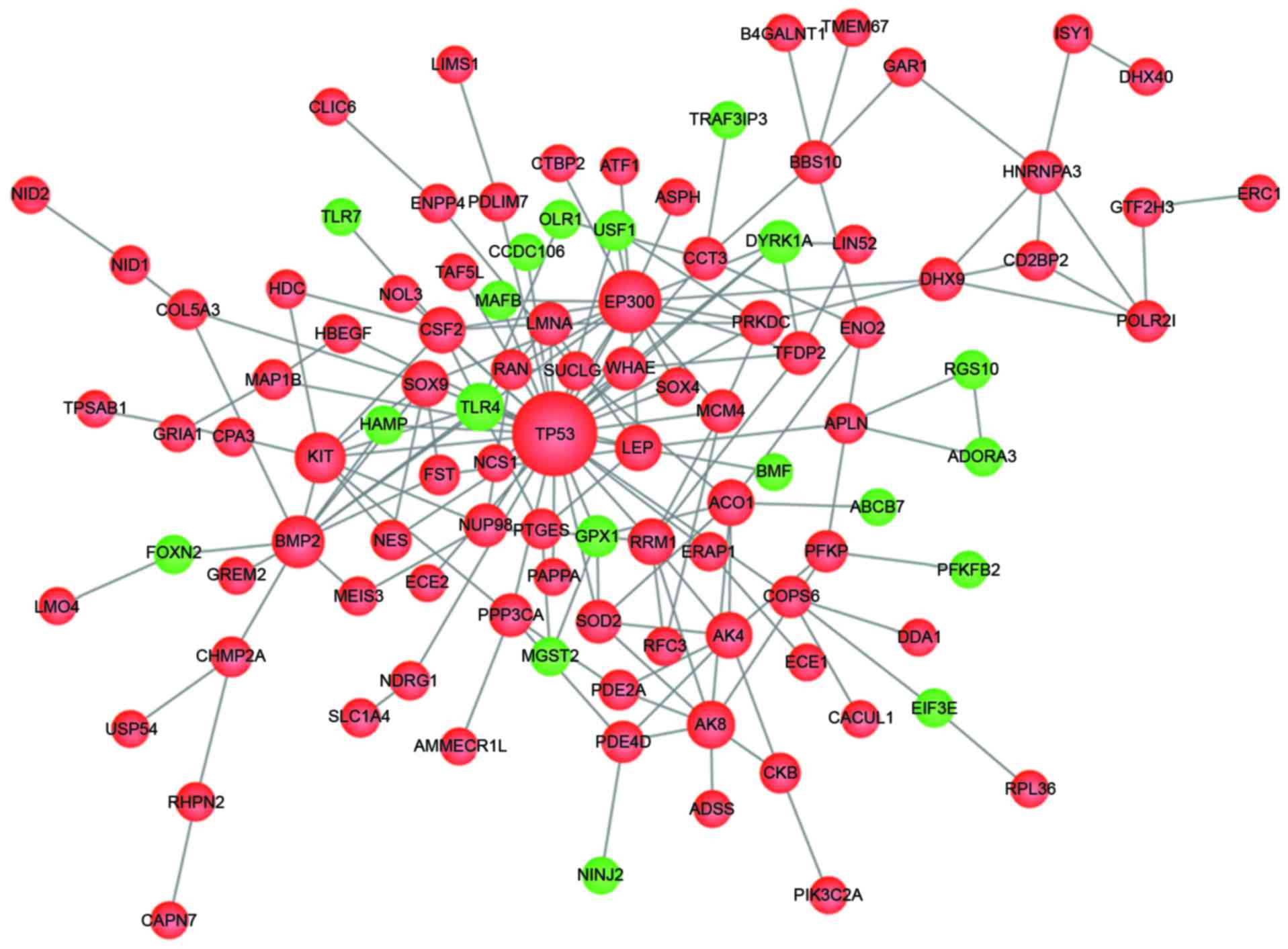
A total of 189 PPI pairs were obtained from the
STRING database. After wiping out the pairs isolated from the major
network, the PPI network composed of 172 edges and 105 nodes
(Fig. 4). In the network, nodes
represent genes and edges represent the interaction between genes.
The PPI network of DEGs was in a state of high aggregation, which
is an essential property of biological networks. With a threshold
degree ≥3, a total of 43 genes were selected (Table II). As shown in Fig. 4, tumor protein p53 (degree, 33) and
E1A binding protein P300 (degree, 18) had more degrees than the
other DEGs.
 | Table II.DEGs with degree ≥3. |
Table II.
DEGs with degree ≥3.
| Gene symbol | Gene name | P-value | logFC | Degree |
|---|
| CKB | creatine kinase
B | 0.0011 | 0.957 | 3 |
| KIT | KIT proto-oncogene
receptor tyrosine kinase | 0.0018 | 0.6283 | 10 |
| AK4 | adenylate kinase
4 | 0.0042 | 0.4736 | 7 |
| POLR2I | RNA polymerase II
subunit I | 0.0053 | 0.2292 | 4 |
| APLN | apelin | 0.0092 | 0.5261 | 3 |
| RRM1 | ribonucleotide
reductase catalytic subunit M1 | 0.0094 | 0.2653 | 7 |
| CSF2 | colony stimulating
factor 2 | 0.0097 | 0.2979 | 7 |
| ENO2 | enolase 2 | 0.0143 | 0.5576 | 4 |
| NUP98 | nucleoporin 98 | 0.0174 | 0.1405 | 5 |
| TLR4 | toll like receptor
4 | 0.0179 | −0.422 | 8 |
| NES | nestin | 0.0189 | 0.481 | 3 |
| CD2BP2 | CD2 cytoplasmic
tail binding protein 2 | 0.019 | 0.1659 | 3 |
| PPP3CA | protein phosphatase
3 catalytic subunit α | 0.019 | 0.2495 | 5 |
| COPS6 | COP9 signalosome
subunit 6 | 0.0196 | 0.2328 | 4 |
| FST | follistatin | 0.0199 | 0.7444 | 3 |
| EP300 | E1A binding protein
p300 | 0.0202 | 0.2351 | 18 |
| BBS10 | Bardet-Biedl
syndrome 10 | 0.0211 | 0.2222 | 5 |
| MAP1B | microtubule
associated protein 1B | 0.0215 | 0.1588 | 3 |
| HNRNPA3 | heterogeneous
nuclear ribonucleoprotein A3 | 0.022 | 0.2015 | 5 |
| PDE4D | phosphodiesterase
4D | 0.023 | 0.2095 | 4 |
| SOD2 | superoxide
dismutase 2, mitochondrial | 0.0236 | 0.3115 | 5 |
| PRKDC | protein kinase,
DNA-activated, catalytic polypeptide | 0.0252 | 0.3726 | 6 |
| TFDP2 | transcription
factor Dp-2 | 0.026 | 0.232 | 5 |
| GPX1 | glutathione
peroxidase 1 | 0.0261 | −0.7343 | 4 |
| PTGES | prostaglandin E
synthase | 0.0275 | 0.2743 | 3 |
| ACO1 | aconitase 1 | 0.0303 | 0.3555 | 7 |
| RAN | RAN, member RAS
oncogene family | 0.0306 | 0.3282 | 3 |
| COL5A3 | collagen type V α3
chain | 0.0316 | 0.1173 | 3 |
| USF1 | upstream
transcription factor 1 | 0.0332 | −0.4322 | 3 |
| SOX9 | SRY-box 9 | 0.0345 | 0.2414 | 7 |
| PDE2A | phosphodiesterase
2A | 0.0356 | 0.1566 | 3 |
| CHMP2A | charged
multivesicular body protein 2A | 0.0361 | 0.1974 | 3 |
| AK8 | adenylate kinase
8 | 0.0362 | 0.2411 | 8 |
| DHX9 | DEAH-box helicase
9 | 0.0367 | 0.1559 | 5 |
| LEP | leptin | 0.0373 | 0.7399 | 7 |
| TP53 | tumor protein
p53 | 0.038 | 0.1518 | 33 |
| YWHAE | tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
epsilon | 0.0383 | 0.1716 | 5 |
| MCM4 | minichromosome
maintenance complex component 4 | 0.0393 | 0.2495 | 5 |
| PFKP |
phosphofructokinase, platelet | 0.0395 | 0.3782 | 4 |
| LMNA | lamin A/C | 0.0446 | 0.4168 | 5 |
| CCT3 | chaperonin
containing TCP1 subunit 3 | 0.0446 | 0.2518 | 5 |
| BMP2 | bone morphogenetic
protein 2 | 0.0458 | 0.1583 | 11 |
| DYRK1A | dual specificity
tyrosine phosphorylation regulated kinase 1A | 0.0495 | −0.2047 | 5 |
GO functional analysis
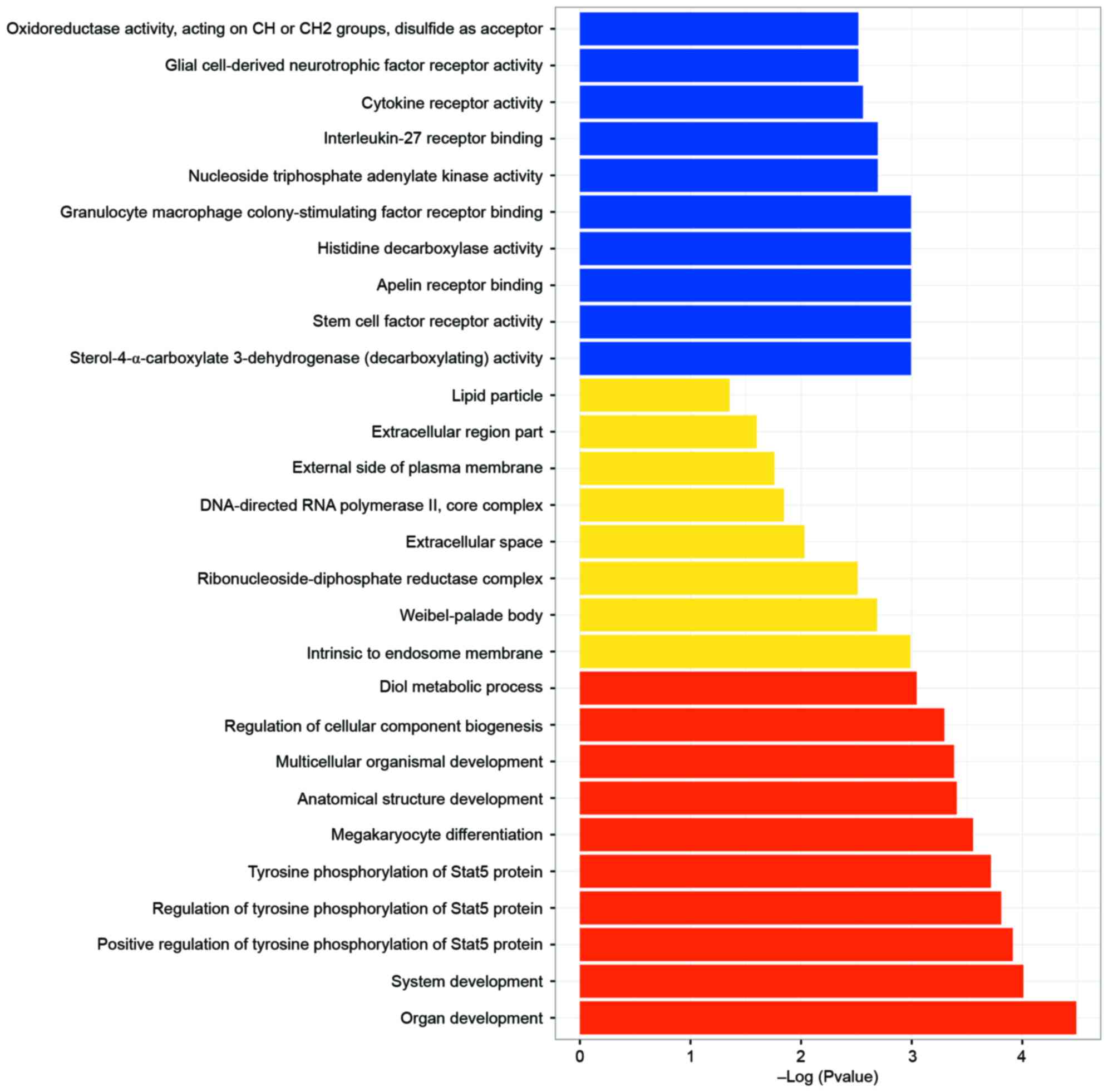
Using a threshold of P<0.05, a total of 401
significant GO terms were enriched and the top 10 enriched terms
for each category are presented in Fig. 5; only 8 terms were enriched in
cellular components. The most enriched GO terms of the DEGs were
mainly associated with tyrosine phosphorylation of signal
transducer and activator of transcription-5 (Stat5) protein, which
is closely associated with the activation of T cells.
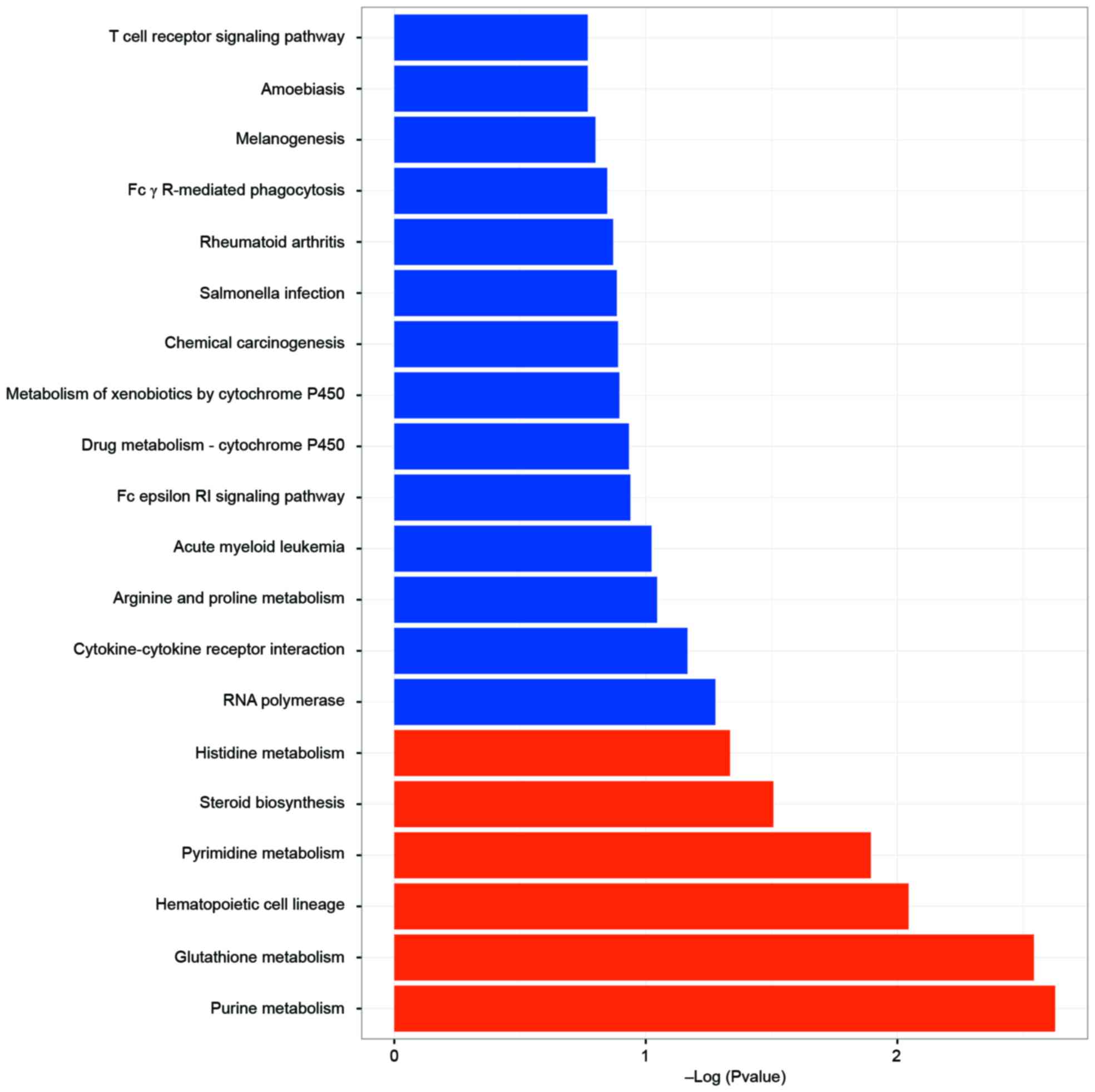
KEGG pathway enrichment analysis
Using a threshold of P<0.05, a total of 6
significant KEGG pathways were enriched (Fig. 6). The most enriched KEGG pathways
of the DEGs were mainly associated with the pharmacological
functioning of MMF in activated T cells, including purine
metabolism, glutathione metabolism and pyrimidine metabolism.
Discussion
The treatment of CAS includes three modalities:
Medical management, carotid artery angioplasty and stenting, and
CEA (26). However, there are
doubts regarding the efficacy of carotid artery angioplasty and
stenting, and CEA, in specific patients (27–29).
Previous studies have aimed to identify novel treatments and
medications for atherosclerosis. Following construction of the
hypercholesterolaemic apolipoprotein E-deficient murine model, Chen
et al (30) demonstrated
that preimplantation factor could prevent atherosclerosis via its
immunomodulatory effects without affecting serum lipids. In
addition, Sun et al (31)
developed trifunctional Simian virus 40 (SV40)-based nanoparticles
for in vivo targeting and imaging of atherosclerotic
plaques, and targeted SV40 virus-like nanoparticles were revealed
to deliver a greater concentration of the anticoagulant drug
Hirulog to atherosclerotic plaques.
It is well known that MMF is a T cell suppressor,
which is able to reduce synthesis of guanine via IMPDH inhibition,
resulting in the suppression of T cell proliferation (32). van Leuven et al (19) demonstrated that treatment with MMF
was able to reduce inflammatory cell infiltration, with a
concomitant decrease in proinflammatory gene expression in patients
with SCAS. The present study downloaded and analyzed a GEO mRNA
expression profile uploaded by van Leuven et al (19). A total of 210 DEGs were identified
between MMF-treated and placebo-treated groups. Subsequently, the
19 most significant DEGs were selected to undergo GO functional
analysis and KEGG pathway enrichment analysis. The results revealed
that the most enriched KEGG pathway was purine metabolism,
indicating that suppression of inflammatory activity served an
important role in the MMF treatment of patients with SCAS.
Furthermore, with a threshold of degree ≥3 and |logFC|≥0.5, three
genes, APLN, creatine kinase B (CKB) and KIT, were selected as the
key genes.
APLN is a peptide, which was initially identified by
Tatemoto et al in 1998 (33), that functions as an endogenous
ligand for the orphaned G-protein-coupled receptor (APJ) (34). Previous studies have revealed that
APJ deficiency can prevent oxidative stress-associated
atherosclerosis and that the APLN-APJ system is a mediator of
oxidative stress in vascular tissue (33,35–37).
In the present study, expression of APLN was significantly
different between MMF-treated and placebo-treated groups, and APLN
was a key gene in the PPI network, indicating that APLN may be a
target of MMF for the treatment of SCAS. According to the results
of the KEGG analysis, purine metabolism was the most enriched
signaling pathway, indicating that MMF modified the atherosclerotic
plaque by suppressing activated T cells. Recent studies have also
reported that APLN is involved in the immune response (38,39).
By analyzing the expression and function of the APLN-APJ system in
tumor vasculature, Kidoya et al (38) indicated that the APLN-APJ system
could induce maturation of tumor vasculature and improve the
efficiency of immune therapy. The results of the GO analysis
suggested that terms associated with Stat5 were the most enriched,
including tyrosine phosphorylation of Stat5 protein, regulation of
tyrosine phosphorylation of Stat5 protein and positive regulation
of tyrosine phosphorylation of Stat5 protein (Table III). Previous studies have
indicated that Stat5 has a strong association with T cells. Lindahl
et al (40) provided
evidence to suggest that microRNA-21 is expressed in situ in
cutaneous T cell lymphomas skin lesions, as induced by IL-2 and
IL-15 cytokines, and is regulated by Stat5 in malignant T cells.
The APLN-APJ system is also involved in the immune response and
Stat3. Han et al (41)
indicated that binding of phosphorylated-Stat3 to the APLN promoter
is the final step underlying proinflammatory cytokine-induced
enteric APLN expression during intestinal inflammation. However,
whether APLN is involved in the Stat5 signaling remains to be
elucidated. It may be hypothesized that APLN affects T cells
through the Stat5 signaling pathway; however, further studies are
required.
 | Table III.Most significantly enriched GO terms
(P<0.001). |
Table III.
Most significantly enriched GO terms
(P<0.001).
| GO ID | GO term | Associated
genes |
|---|
| GO:0048513 | Organ
development | AK4, APLN, CKB,
CSF2, ECE1, GPM6B, KIT, LMO4, NSDHL, SCIN |
| GO:0048731 | System
development | AK4, APLN, CKB,
CSF2, ECE1, GFRA2, GPM6B, KIT, LMO4, NSDHL, SCIN |
| GO:0042523 | Positive regulation
of tyrosine phosphorylation of Stat5 protein | CSF2, KIT |
| GO:0042522 | Regulation of
tyrosine phosphorylation of Stat5 protein | CSF2, KIT |
| GO:0042506 | Tyrosine
phosphorylation of Stat5 protein | CSF2, KIT |
| GO:0030219 | Megakaryocyte
differentiation | KIT, SCIN |
| GO:0048856 | Anatomical
structure development | AK4, APLN, CKB,
CSF2, ECE1, GFRA2, GPM6B, KIT, LMO4, NSDHL, SCIN |
| GO:0007275 | Multicellular
organismal development | AK4, APLN, CKB,
CSF2, ECE1, GFRA2, GPM6B, KIT, LMO4, NSDHL, SCIN |
| GO:0044087 | Regulation of
cellular component biogenesis | CSF2, GPM6B, LMO4,
SCIN |
| GO:0034311 | Diol metabolic
process | AK4, HDC |
Mast/stem cell growth factor receptor, also known as
proto-oncogene c-Kit, tyrosine-protein kinase Kit, or cluster of
differentiation 117 is a receptor tyrosine kinase protein that in
humans is encoded by the KIT gene (42). The GO analysis results revealed
that KIT was closely associated with tyrosine phosphorylation of
Stat5 protein, thus indicating that dysregulation of Stat5 may be
important in the process of SCAS.
Brain-type creatine kinase is a creatine kinase
encoded by the CKB gene in humans, which is associated with
creatine kinase activity and cellular monovalent inorganic anion
homeostasis (43). However, to the
best of our knowledge, the involvement of CKB in the process of
SCAS has not yet been reported.
In conclusion, APLN and KIT may serve important
roles in the MMF treatment of SCAS. The results of the present
study suggested that MMF may upregulate APLN to inhibit the
proliferation of T cells through the Stat5 signaling pathway.
Further investigation of the function of APLN and KIT in
atherosclerosis is urgently required.
Acknowledgements
The present study was supported by the Youth
Innovation Fund Projects of Inner Mongolia Medical University
(grant no. YKD2013QNCX023), the Health Department Medical
Scientific Research Projects of the Inner Mongolia Autonomous
Region (grant no. 201302089) and the Health Department Medical
Scientific Research Projects of the Inner Mongolia Autonomous
Region (grant no. 201302090).
References
|
1
|
Murphy SL, Xu J and Kochanek KD: Deaths:
Final data for 2010. National vital statistics reports: From the
Centers for Disease Control and Prevention, National Center for
Health Statistics, national vital statistics system. 61:1–117.
2013.PubMed/NCBI
|
|
2
|
Kochanek KD, Xu J, Murphy SL, Minino AM
and Kung HC: Deaths: Final data for 2009. National vital statistics
reports: From the Centers for Disease Control and Prevention,
National Center for Health Statistics, national vital statistics
system. 59:1–116. 2011.
|
|
3
|
Grotta JC: Carotid stenosis. N Engl J Med.
369:2360–2361. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mozaffarian D, Benjamin EJ, Go AS, Arnett
DK, Blaha MJ, Cushman M, de Ferranti S, Després JP, Fullerton HJ,
Howard VJ, et al: Heart disease and stroke statistics-2015 update:
a report from the American Heart Association. Circulation.
131:e29–322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sacco RL, Adams R, Albers G, Alberts MJ,
Benavente O, Furie K, Goldstein LB, Gorelick P, Halperin J,
Harbaugh R, et al: Guidelines for prevention of stroke in patients
with ischemic stroke or transient ischemic attack: A statement for
healthcare professionals from the American Heart
Association/American Stroke Association Council on Stroke:
Co-sponsored by the Council on Cardiovascular Radiology and
Intervention: The American Academy of Neurology affirms the value
of this guideline. Circulation. 37:e409–e449. 2006.
|
|
6
|
Kistler JP and Furie KL: Carotid
endarterectomy revisited. N Engl J Med. 342:1743–1745. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davies MJ and Thomas A: Thrombosis and
acute coronary-artery lesions in sudden cardiac ischemic death. N
Engl J Med. 310:1137–1140. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Z, Hou P, Fan D, Dong M, Ma M, Li H,
Yao R, Li Y, Wang G, Geng P, et al: The degradation of EZH2
mediated by lncRNA ANCR attenuated the invasion and metastasis of
breast cancer. Cell Death Differ. 24:59–71. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Libby P: Inflammation in atherosclerosis.
Arterioscler Thromb Vasc Biol. 32:2045–2051. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rocha VZ and Libby P: Obesity,
inflammation, and atherosclerosis. Nat Rev Cardiol. 6:399–409.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sparrow CP and Olszewski J: Cellular
oxidation of low density lipoprotein is caused by thiol production
in media containing transition metal ions. J Lipid Res.
34:1219–1228. 1993.PubMed/NCBI
|
|
12
|
Topper JN and Gimbrone MA Jr: Blood flow
and vascular gene expression: Fluid shear stress as a modulator of
endothelial phenotype. Mol Med Today. 5:40–46. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
De Caterina R, Libby P, Peng HB,
Thannickal VJ, Rajavashisth TB, Gimbrone MA Jr, Shin WS and Liao
JK: Nitric oxide decreases cytokine-induced endothelial activation.
Nitric oxide selectively reduces endothelial expression of adhesion
molecules and proinflammatory cytokines. J Clin Invest. 96:60–68.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Castellani ML, Anogeianaki A, Felaco P,
Toniato E, De Lutiis MA, Shaik B, Fulcheri M, Vecchiet J, Tetè S,
Salini V, et al: IL-35, an anti-inflammatory cytokine which expands
CD4+CD25+ Treg Cells. J Biol Regul Homeost Agents. 24:131–135.
2010.PubMed/NCBI
|
|
15
|
Oršolić N: Bee venom in cancer therapy.
Cancer Metastasis Rev. 31:173–194. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma Y, Liu X, Wei Z, Wang X, Xu D, Dai S,
Li Y, Gao M, Ji C, Guo C, et al: The expression of a novel
anti-inflammatory cytokine IL-35 and its possible significance in
childhood asthma. Immunol Lett. 162:11–17. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang Y, Lin YZ, Shi Y and Ji QW: IL-35: A
potential target for the treatment of atherosclerosis. Pharmazie.
68:793–795. 2013.PubMed/NCBI
|
|
18
|
Allison AC and Eugui EM: Mechanisms of
action of mycophenolate mofetil in preventing acute and chronic
allograft rejection. Transplantation. 80 2 Suppl:S181–S190. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Leuven SI, van Wijk DF, Volger OL, de
Vries JP, Van Der Loos CM, de Kleijn DV, Horrevoets AJ, Tak PP, Van
Der Wal AC, de Boer OJ, et al: Mycophenolate mofetil attenuates
plaque inflammation in patients with symptomatic carotid artery
stenosis. Atherosclerosis. 211:231–236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
25
|
Hosack DA, Dennis G Jr, Sherman BT, Lane
HC and Lempicki RA: Identifying biological themes within lists of
genes with EASE. Genome Biol. 4:R702003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bae C, Szuchmacher M and Chang JB:
Comparative review of the treatment methodologies of carotid
stenosis. Int J Angiol. 24:215–222. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tsukahara T, Hatano T, Nakakuki T, Tsuji
Y, Aoyama T and Ogata H: Combined treatment using CEA and CAS for
carotid arterial stenosis. Acta Neurochir Suppl. 103:109–112. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
De Borst GJ: Recruiting RCTs comparing
CAS, CEA and best medical treatment for asymptomatic carotid
stenosis. J Cardiovasc Surg (Torino). 56:837–844. 2015.PubMed/NCBI
|
|
29
|
Lal BK: Recurrent carotid stenosis after
CEA and CAS: Diagnosis and management. Semin Vasc Surg. 20:259–266.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen YC, Rivera J, Fitzgerald M, Hausding
C, Ying YL, Wang X, Todorova K, Hayrabedyan S, Barnea ER and Peter
K: PreImplantation factor prevents atherosclerosis via its
immunomodulatory effects without affecting serum lipids. Thromb
Haemost. 115:1010–1024. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun X, Li W, Zhang X, Qi M, Zhang Z, Zhang
XE and Cui Z: In Vivo targeting and imaging of atherosclerosis
using multifunctional virus-like particles of simian virus 40. Nano
Lett. 16:6164–6171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Allison AC and Eugui EM: Purine metabolism
and immunosuppressive effects of mycophenolate mofetil (MMF). Clin
Transplant. 10:77–84. 1996.PubMed/NCBI
|
|
33
|
Tatemoto K, Hosoya M, Habata Y, Fujii R,
Kakegawa T, Zou MX, Kawamata Y, Fukusumi S, Hinuma S, Kitada C, et
al: Isolation and characterization of a novel endogenous peptide
ligand for the human APJ receptor. Biochem Biophys Res Commun.
251:471–476. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Falcão-Pires I and Leite-Moreira AF:
Apelin: A novel neurohumoral modulator of the cardiovascular
system. Pathophysiologic importance and potential use as a
therapeutic target. Rev Port Cardiol. 24:1263–1276. 2005.PubMed/NCBI
|
|
35
|
Kleinz MJ, Skepper JN and Davenport AP:
Immunocytochemical localisation of the apelin receptor, APJ, to
human cardiomyocytes, vascular smooth muscle and endothelial cells.
Regul Pept. 126:233–240. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kleinz MJ and Davenport AP:
Immunocytochemical localization of the endogenous vasoactive
peptide apelin to human vascular and endocardial endothelial cells.
Regul Pept. 118:119–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hashimoto T, Kihara M, Imai N, Yoshida S,
Shimoyamada H, Yasuzaki H, Ishida J, Toya Y, Kiuchi Y, Hirawa N, et
al: Requirement of apelin-apelin receptor system for oxidative
stress-linked atherosclerosis. Am J Pathol. 171:1705–1712. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kidoya H, Kunii N, Naito H, Muramatsu F,
Okamoto Y, Nakayama T and Takakura N: The apelin/APJ system induces
maturation of the tumor vasculature and improves the efficiency of
immune therapy. Oncogene. 31:3254–3264. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Adam F, Khatib AM, Lopez JJ, Vatier C,
Turpin S, Muscat A, Soulet F, Aries A, Jardin I, Bobe R, et al:
Apelin: An antithrombotic factor that inhibits platelet function.
Blood. 127:908–920. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lindahl LM, Fredholm S, Joseph C, Nielsen
BS, Jønson L, Willerslev-Olsen A, Gluud M, Blümel E, Petersen DL,
Sibbesen N, et al: STAT5 induces miR-21 expression in cutaneous T
cell lymphoma. Oncotarget. 7:45730–45744. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han S, Wang G, Qi X, Englander EW and
Greeley GH Jr: Involvement of a Stat3 binding site in
inflammation-induced enteric apelin expression. Am J Physiol
Gastrointest Liver Physiol. 295:G1068–G1078. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Andre C, Hampe A, Lachaume P, Martin E,
Wang XP, Manus V, Hu WX and Galibert F: Sequence analysis of two
genomic regions containing the KIT and the FMS receptor tyrosine
kinase genes. Genomics. 39:216–226. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mariman EC, Schepens JT and Wieringa B:
Complete nucleotide sequence of the human creatine kinase B gene.
Nucleic Acids Res. 17:63851989. View Article : Google Scholar : PubMed/NCBI
|