Introduction
Esophageal cancer refers to upper gastrointestinal
tract tumors of epithelial cellular origin. It is the 6th most
common cause of cancer-associated mortality, and the eighth most
common malignancy worldwide (1,2). In
China, it is the 5th most common cancer and the 4th most common
cause of cancer-associated mortality (3,4).
Early detection of the disease is essential to improve the survival
of patients with esophageal cancer.
MicroRNAs (miRNAs/miRs) are endogenous, single-stran
ded non-coding RNAs with 19–25 nucleotides, acting as important
mediators in the regulation of gene expression, cell
differentiation, the cell cycle and apoptosis (5,6).
miRNA microarray profiling of human tumors has demonstrated that
certain groups of miRNAs may offer opportunities in the
identification of novel biomarkers and therapeutic targets
(7,8). A number of miRNAs, including miR-21,
miR-373 and miR-483, have been reported to be potential biomarkers
of esophageal cancer. miR-483 is an intronic miRNA located within
the insulin like growth factor 2 (Igf2) gene locus in mammalian
cells (9), and has been associated
with a diverse set of human pathologies, including cancer (10–12).
A polymorphism at the miR-483-5p binding site in the
3′-untranslated region of the basigin gene has been demonstrated to
be associated with increased susceptibility to esophageal cancer in
a Chinese population (13).
However, to the best of the authors' knowledge, there has been no
study regarding the mechanism underlying the regulatory role of
miR-483-5p in the development of esophageal cancer.
Epigenetic alterations have been a subject of
research, due to their involvement in malignant transformation and
tumor progression. There has been an increase in basic and applied
research into the field of the epigenetic regulation of esophageal
cancer development (14–16). The aim of the present study was to
clarify the association between miR-483-5p expression in serum and
tissues from patient with esophageal cancer, with epigenetic
alterations in the Igf2 promoter, in addition to the effect of
imiR-483-5p on target gene expression.
Materials and methods
Patients
The study protocol was approved by the Ethics
Committee of the Xinxiang Central Hospital (Xinxiang, China). A
total of 23 patients (the age range, 25–60 years; median age, 46;
17 males and 6 females.) with esophageal squamous cell carcinoma
(ESCC) from the Xinxiang Central Hospital and 50 healthy subjects
were recruited to the present study between January 2014 and
February 2015. All participants were genetically unrelated ethnic
Han Chinese from the same geographic region (Henan, China). The
diagnosis of ESCC was confirmed by histopathology in all patients.
Written informed consent was obtained from all participants prior
to the study. ESCC tissues and adjacent non-cancerous esophageal
tissues (at least 5 cm away from the tumor) from all 23 patients
were collected. A total of 3 ml peripheral blood was collected from
each participant (including 50 healthy persons and 23 patients with
ESCC; patient blood samples were collected prior to surgery and at
7 days post-surgery).
Tissue and serum sample processing and
RNA isolation
All tissue samples were collected during surgery,
immediately snap-frozen in liquid nitrogen, and stored at −80°C
until RNA extraction. Total RNA was isolated using
TRIzol™ (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to the manufacturer's
instructions.
Peripheral blood was collected in tubes containing
separating gel and clot activator, placed in a water bath for 20
min at 37°C, and centrifuged at 3,500 × g for 10 min at room
temperature. The supernatants were transferred to Eppendorf tubes.
A second centrifugation at 12,000 × g for 10 min at 4°C was
performed to completely remove all cellular components. The serum
was subsequently aliquoted and stored at −80°C until RNA
extraction. All blood samples were processed within 3 h following
collection. Total serum RNA was isolated from 100 µl serum and
eluted in 300 µl RNase-free water using TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
qPCR for individual miRNAs was performed on
independent sets of serum or tissue using a two-step procedure.
qPCR for miRNA Stem-Loop™ RT primers for miR-483-5p and
miR-16-5p were synthesized by Applied Biosystems (Thermo Fisher
Scientific, Inc.) (Table I). A
PrimeScript™ RT reagent kit (Perfect Real Time; Takara
Biotechnology Co., Ltd., Dalian, China) was used to reverse
transcribe the total RNA. A SYBR Green (Takara Biotechnology Co.,
Ltd.) qPCR assay kit was used to detect the expression of
miR-483-5p and miR-16-5p. The qPCR reaction was performed over 45
cycles (95°C, 10 sec; 60°C, 30 sec) following an initial
denaturation step (95°C, 5 min) on the CFX96 system using Bio-Rad
CFX Manager 2.0 Software (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The expression levels of miRNA were calculated and quantified
using the 2−ΔΔCq method (17). miR-16-5p was used as the internal
control. All reactions were performed in triplicate.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer | Sequence |
|---|
| miR-483-5p-RT |
GTCGTATCCATGGCAGGGTCCGAG |
|
|
GTATTCGCCATGGATACGACCTCCCT |
| miR-483-5p-F |
GCAAGACGGGAGGAAAGAAGGGA |
| universal
reverse | TGGCAGGGTCCGAGGT |
| GAPDH-F |
GCACCGTCAAGGCTGAGAAC |
| GAPDH-R |
TGGTGAAGACGCCAGTGGA |
| Socs3-F |
CAGGAATGTAGCAGCGATGGAA |
| Socs3-R |
CCTGTCCAGCCCAATACCTGA |
| ALCAM-F |
CCTTGTTGCTGGTGTCGTCTACT |
| ALCAM-R |
ATTACCGAGGTCCTTGTTTACATGT |
| ARHGDIA-F |
AACCGAGAGATAGTGTCCGGC |
| ARHGDIA-R |
TCTTGACGCCTTTCCTGTACG |
| Igf2-F |
CCGTGCTTCCGGACAACTT |
| Igf2-R |
CTGCTTCCAGGTGTCATATTGG |
| miR-16-RT |
GTCGTATCCATGGCAGGGTCCGAGGT |
|
|
ATTCGCCATGGATACGACCGCCAAT |
| miR-16-F |
GCGGTAGCAGCACGTAAATATT |
| Igf2-MF3 |
AGCGGTTTCGGTGTCGTTATC |
| Igf2-MR3 |
CGAACGCCCAACTCGATT |
| Igf2-UF3 |
GGATTGTGGGTGTTTAGTTTGGTT |
| Igf2-UR3 |
CCTTTCCACACTACATCCCAAAA |
Prediction of miR-483-5p target
genes
miR-483-5p target genes were predicted using miRBase
(www.mirbase.org), Target Scan (www.targetscan.org), and PicTar (pictar.mdc-berlin.de).
Genomic DNA isolation and methylation
analysis
Genomic DNA was extracted using an EZ DNA
Methylation-Gold™ kit (Qiagen GmbH, Hilden, Germany).
The methylation status of the Igf2 gene was determined using the
methylation-specific PCR (MSP) method on bisulfate-treated genomic
DNA. The primers specific for either unmethylated or methylated
alleles are listed in Table I. As
an internal control, all purified genomic DNA samples were
successfully tested by PCR with a Takara EpiTaq™ HS kit
(for bisulfite-treated DNA; Takara Biotechnology Co., Ltd.).
Methylated and unmethylated DNAs of normal human peripheral
lymphocytes were used as a positive control for the methylated and
as a negative control for unmethylated genes, respectively. Samples
with H2O2 instead of DNA were included for
each PCR set. PCR products were analyzed on a 1% agarose gel,
stained with ethidium bromide, and visualized under ultraviolet
light (DL 2000 Marker; Genstar, Beijing, China). Each MSP was
repeated at least once to confirm the results.
Statistical analysis
Data were reported as mean ± standard deviation for
quantitative variables. The difference in mRNA or miRNA expression
levels between paired tissue samples was calculated using the
Wilcoxon matched-pairs test. Correlations between independent
samplings and qPCR analysis of Igf2 and miRNA were determined by
the Spearman correlation test. The Mann-Whitney test was performed
to determine the significance of serum miRNA levels. The area under
the curve (AUC) for tissue and serum miRNAs was determined using
receiver operator characteristic (ROC) analysis. P<0.05 was
considered to indicate a statistically significant difference. The
statistical analysis was performed using SPSS 16.0 software (SPSS,
Inc., Chicago, IL, USA).
Results
Analysis of the expression levels of
miR-483-5p in ESCC
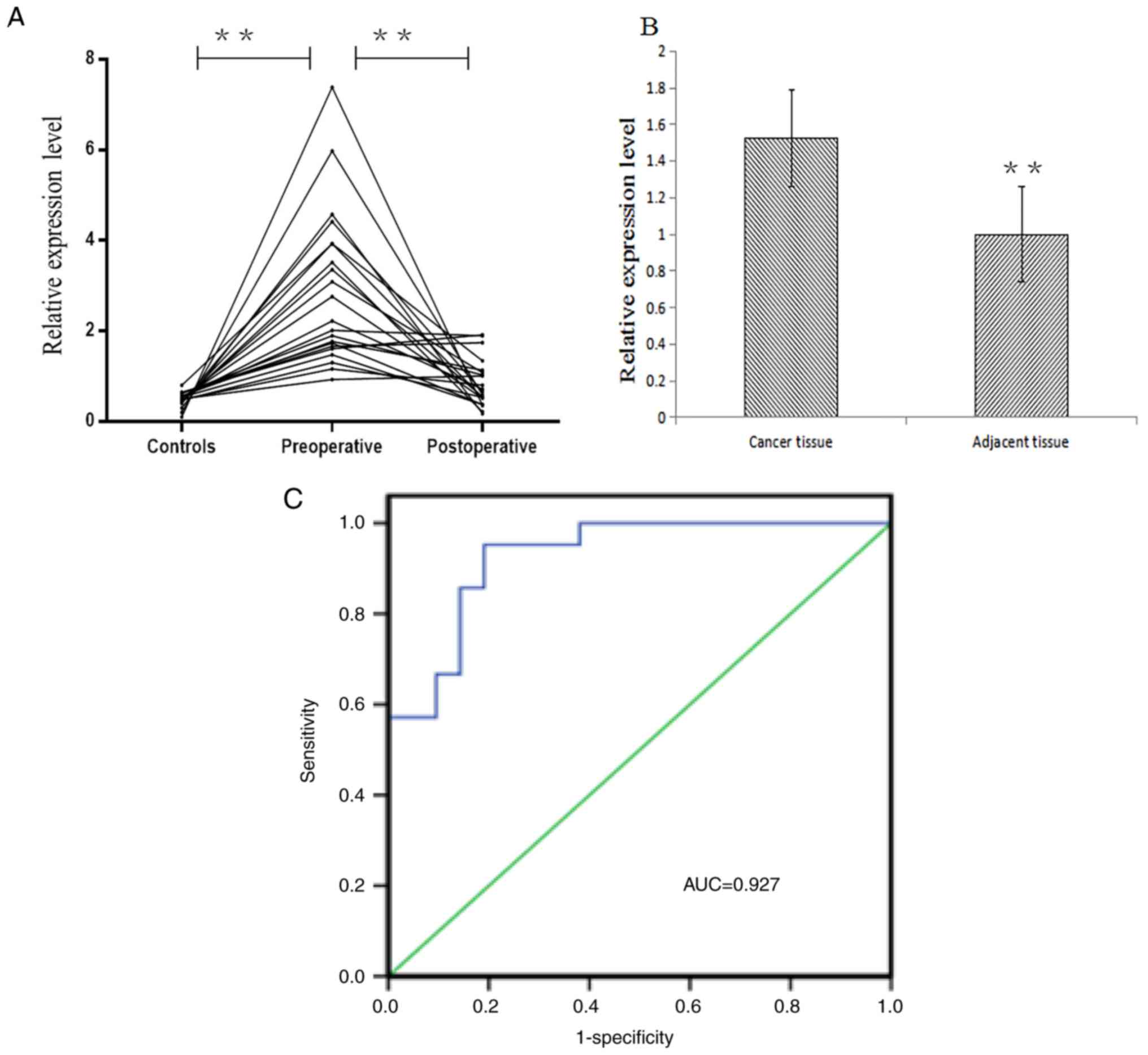
To investigate the role of miR-483-5p in ESCC, the
miR-483-5p expression levels were evaluated in the serum from
patients with ESCC patients prior to and following surgery, and
from ESCC-free subjects. The results demonstrated that miR-483-5p
was highly expressed in the serum prior to surgery in patients with
ESCC, which was significantly increased compared with those
following surgery in patients with ESCC and normal subjects
(P<0.05; Fig. 1A).
The expression level of miR-483-5p in cancer tissues
of patients with ESCC was significantly increased compared with
those in adjacent non-cancerous tissues (P<0.01), and the
difference between cancer tissues was similar to that between the
serum samples (Fig. 1B). The
expression level of miR-483-5p was positively correlated with the
clinical tumor, node, metastasis (TNM) staging of patients with
ESCC (P<0.05), and with the degree of lymph node metastasis
(P<0.05; Table II).
 | Table II.Correlation of miR-483-5p with
clinical features in patients with esophageal squamous cell
carcinoma. |
Table II.
Correlation of miR-483-5p with
clinical features in patients with esophageal squamous cell
carcinoma.
|
|
| miR-483-5p |
|---|
|
|
|
|
|---|
| Characteristic | No. cases | % | P-value |
|---|
| TNM stage |
|
| 0.033 |
| I | 3 | 11.5 |
|
| II | 8 | 30.8 |
|
| III | 13 | 50.0 |
|
| IV | 2 | 7.7 |
|
| Lymph node
metastasis |
|
| 0.048 |
| No | 11 | 42.3 |
|
|
Yes | 15 | 57.7 |
|
In order to analyze the diagnostic potential of
serum miRNA in ESCC, the ROC curve and AUC value were analyzed to
further assess the reliability of serum miR-483-5p expression
levels examined by the qPCR. ROC curve analysis demonstrated that
the AUC value of miR-483-5p was 0.927 (95% confidence interval,
0.85–1.00). When the threshold value was 0.762, the sensitivity was
95.2% and the specificity was 81% (Fig. 1C). AUC may be used as an indicator
for the accurate evaluation of certain diagnostic methods, and used
for clinical diagnostic tests. A larger AUC value indicates a
greater diagnostic value. The closer the AUC is to 1, the higher
its accuracy is, suggesting that serum miR-483-5p may be considered
to be a diagnostic marker in ESCC.
Analysis of Igf2 gene expression and
promoter region methylation
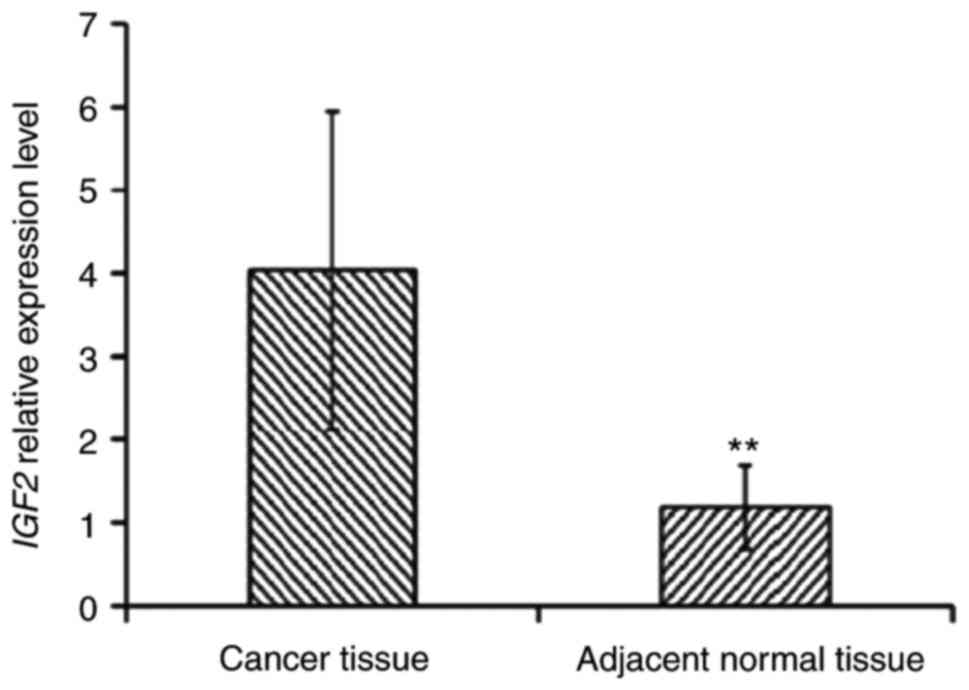
It has been reported in the literature that miR-483
is located in the second intron of Igf2, and that the expression
levels of Igf2 directly affect the expression of miR-483.
Therefore, the expression of the Igf2 gene and the methylation
levels in its promoter region were examined. The experimental
results demonstrated that the expression levels of Igf2 in cancer
tissues of patients with ESCC were significantly increased compared
with those in paracancerous tissues (P<0.01; Fig. 2). The methylation level of the Igf2
promoter region was decreased in tumor tissues (31.82%) compared
with adjacent non-cancerous tissues (54.55%; Fig. 3).
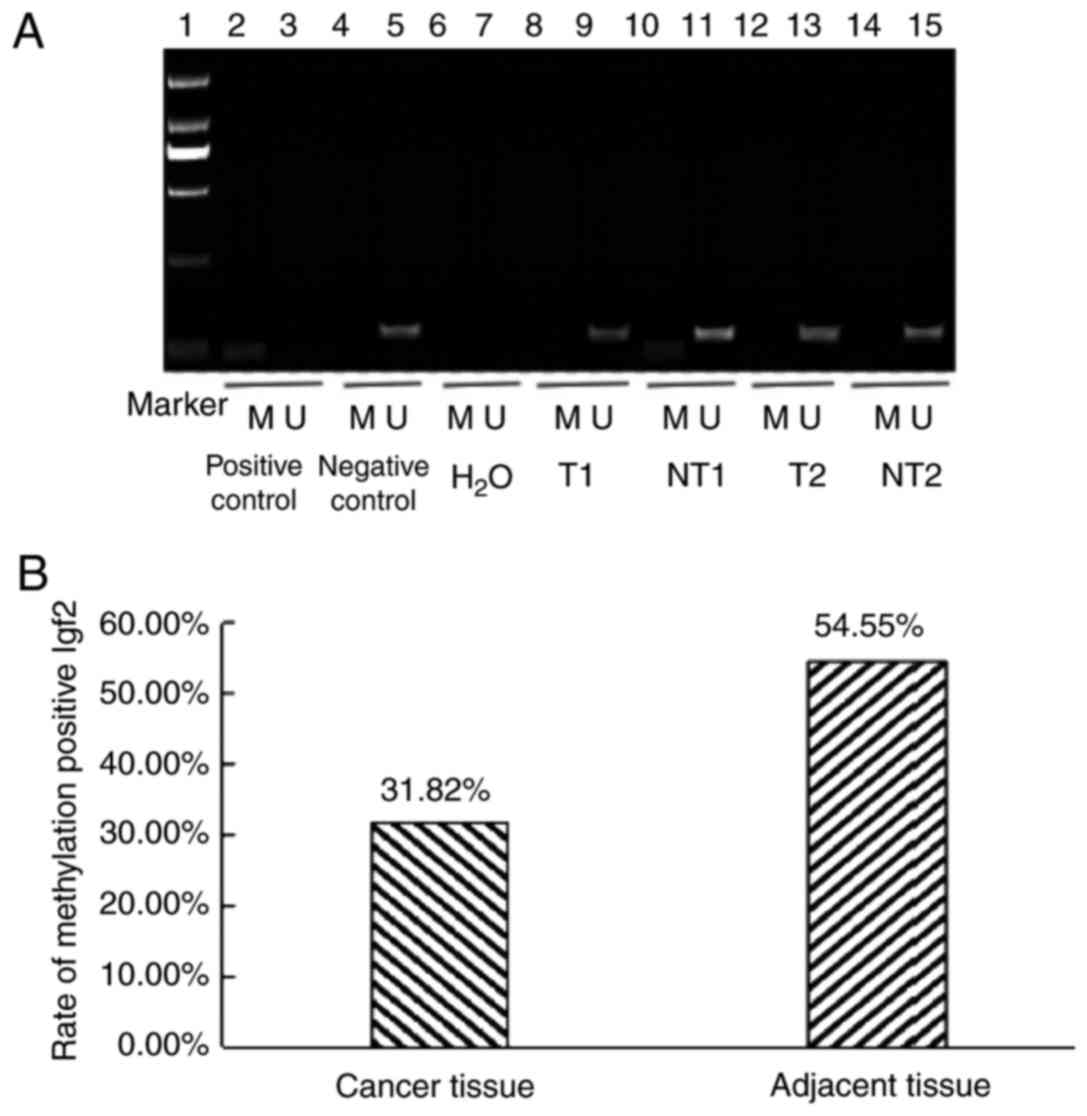 | Figure 3.DNA promoter methylation analysis. (A)
Detection of methylation status of the IGF2 gene promoter region
(Marker DL 2,000). Lane 1, marker (DL 2,000); lane 2, positive
control of DNA methylation in peripheral blood lymphocytes from
healthy people; lane 3, negative control of DNA methylation in
peripheral blood lymphocytes from healthy people; lane 4, positive
control of DNA nonmethylation in peripheral blood lymphocytes from
healthy people; lane 5, negative control of DNA nonmethylation in
peripheral blood lymphocytes from healthy people; lane 6, distilled
water as the negative control of the methylated template; lane 7,
distilled water as a template for the negative control of
nonmethylation; lane 8, methylation of cancer tissue; lane 9,
nonmethylation of cancer tissue; lane 10, methylation of adjacent
normal tissue; lane 11, nonmethylation of adjacent normal tissue;
lane 12, methylation of cancer tissue; lane 13, nonmethylation of
cancer tissue; lane 14, methylation of adjacent normal tissue; and
lane 15, nonmethylation of adjacent normal tissue. (B) Methylation
positive rate of the IGF2 gene in cancer tissue and adjacent normal
tissue from patients with esophageal squamous cell carcinoma. IGF2,
insulin-like growth factor 2; M, methylated; U, unmethylated; T,
tumor tissues; NT, normal tissues. |
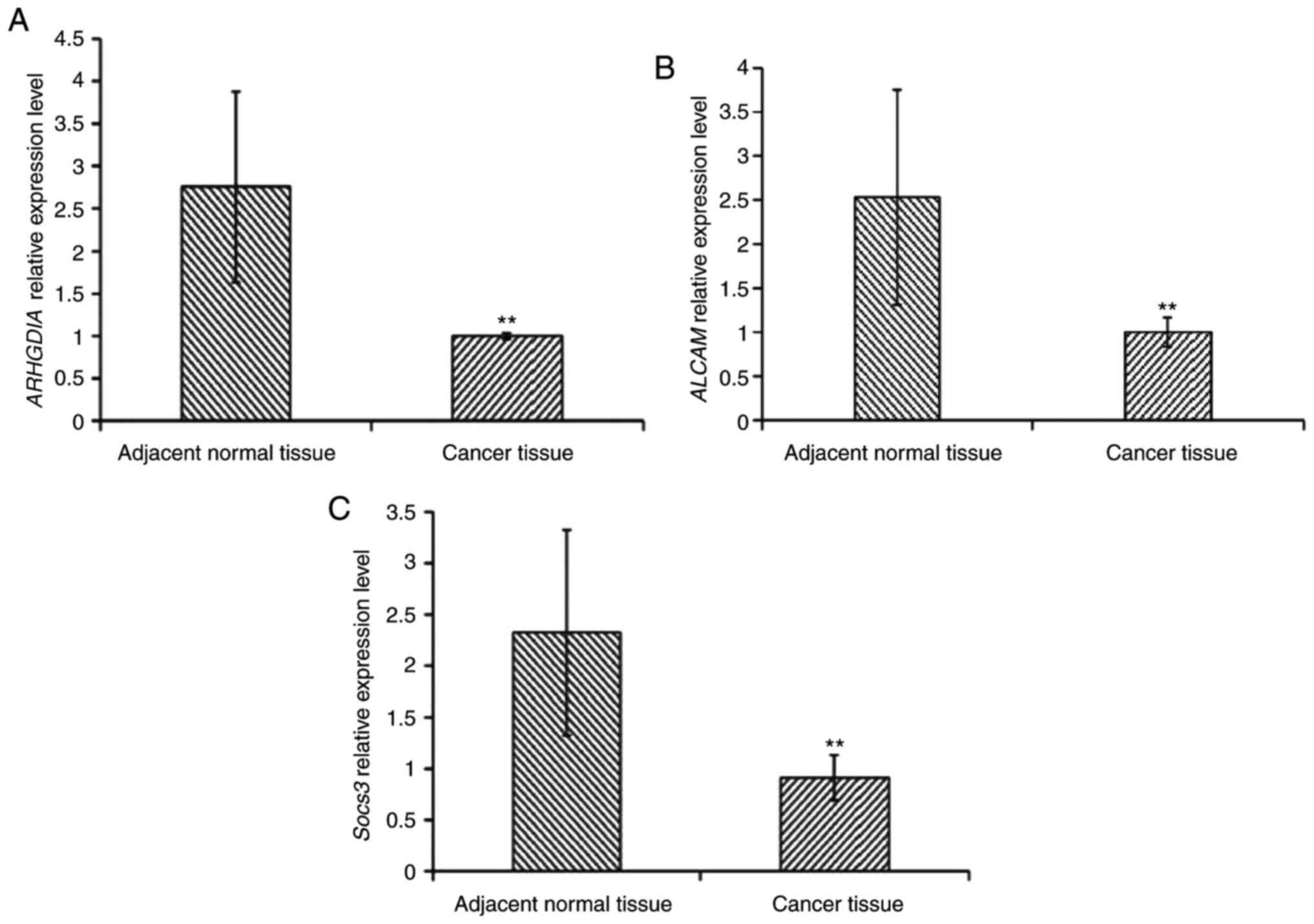
Analysis of the expression of
miR-483-5p target genes
miRNAs exert their functions primarily by affecting
the expression of their target genes. A bioinformatical analysis
was performed for the target genes of miR-483-5p (Fig. 4). A total of three miR-483-5p
target genes including Rho GDP dissociation inhibitor α (ARHGDIA),
activated leukocyte cell adhesion molecule (ALCAM) and suppressor
of cytokine signaling 3 (Socs3) were selected for further analysis.
The results demonstrated that the expression levels of the three
target genes in cancer tissues was significantly decreased compared
with adjacent non-cancerous tissues, implying that miR-483-5p may
influence the expression of these genes (Fig. 5).
Discussion
The study of miRNAs has been extended into numerous
types of tumor. The expression of miR-483 has been demonstrated to
be upregulated in approximately one-half of human tumors (18), including adrenocortical carcinoma
and hepatocellular carcinoma (11,19),
and its oncogenic targets, including cellular tumor antigen p53,
BCL2 binding component 3, catenin β1, and insulin-like growth
factor 1 receptor have been identified (20).
The degree of methylation of the promoter region
affects the regional DNA structure and influences gene
transcription. The results of the present study demonstrated that
the differences in Igf2 promoter methylation resulted in the
differential expression of Igf2 between cancer tissues and
paracancerous tissues in patients with ESCC. The methylation level
of the Igf2 promoter region in cancer tissues was low, although
Igf2 gene expression was increased. The methylation of the promoter
region of Igf2 in adjacent non-cancerous tissues was high, while
the Igf2 expression level was decreased. miR-483-5p is coexpressed
with Igf2 (21); thus, the
expression of miR-483-5p is enhanced when the expression of Igf2 is
increased. Therefore, the extent of methylation in the host gene
promoter region influences miRNA expression, indicating that
epigenetic modification serves an important role in the regulation
of miRNA expression.
It has been demonstrated that miRNAs are able to
bind to their complementary mRNA sites through base-pairing to
regulate gene expression (22).
Each miRNA has hundreds of evolutionarily conserved or
non-conservative target genes. Therefore, appraisal of the miRNA
target genes has become a challenge. In the present study, the mRNA
levels of miR-483-5p target genes, including ARHGDIA, ALCAM and
Socs3, were detected, which demonstrated that the levels of these
genes were negatively-associated with the expression of miR-483-5p.
However, the expression of these genes was low in cancer tissues,
and high in adjacent non-cancerous tissues, suggesting that
miR-483-5p may mediate its potential the expression of the target
genes ARHGDIA, ALCAM and Socs3, resulting in decreased expression
in ESCC.
A recent study demonstrated that ARHGDIA may be a
candidate tumor suppressor, and that it was downregulated in
hepatoma and mammary cancer (23).
Downregulation of ARHGDIA may reverse the activity of Rac family
small GTPase 1 and cell division cycle 42, and increase cell
migration and invasion to promote tumor metastasis (24). In the present study, the expression
levels of miR-483-5p correlated with TNM stage and lymph node
metastasis, suggesting that miR-483-5p may promote the development
of ESCC by downregulating the target gene ARHGDIA.
ALCAM is involved in homotypic or heterotypic
cellular adsorption. The expression levels of ALCAM vary in
distinct cancer tissues or at distinct stages of tumor progression
(25–27). Olson et al (18) reported a negative correlation
between ALCAM levels and the degree of tumor malignancy, and ALCAM
expression is elevated in early ESCC (25). Therefore, the reduction of ALCAM
expression may be due to the fact that the majority of samples in
the present study were advanced ESCC.
The Socs3 gene belongs to the cytokine signaling
inhibitor protein family. The Socs3 protein is able to negatively
regulate the signaling processes of insulin and a number of
cytokines to regulate immune reactions, inflammation and lymphocyte
differentiation (28). Similarly,
miR-483 negatively regulates the target gene Socs3 to regulate
liver cancer cell proliferation and development (29). The results of the present study
demonstrated that miR-483-5p exhibited high expression, although
Socs3 exhibited low expression, in ESCC cancer tissues, indicating
that Socs3 may serve a role in ESCC pathogenesis.
In conclusion, the results of the present study
suggested that miR-483-5p may be involved in ESCC pathogenesis, and
that low methylation of the Igf2 gene promoter region led to
increased expression of Igf2 and miR-483-5p in ESCC. As a result,
the decrease in the ARHGDIA, ALCAM and Socs3 expression levels may
cause the upregulation of oncogenes and downregulation of tumor
suppressors, thereby inducing ESCC. Further studies are required to
investigate the detailed mechanism and function of miR-483-5p in
ESCC.
Acknowledgements
The present study was supported by the Key Projects
of Science and Technology in Henan Province (grant no.
172102310407) and the Key Research Projects of Henan Higher
Education Institutions (grant no. 16A180028).
References
|
1
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wen SW, Zhang YF, Li Y, Liu ZX, Lv HL, Li
ZH, Xu YZ, Zhu YG and Tian ZQ: Characterization and effects of
miR-21 expression in esophageal cancer. Genet Mol Res.
14:8810–8818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, He Y, Zheng R, Zhang S, Zeng H,
Zou X and He J: Esophageal cancer incidence and mortality in China,
2009. J Thorac Dis. 5:19–26. 2013.PubMed/NCBI
|
|
4
|
Peng JZ, Xue L, Liu DG and Lin YH:
Association of the p53 Arg72Pro polymorphism with esophageal cancer
in Chinese populations: A meta-analysis. Genet Mol Res.
14:9024–9033. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garzon R and Croce CM: MicroRNAs and
cancer: Introduction. Semin Oncol. 38:721–723. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mulrane L, Klinger R, McGee SF, Gallagher
WM and O'Connor DP: microRNAs: A new class of breast cancer
biomarkers. Expert Rev Mol Diagn. 14:347–363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma J and Li X: MicroRNAs are involved in
the toxicity of microcystins. Toxin Rev. 36:165–175. 2017.
View Article : Google Scholar
|
|
9
|
Fu H, Tie Y, Xu C, Zhang Z, Zhu J, Shi Y,
Jiang H, Sun Z and Zheng X: Identification of human fetal liver
miRNAs by a novel method. FEBS Lett. 579:3849–3854. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
De-Ugarte L, Yoskovitz G, Balcells S,
Güerri-Fernández R, Martinez-Diaz S, Mellibovsky L, Urreizti R,
Nogués X, Grinberg D, García-Giralt N and Díez-Pérez A: miRNA
profiling of whole trabecular bone: Identification of
osteoporosis-related changes in miRNAs in human hip bones. BMC Med
Genomics. 8:752015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Veronese A, Lupini L, Consiglio J, Visone
R, Ferracin M, Fornari F, Zanesi N, Alder H, D'Elia G, Gramantieri
L, et al: Oncogenic role of miR-483-3p at the IGF2/483 locus.
Cancer Res. 70:3140–3149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang C, Sun Y, Wu H, Zhao D and Chen J:
Distinguishing adrenal corticalcarcinomas and adenomas: A study of
clinicopathological features and biomarkers. Histopathology.
64:567–576. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li HY, Liu YC, Bai YH, Sun M, Wang L,
Zhang XB and Cai B: SNP at miR-483-5p-binding site in the
3′-untranslated region of the BSG gene is associated with
susceptibility to esophageal cancer in a Chinese population. Genet
Mol Res. 15:1–10. 2016.
|
|
14
|
Baba Y, Watanabe M and Baba H: A review of
the alterations in DNA methylation in esophageal squamous cell
carcinoma. Surgery today. 43:1355–1364. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Toh Y, NEgashira A and Yamamoto M:
Epigenetic alterations and their clinical implications in
esophageal squamous cell carcinoma. Gen Thorac Cardiovasc Surg.
61:262–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang K, Johnson A, Ali SM, Klempner SJ,
Bekaii-Saab T, Vacirca JL, Khaira D, Yelensky R, Chmielecki J,
Elvin JA, et al: Comprehensive genomic profiling of advanced
esophageal squamous cell carcinomas and esophageal adenocarcinomas
reveals similarities and differences. Oncologist. 20:1132–1139.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Olson P, Lu J, Zhang H, Shai A, Chun MG,
Wang Y, Libutti SK, Nakakura EK, Golub TR and Hanahan D: MicroRNA
dynamics in the stages of tumorigenesis correlate with hallmark
capabilities of cancer. Genes Dev. 23:2152–2165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Soon PS, Tacon LJ, Gill AJ, Bambach CP,
Sywak MS, Campbell PR, Yeh MW, Wong SG, Clifton-Bligh RJ, Robinson
BG and Sidhu SB: miR-195 and miR-483-5p identified as predictors of
poor prognosis in adrenocortical cancer. Clin Cancer Res.
15:7684–7692. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li F, Ma N, Zhao R, Wu G, Zhang Y, Qiao Y,
Han D, Xu Y, Xiang Y, Yan B, et al: Overexpression of miR-483-5p/3p
cooperate to inhibit mouse liver fibrosis by suppressing the TGF-β
stimulated HSCs in transgenic mice. Cell Mol Med. 18:966–974. 2014.
View Article : Google Scholar
|
|
21
|
Ma N, Wang X, Qiao Y, Li F, Hui Y, Zou C,
Jin J, Lv G, Peng Y, Wang L, et al: Coexpression of an intronic
microRNA and its host gene reveals a potential role for miR-483-5p
as an IGF2 partner. Mol Cell Endocrinol. 333:96–101. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Srinivasan S, Selvan ST, Archunan G,
Gulyas B and Padmanabhan P: MicroRNAs-the next generation
therapeutic targets in human diseases. Theranostics. 3:930–942.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song Q, Xu Y, Yang C, Chen Z, Jia C, Chen
J, Zhang Y, Lai P, Fan X, Zhou X, et al: miR-483-5p promotes
invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1
and ALCAM. Cancer Res. 74:3031–3042. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tovar V, Alsinet C, Villanueva A, Hoshida
Y, Chiang DY, Solé M, Thung S, Moyano S, Toffanin S, Mínguez B, et
al: IGF activation in a molecular subclass of hepatocellular
carcinoma and pre-clinical efficacy of IGF-1R blockage. Hepatol.
52:550–559. 2010. View Article : Google Scholar
|
|
25
|
Verma A, Shukla NK, Deo SV, Gupta SD and
Ralhan R: MEMD/ALCAM: A potential marker for tumor invasion and
nodal metastasis in esophageal squamous cell carcinoma. Oncology.
68:462–470. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eskandari L, Akbarzadeh A, Zarghami N and
Rahmati-Yamchi M: Gold nanoprobe-based method for sensing activated
leukocyte cell adhesion molecule (ALCAM) gene expression, as a
breast cancer biomarker. Artif Cells Nanomed Biotechnol.
45:277–282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hui B, Chen X, Hui L, Xi R and Zhang X:
Serum miRNA expression in patients with esophageal squamous cell
carcinoma. Oncol Lett. 10:3008–3012. 2015.PubMed/NCBI
|
|
28
|
Shi L, Liu S, Zhao W and Shi J: miR-483-5p
and miR-486-5p are down-regulated in cumulus cells of metaphase II
oocytes from women with polycystic ovary syndrome. Reprod Biomed
Online. 31:565–572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jorgensen SB, O'Neill HM, Sylow L,
Honeyman J, Hewitt KA, Palanivel R, Fullerton MD, Öberg L,
Balendran A and Galic S: Deletion of skeletal muscle SOCS3 prevents
insulin resistance in obesity. Diabetes. 62:56–64. 2013. View Article : Google Scholar : PubMed/NCBI
|