Introduction
Glioma is a pernicious tumor in the central nervous
system. At present, about 70% of all brain tumors results from
glioma each year (1). The
patients' survival rates strongly depend upon the tumors'
histological grades. Patients afflicted with this serious malignant
form survive on average less than one year (2). Although progress has been made in
surgery, radiotherapy and chemotherapy regimens, the prognosis of
patients with glioma is still poor because of its fast and invasive
growth, its genetic heterogeneity, and our poor understanding on
its underlying molecular mechanisms (3). Researchers are working on new
treatments. (4,5). The previous research confirmed that
VM is related with tumor blood supply and metastasis (6). In glioma, the VM positive rate
resulted from the glioma's grades and responsible of poor
prognosis, the glioblastoma (GBM) (grade IV of glioma) has the
strongest ability of VM formation. As a novel blood supply's form,
the inhibition of VM could serve as an alternative therapeutic
target for gliomas (7). Along
these years, it's the vascular-targeted therapy that has gradually
been more and more accepted.
MicroRNAs (miRNAs), which are approximately 22
nucleotide-long RNAs without coding endogenous can regulate gene
expression of target at post-transcriptional level. They can bind
with the 3′-untranslated regions (3′-UTRs) of mRNAs and, by doing
so, repress their translation or cause their degradation (8,9).
Anomalous miRNA expression has been found to have some effects on
the developing process of a wide range of tumors and, as such,
miRNAs may be used for their diagnosis, prognosis and treatment
(10,11). Various miR-200 family members,
i.e., miR-141, 200c, 200b, 200a and 429, have been reported to play
important roles in a variety of cancers (12,13),
including proliferation, invasion and apoptosis (14,15).
But, so far, the expression and function of miR-141 in the
developing process of human glioma has remained unclear. EphA2 gene
belongs to the ephrin receptor subfamily of the protein-tyrosine
kinase family, it has been involved in mediating the development of
events, especially in the nervous system (16).
Here, the expression of miR-141 in primary human
glioma tissues and glioma-derived cell lines was studied, also with
the effect of exogenous miR-141 expression on glioma tumor
progression, including cell proliferation, migration, invasion and
vasculogenic mimicry (VM) both in vitro and in vivo.
By using a luciferase reporter assay, direct interactions between
miR-141 and the 3′UTR of its predicted target EphA2 were
assessed.
Materials and methods
Clinical samples
The normal brain tissues samples came from
intracranial trauma cranial decompression surgery, and glioma
tissues samples came from the pathologic glioma surgeries. Cases
inclusion/exclusion criteria: i) patients underwent surgical
treatment, without radiotherapy and chemotherapy before surgery;
ii) postoperative pathology was diagnosed as glioblastoma, and iii)
complete patient information can be obtained. All glioma samples
were classified to 4 grades, according to criteria from the World
Health Organization (WHO): Primary grade pilocytic astrocytoma (WHO
I), grade II astrocytoma (WHO II), grade III anaplastic astrocytoma
(WHO III) and grade IV glioblastoma multiforme (WHO IV). Ten normal
brain tissue samples and forty primary glioma tissue samples were
selected for this study. Of each grade 10 cases were included. All
samples were freshly frozen in liquid nitrogen and then stored at
−80°C until RNA extraction. At the same time, this research work
was permitted by the Institutional Review Boards of Southern
Medical University and the Zhujiang Hospital, and all participants
provided written informed consent.
Immunohistochemical assay
The glioma tissues and normal brain tissues were
fixed for 1 h in 4% paraformaldehyde in phosphate buffered saline
(PBS), and sliced into 3–5 µm sections. Then operated the
deparaffinization and hydration, which means the slides were
treated with peroxide after blocked by serum (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The anti-EphA2
antibodies (ab78002; Abcam, Cambridge, MA, USA) were added and
incubated overnight. After washed for five times with PBS, the
slides were added and incubated for 30 min at 37°C with
diaminobenzidine (DAB) solution (secondary antibody: HRP conjugated
goat anti-rabbit antibody). At last, the lightly counterstained
process with hematoxylin, dehydrated process and mounted process
were treated on these slides, respectively. Visual analysis was
performed with Olympus fluorescence microscope (CX71; Olympus
Corporation, Tokyo, Japan).
Cell culture and transfection
The GBM cell lines (A172, code: TCHu171 cell and
U251, code: TCHu 58 cell) and the normal glial cell line (HEB cell)
were obtained from Shanghai Cell Collection (http://www.cellbank.org.cn/, Shanghai, China). The
cells' culture condition was: 10% fetal bovine serum (FBS) in
Dulbecco's modified Eagle's medium (DMEM) and at 37°C in a 5%
CO2 humidified atmosphere. All cells were harvested in
their log phase. miRNAs were bought from GenePharm (Shanghai,
China) and then transfected into the cells using a Lipofectamine
2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) by
following the protocol in manuals. The respective miRNA sequences
are listed in Table I. After
transfection, reverse transcription-quantitative polymerase chain
reaction (RT-qPCR; 48 h after transfection) and western blotting
(72 h after transfection) analyses were employed to assess these
transfection efficiencies.
 | Table I.Sequences of the microRNA-141 mimic
and microRNA-Negative Control mimic control. |
Table I.
Sequences of the microRNA-141 mimic
and microRNA-Negative Control mimic control.
| Genes | Names | Primers (5′ to
3′) |
|---|
| miR-141 | miR-141 mimic | UAACACUGUCUGGUA
AAGAUGG |
| Mimic control | miR-141-NC | GGGAGUGAAGACACG
GAGCCAGA |
Assay of cell viability
The A172 and U251 cells transfected were seeded in
96-wells plates (1×103 cells/well) in triplicate and
incubated. After this incubation, a 3-(4,5-dimethyl-2-thiazolyl)-2,
5-diphenyl-2-H-tetrazolium bromide (MTT) working solution was added
into the medium after which the cells were incubated for another 4
h. Next, the medium was discarded and dimethyl sulphoxide (DMSO)
(150 µl) was added in order to dissolve the formazan crystals. Cell
viabilities were evaluated at 24, 48 and 72 h by measuring the 490
nm absorbance by employing a microplate reader (Bio-Rad 680
Microplate Reader; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The assays were repeated thrice.
Wound healing assay
Transfected A172 and U251 cells (1×106
cells/well) were seeded into 6-well plates and up to confluence
during 24–48 h. Next, cells from each well were divided into 2–3
grids. After attachment, artificial, homogenous wounds were created
by scratching the monolayers with 200 µl pipette tips and washed
the cells for twice with serum-free medium. Next, the cells
continued culturing in medium with serum-free for another 24 h.
Once a wound was inflicted and after 24 h, microscopic images of
the same area were immediately captured. Finally, the following
formula was employed to calculate the cell migration distances:
(migration distance=initial distance-final distance).
Transwell invasion assay
Transfected A172 and U251 cells were resuspended in
medium with serum-free until a density of 5×105
cells/ml. Three hundred microliters of these cell suspension were
harvested and added into the upper chamber of each well containing
a 24-well polycarbonate Transwell membrane insert (BD Biosciences,
Franklin Lakes, NJ, USA) and coated with 30 mg/cm2
matrigel (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). After
incubation for 24 h at 37°C, and cells on the upper membrane
surface were gently removed with a cotton swab. Next, fixed the
filters for 30 min in 95% ethanol and stained for 30 min with a
0.2% crystal violet solution, after which the cells that invaded
the matrigel and adhered to the lower surface of the filter were
counted (five high-power fields per chamber) using an inverted
microscope.
Cell cycle assay
Transfected A172 and U251 cells were collected,
washed by PBS and fixed for 12 h in 70% ethanol at 4°C. After
washing by PBS, 2×106 cells were added and incubated for
30 min with the DNA-binding dye propidium iodide (PI, 50 µg/ml) and
RNase (1.0 mg/ml) at 37°C in dark. At last, the cells were washed
and red fluorescence was readed and analyzed by a flow cytometer
(Accuri™ C6; BD Biosciences). The cell cycle analyses were
performed in triplicate and repeated thrice.
Apoptosis assay
Apoptosis was determined by an FITC Annexin V
apoptosis detection kit (556547; BD Biosciences) by following the
manufacturer's instructions. Briefly, collected cells 48 h after
transfection, washed twice in PBS and re-suspended in binding
buffer with a density of 1×103 cells/ml. Next, the cells
were simultaneously incubated for 20 min with FITC-labeled Annexin
V and PI and analyzed by a flow cytometer (Accuri™ C6; BD
Biosciences).
RNA extraction and RT-qPCR
Total RNA from 2×106 glioma cells was
extracted by operating TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) after which a cDNA Synthesis Kit was used to
synthesize cDNA by following the manufacturer's instructions
(Takara Biotechnology Co., Ltd., Dalian, China). RT-qPCR was
performed to assess miRNA and mRNA expression levels using a
LightCycler 480 detection system (Roche Diagnostics, Indianapolis,
IN, USA) and SYBR-Green dye. The primers used are shown in Table II. β-actin mRNA and U6 snRNA
levels were employed for normalization. DNA was amplified with an
initial denaturation at 95°C for 3 min, followed by 45 cycles of
95°C for 20 sec and 60°C for 1 min. RT-qPCR data were used to
analyze and express as relative mRNA or miRNA levels by CT values
and then subsequently convert to fold changes.
 | Table II.Primers used in the present
study. |
Table II.
Primers used in the present
study.
| Genes | Names | Primers (5′ to
3′) |
|---|
| miR-141 | miR-141-RT |
GTCGTATCCAGTGCGTGTCGT
GGAGTCGGCAATTGCACTGGA TACGACTCCAACA |
|
| miR-141-F |
GGCATCTTCCAGTACAGTGT |
|
| miR-141-R |
CAGTGCGTGTCGTGGAGT |
| U6 | U6-RT |
GTCGTATCCAGTGCAGGGTCC
GAGGTATTCGCACTGGATACG ACAAAAAT |
|
| U6-F |
CAAATTCGTGAAGCGTTCCATA |
|
| U6-R |
AGTGCAGGGTCCGAGGTATTC |
| EphA2 | EphA2-F |
GCCCCACATGAACTACACCT |
|
| EphA2-R |
GGCTCTGTCTGGTTGATGCT |
| β-actin | β-actin-F |
CCTGTACGCCAACACAGTGC |
|
| β-actin-R |
ATACTCCTGCTTGCTGATCC |
Western blot analysis
2×106 cells were lysed by a
radioimmunoprecipitation assay buffer which contains a protease
inhibitor cocktail (Roche Diagnostics) using standard procedures.
Proteins' concentrations were measured by using a Micro BCA protein
assay kit (Pierce; Thermo Fisher Scientific, Inc.). Total cell
lysates (50 µg) were loaded in each lane and resolved by sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE;
Invitrogen; Thermo Fisher Scientific, Inc.), and then transferred
to polyvinylidene difluoride (PVDF) membranes. The PVDF membranes
were blocked with 7% non-fat milk for 1 h at 37°C, followed by
immunoblot detection and visualization with ECL western blotting
detection reagents (Pierce; Thermo Fisher Scientific, Inc.).
Immunoblotting was performed using anti-EphA2 antibodies (ab185156,
dilution: 1/1,000; Abcam), anti-Bax antibodies (ab32503, dilution:
1/1,000; Abcam) and anti-Bcl-2 antibodies (ab32124, dilution:
1/1,000; Abcam) at 37°C for 2 h respectively, followed by
incubation with the appropriate horseradish-peroxidase-conjugated
IgG secondary antibodies (ab97023, dilution: 1/5,000; Abcam) for 1
h at room temperature. GAPDH (ab8245, dilution: 1/1,000; Abcam)
levels were used for normalization. The protein bands were scanned
and quantified using a ChemiDoc image analysis system (Bio-Rad
Laboratories, Inc.).
Luciferase assay
Wild-type and mutant EphA2 3′UTR reporter and
control constructs were bought and obtained from Shanghai GeneChem
(Shanghai, China). For dual luciferase assays, 150 nM pre-miR-141
or a negative precursor control were transfected into
5×105 cells by employing Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). After 24 h, both Renilla
luciferase reporter (0.35 ng) and firefly luciferase reporter (1.5
mg) were simultaneously transfected into cells in 24-well plates.
After 24 h, firefly luciferase activities were detected by
employing a dual luciferase reporter assay kit (Promega
Corporation, Madison, WI, USA), whose results were normalized into
Renilla luciferase activities by following the manual's
protocol.
In vivo xenografting
Male Fisher 344 rats weighing 200–220 g were
obtained from the Animal Research Center, which belongs to Southern
Medical University, China, and vthen divided randomly into 2 groups
(four rats per group). The rats were maintained on a 12 h light/12
h dark cycle under room temperature (23±1°C) and humidity (55±5%)
and fed with standard forage and clean water. miR-141 and miR-NC
sequences were inserted into a pcDNA6.2-GW/EmGFP-miR vector. After
transfection into A172 cells, positive clones were selected using 1
µg/ml puromycin and propagated further. After verification of
miR-141 expression, the stably transfected cells were suspended in
5 µl medium with serum-free (2×108 cells/ml) and
intracranially injected into the rats as Yang et al
(17), report in 2012, with a few
difference. Briefly, 3.5% (w/v) chloral hydrate (10 ml/kg)
anesthetized into animals and put in sterile conditions. Aseptic
surgical techniques were used to perform a midline incision and to
open the scalp to expose the frontal and temporalis bones. A burr
hole was generated through the skull at an appropriate location
(2.0 mm posterior to the bregma and 1.0 mm right to the midline)
without breaking the dura. Next, a 26-gauge needle was inserted 3.0
mm ventral to the dura and retreated 0.5 mm, after which the cells
were implanted using a 10-µl micro-syringe at an infusion rate of 1
µl/min. A total of 2×105 cells were inoculated into the
brain. After the infusion, the needle was kept in place for 10 min
(in order to balance the pressure of the cranial vault). Next,
removed needle slowly, and immediately sealed the hole with sterile
bone-wax to prevent the solution from leaking. Finally, the animals
were returned to the animal care facilities. The rats were given a
daily physical examination. After 3 weeks (determined in a
preliminary experiment), the rats were anesthetized using 1% sodium
pentobarbital (40 mg/kg per rat), and then were sacrificed by
exsanguination. The tumor samples were carefully removed and
weighed. Western blotting was employed and operated to determine
the apoptosis-related proteins' expression.
Assay of VM
VM experiments followed the Li et al
(18) report, in 2014, in
vitro, with a few modifications. Briefly, 50 µl ECM matrigel
(Sigma-Aldrich; Merck KGaA) was added and dropped onto 18 mm glass
coverslips in 6-well plates and incubated for 1 h at 37°C. Next,
A172 and U251 cells transfected with miR-141 mimic and miR-NC were
seeded onto the coverslips coated. After incubation for 24–48 h,
the development of VM was evaluated by periodic acid-Schiff (PAS)
staining. To this end, the cells were fixed for 10 min in 4%
paraformaldehyde in PBS, oxidized for 5 min in a 0.5% periodic acid
solution, washing with PBS, and placed for 15 min in Schiff
reagent, after which the coverslips were immediately rinsed with
PBS. Finally, dried the coverslips at room temperature and the PAS
signals were imaged at a 400× magnification. In vivo, the
glioma tumor tissues were sliced into 3–5 µm sections, and the
following steps of PAS staining as previously.
Statistical analyses
SPSS v17.0 statistical analysis software (SPSS,
Inc., Chicago, IL, USA) was employed for the statistical analyses.
The significance of the differences was determined by Student's
t-test, or one-way analysis of variance with the least significant
difference post hoc test when equal variances were assumed or with
Dunnett's T3 post hoc test when equal variances were not assumed.
The data are presented as the mean ± standard error mean and
P<0.05 was considered to indicate a statistically significant
difference.
Results
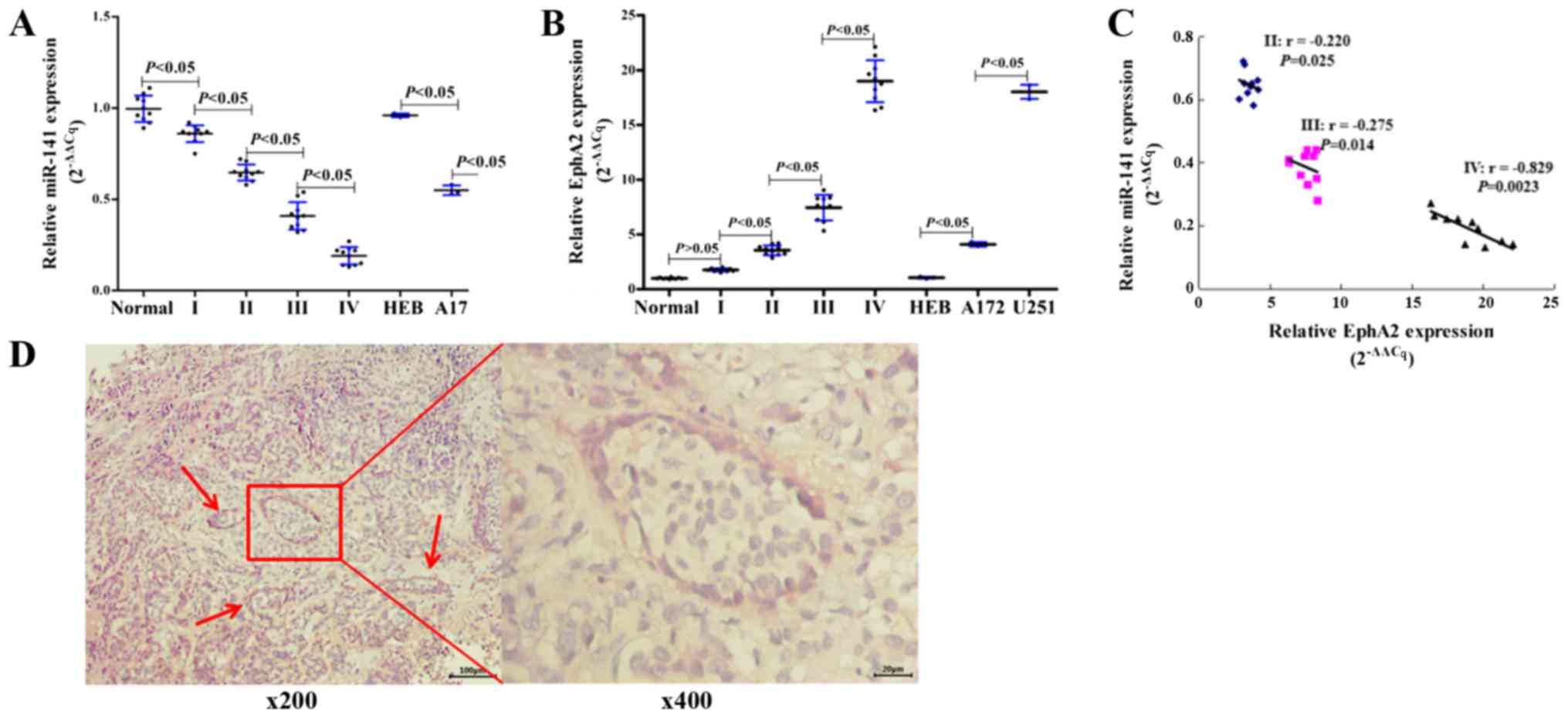
miR-141 is downregulated and EphA2 is
upregulated in human glioma tissues and cell lines
The expression of miR-141 was assessed using RT-qPCR
in normal brain tissues, primary glioma tissues, and glioma- and
normal glia-derived cell lines. We found that miR-141 was at
relatively high levels expression in normal brain tissues, whereas
its expression was significantly (P<0.05) downregulated in the
glioma samples (grades I, II, III and IV; Fig. 1A). In addition, we also found that
the miR-141's expression steadily decreased as the glioma grades
increased, and the expression of miR-141 was also found down
regulation in the glioma-derived cell lines A172 and U251 (Fig. 1A). In contrast, we found that its
putative target EphA2 was at relatively low levels expression in
normal brain tissues, and whose expression was significantly
(P<0.05) upregulated in glioma samples (grades I, II, III and
IV). We also found that the EphA2 expression steadily increased as
the glioma grades increased. EphA2 expression was also found to be
up-regulated in the glioma-derived cell lines A172 and U251
(Fig. 1B). Furthermore, a Pearson
correlation assay also indicated that a negative correlation
existed in between miR-141 and EphA2 expression levels in glioma
grades II, III and IV (Fig. 1C).
In addition, immunohistochemical assay showed (Fig. 1D) that glioma grade IV tissue forms
the typical VM, and EphA2 was expressed in the epidermis of VM
vessel. We inferred from these results that there may be some sort
of regulation relationship between miR-141 and EphA1 in glioma.
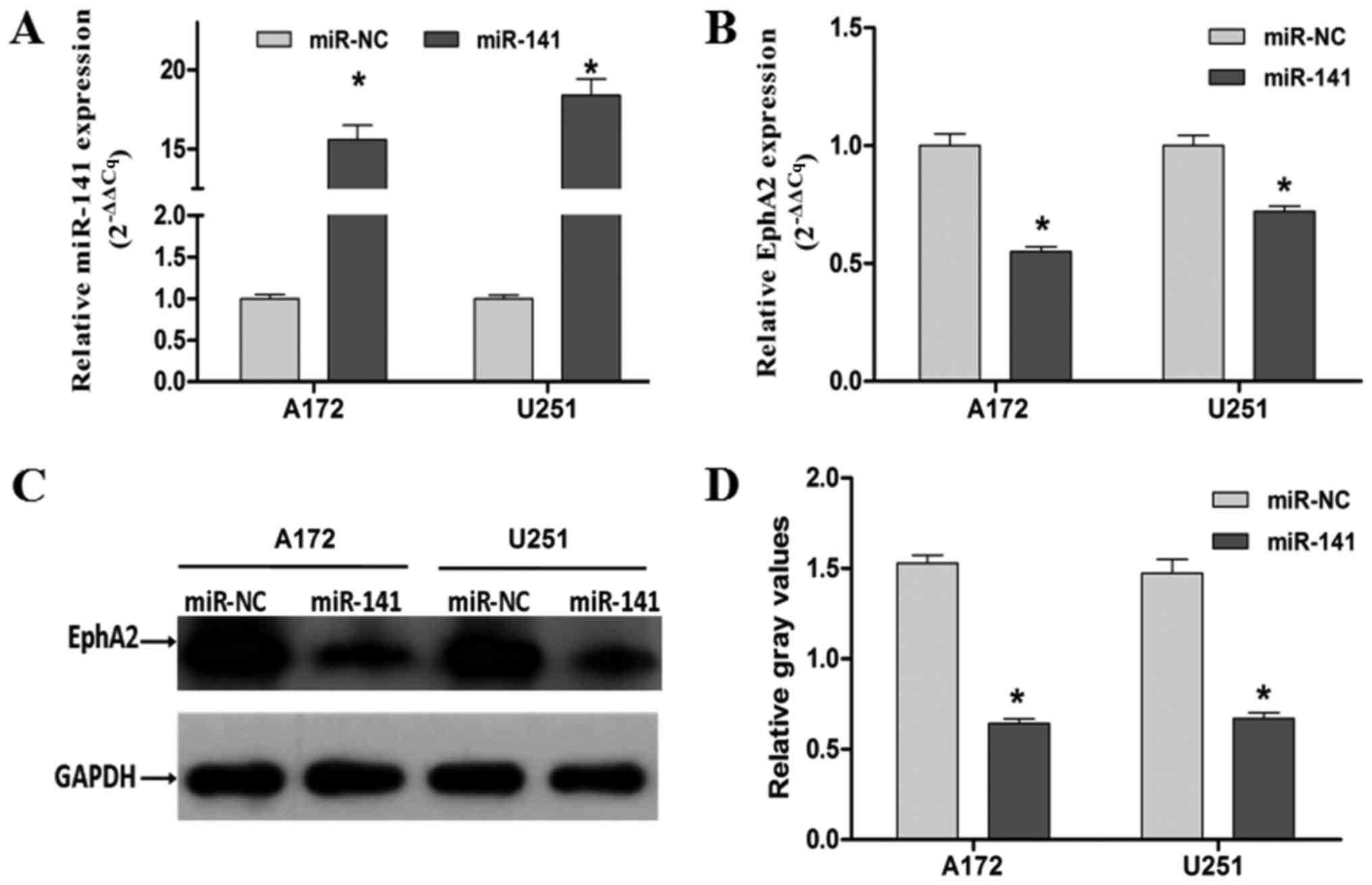
Exogenous miR-141 expression inhibits
glioma cell proliferation, migration and invasion in vitro
To study the miR-141's functional role and its
relationship with EphA2 in human glioma, A172 and U251 cells were
transiently transfected with either a miR-141 mimic or a control
mimic (miR-NC) (Table II). Using
RT-qPCR, we found that compared with the control miR-NC transfected
cells, the miR-141's expression was significantly upregulated in
the miR-141 mimic transfected cells (P<0.05; Fig. 2A). Moreover, we found that in the
miR-141 mimic transfected cells the expression of EphA2 (see below)
was significantly downregulated (Fig.
2B-D, P<0.05) by RT-qPCR and western blotting analyses
(Fig. 2C and D, P<0.05).
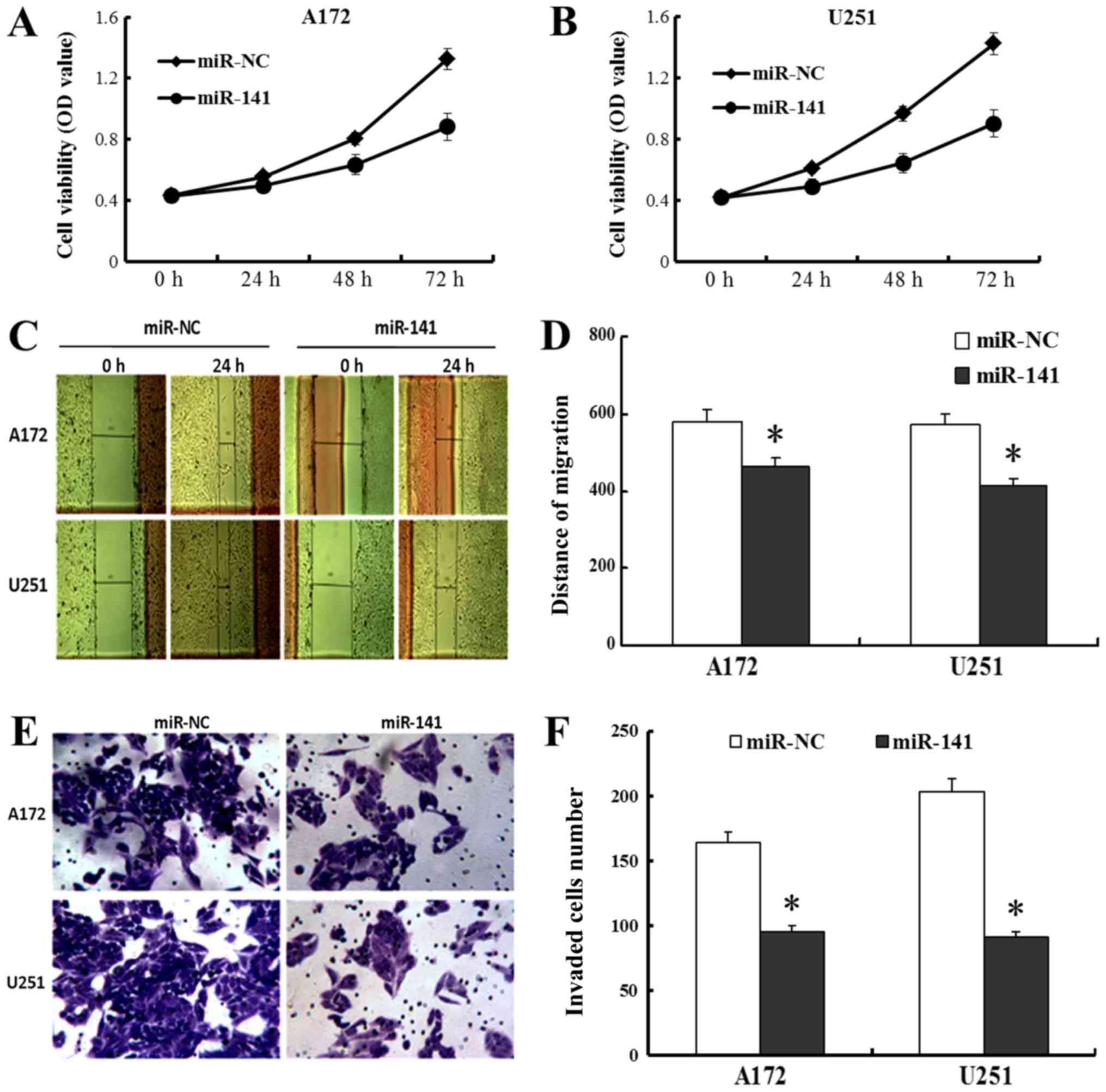
We also found that miR-141's exogenous
overexpression led to decreases in A172 and U251 cell proliferation
using cell viability assay (P<0.05; Fig. 3A and B) and migration using wound
healing assays (P<0.05; Fig. 3C and
D). An in vitro transwell invasion assay was
subsequently employed to examine the invasive capacities of these
cells. By doing so, we found that the invasive capacities were
markedly reduced in the A172 and U251 cells with miR-141 mimic
transfected, i.e., by 42.1 and 55.2%, respectively (P<0.05;
Fig. 3E and F). Based on these
results, we conclude that miR-141's overexpression results in in
vitro inhibition of the proliferation, migration and invasion
of human glioma-derived cells.
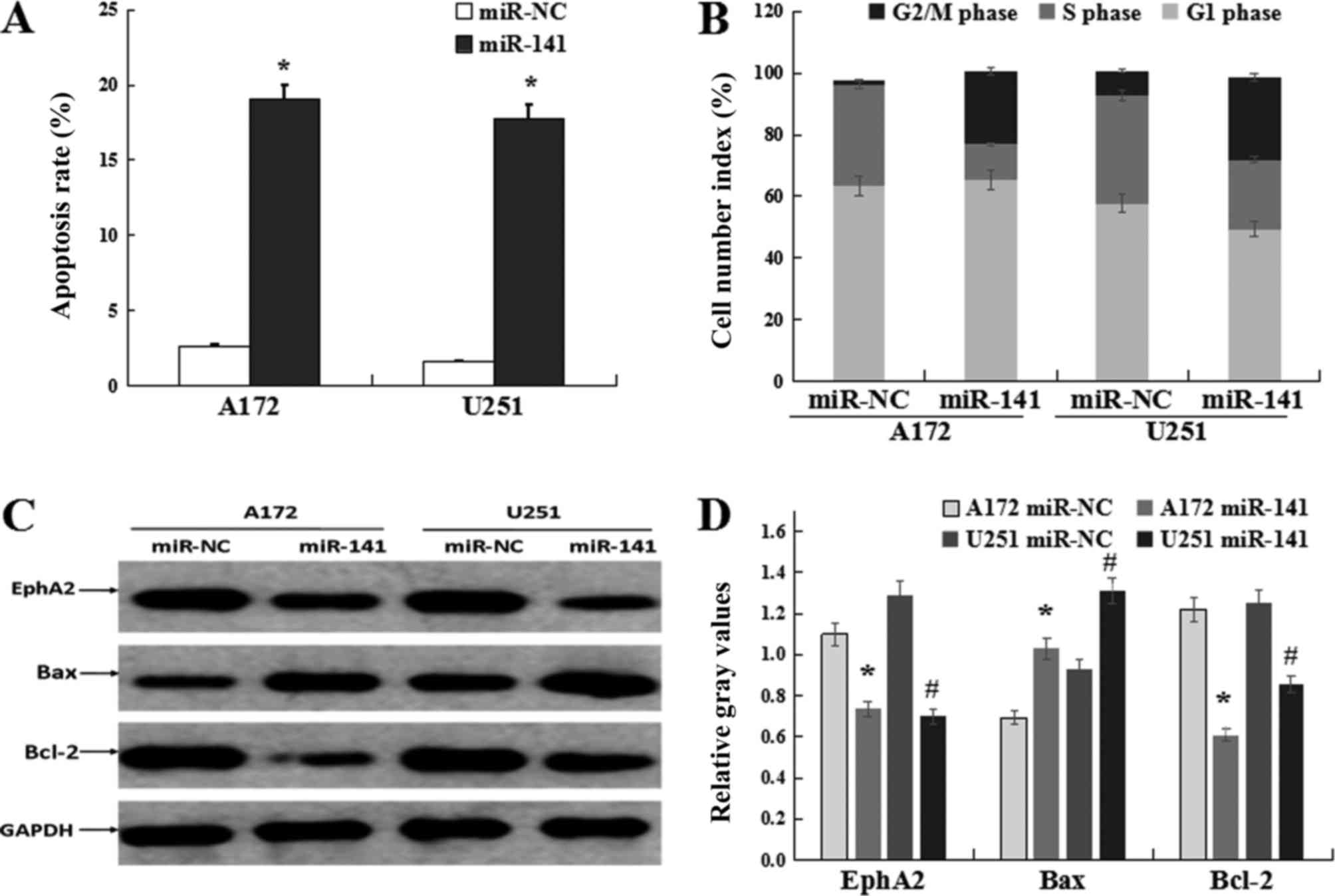
miR-141 induces glioma cell apoptosis
and cell cycle arrest in vitro
Next, we set out to assess the effect of miR-141 on
the regulation of apoptosis and cell cycle progression using A172
and U251 cells which were transfected with either the miR-141 mimic
or the control miR-NC mimic. By doing so, we found that the
apoptotic rates increased in the cells with miR-141 transfected
(19.1% in A172 cells and 17.8% in U251 cells; Fig. 4A). Subsequent cell cycle analyses
revealed that miR-141 mimic transfected cells were arrested at G2/M
phase, i.e., the percentages of G2/M phase cells were found to be
23.48 and 26.61% for the miR-141 transfected A172 and U251 cells,
respectively, while the percentages were 1.34 and 7.87%,
respectively, in the miR-NC group (P<0.05; Fig. 4B). Our additional observation of
the pro-apoptotic protein Bax's up-regulation and the
anti-apoptotic protein Bcl-2's down-regulation in the cells with
miR-141 transfected underscores the notion that miR-141
overexpression induces apoptosis in glioma-derived cells (Fig. 4C and D).
EphA2 is a direct target of miR-141 in
glioma cells
We used the miRecords resource (19) to identify potential miR-141 targets
from three independent prediction tools included on the NCBI
website. Among the putative miR-141 targets identified, we selected
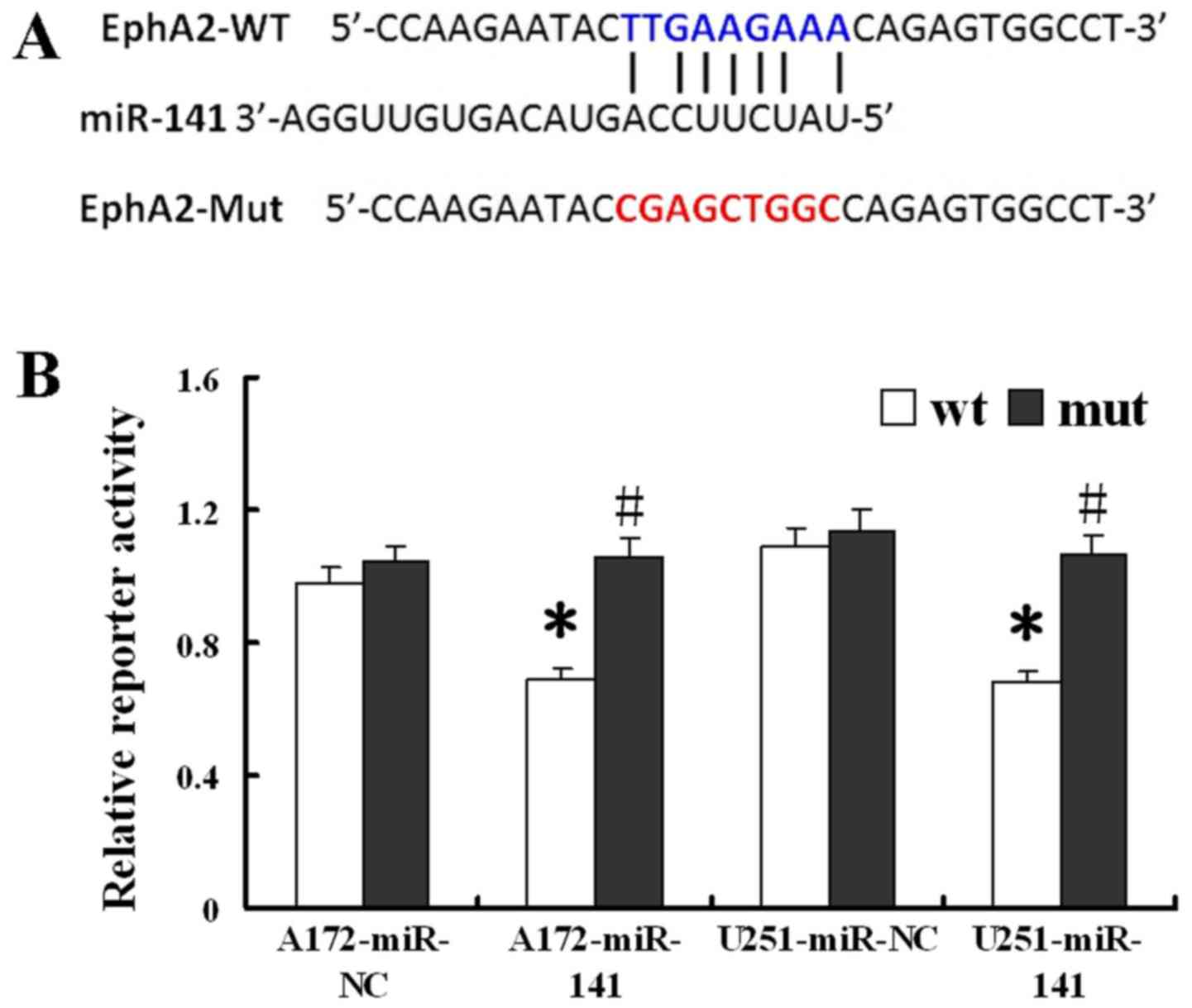
EphA2 for further analysis since the EphA2 3′UTR contains seven
miR-141 potential binding sites (Fig.
5A) and since EphA2 has previously been implicated in tumor
development. Next, an assay of luciferase reporter was performed to
obtain direct evidence that EphA2 serves as a miR-141's genuine
target in glioma cells. By doing so, compared to those
co-transfected with the miR-NC control mimic, we found that cells
simultaneously transfected with both the wild-type reporter and the
miR-141 mimic showed a significant decrease in relative luciferase
activity (P<0.05; Fig. 5B).
However, the cells transfected with both the mutant reporter and
the miR-141 mimic did not show any effect on the reporter
luciferase activity (P<0.05). Based on these data, a conclusion
can be made that EphA2 indeed serves as a direct target of miR-141
in glioma-derived cells.
miR-141 suppresses tumorigenesis in
vivo
Stable miR-141 mimic transfected and miR-NC control
mimic transfected A172 cells were injected intracranially into
rats, after which tumor growth was evaluated. We found that the
tumor weights in the miR-141 mimic group were significantly much
lower than those in the miR-NC group (P<0.05; Fig. 6A and B). Additional western
blotting analyses of the resulting tumors showed that the EphA2
expression levels were also decreased in the miR-141 mimic groups.
In addition, we found that the pro-apoptotic protein Bax expression
levels were increased and that the anti-apoptotic protein Bcl-2
expression levels were decreased in the miR-141 mimic groups in
vivo (Fig. 6C and D). These
data indicate that the apoptotic rates of the miR-141 mimic groups
are increased and, thus, that exogenous miR-141 overexpression in
glioma-derived cells induces apoptosis and suppresses tumorigenesis
in vivo.
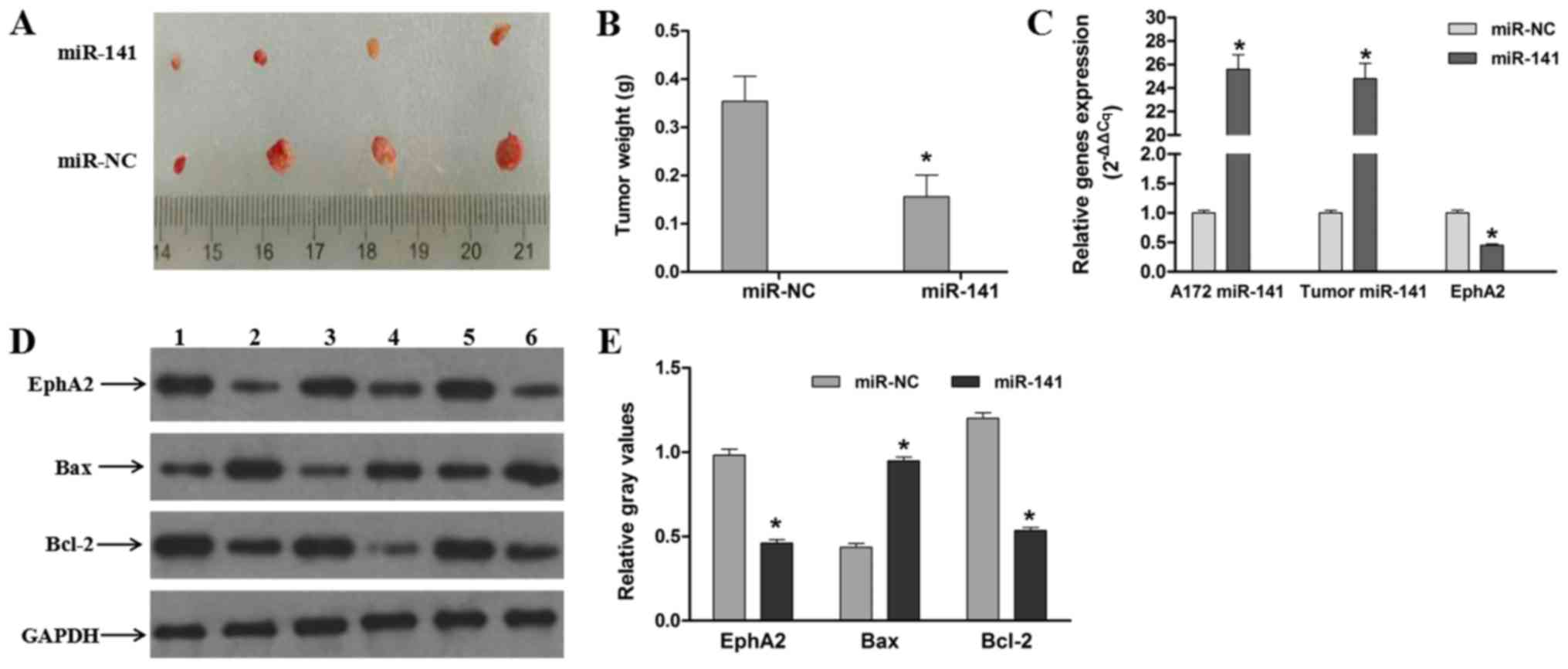 | Figure 6.miR-141 suppresses tumorigenesis
in vivo. (A) Macroscopic appearance of xenotransplanted
tumors. (B) Quantitative analyses of tumor weights. (C) Expression
of miR-141 in stably transfected A172 cells, and expression of
miR-141 and EphA2 in glioma tumor tissues. (D) miR-141 suppresses
EphA2 expression and induces glioma-derived cell apoptosis in
vivo. Lanes 1, 3 and 5 are the miR-NC group; lanes, 2, 4 and 6
are the miR-141 group. (E) Gray scale analyses of the relative
EphA2, Bax and Bcl-2 expression levels. The data are presented as
mean ± standard error mean (n=3). Each bar represents the mean of
three independent experiments performed in triplicate. *P<0.05
vs. miR-NC group. miR, microRNA; NC, negative control; EphA2,
Ephrin type-A receptor 2; Bcl-2, B-cell lymphoma 2; Bax,
Bcl-2-associated X protein. |
miR-141 suppresses the development of
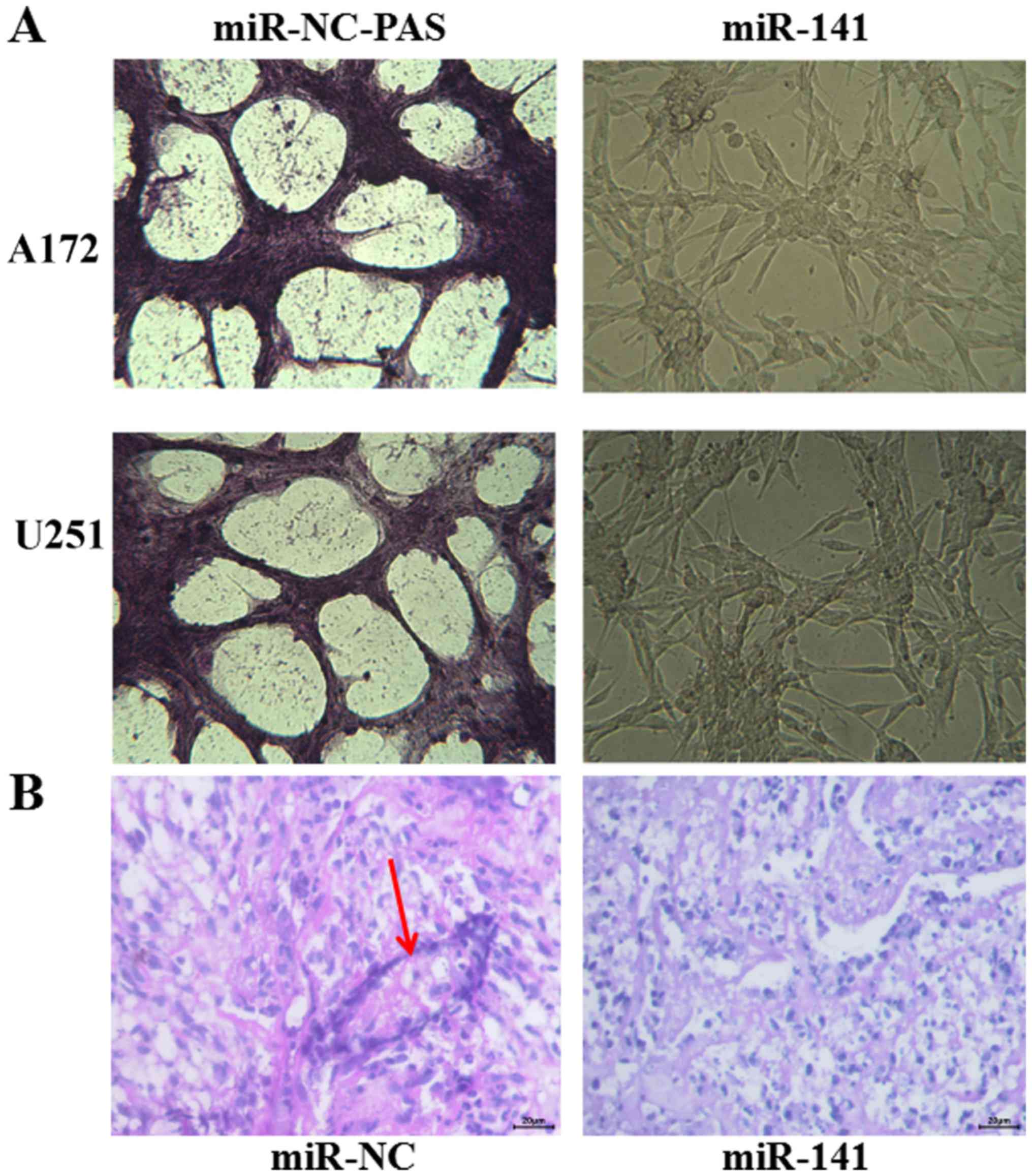
VM in vivo and in vitro
VM is related with tumor blood supply and
metastasis. The number of vessels (nodes) and the remodeling of the
microcirculation are used as histological markers of tumor
progression. Cancers require adequate blood supply for in
vivo growth, and VM serves an alternative pathway for
maintaining this supply. Here, we calculated and analyzed the
vessel numbers of the network channels, which reflect VM
development. We found that A172 and U251 cells, whose levels of
EphA2 are very high, and formed classical VM networks on matrigel
in vitro. After the A172 and U251 cells transient
transfection with miR-141 mimics, and a concomitant down-regulation
of EphA2, we found that the number of vessels decreased from
9.67±0.58 in the miR-NC mimic transfected cells to 5.33±0.58 in the
miR-141 mimic transfected cells (Fig.
7A). In vivo, the tumor tissue, which stably transfected
with miR-141, formed classical VM vessels, compared with miR-NC
group (Fig. 7B). Combined with the
data obtained above, we conclude that miR-141 may be a regulatory
genes, and EphA2 may be a direct regulator for the development of
VM in glioma.
Discussion
By using both western blotting and RT-qPCR, we known
that both the miR-141 and EphA2 expression levels were inversely
correlated in the primary glioma tissues and glioma-derived cell
lines tested. Based on this observation, we hypothesized that EphA2
may serve as a miR-141 target in glioma cells. This hypothesis was
subsequently confirmed using dual luciferase assays. EphA2, a
transmembrane Eph tyrosine kinase receptor, has been reported to
have an important effect on the development of a variety of tumors
(20). EphA2 has frequently been
found to be over-expression and/or deregulation in advanced
cancers, including malignant gliomas (16,21),
and accumulating evidence indicates that the increased expression
of EphA2 may promote cancer progression by inducing its growth and
invasion (22,23). Additional analyses of EphA2
knockout mice have revealed a deficiency ability to support the
implanted tumors' invasion and metastasis (24).
Accumulating evidence indicates that miRNAs may have
key effect on both initiation and progression of tumor (25). In gastric cancer, miR-141 was
decreased, and inhibited gastric cancer cell proliferation, colony
formation, migration and invasion (26,27).
The miR-141's overexpression has been shown to depress the
pancreatic cancer's invasion and migration, also include breast
cancer and so on (28,29). Numerous of reports have shown that
miRNAs could regulate tumor progression through various signaling
or target proteins. In hepatocellular carcinoma, Liu et al
(30) found that miR-141 could
suppress the HCC cells' migration and invasion by targeting Tiam1.
In glioma, Peng et al (31)
found that miR-141 was a tumor inhibitor by targeting TGF-β2.
Furthermore, the miR-141's expression patterns and levels were
different in various tumor types. It's found in ovarian carcinoma
(32), colorectal carcinoma
(33) that miR-141 was up
regulation; while it's down-regulated in breast cancer (29) and gastric cancer (26). These opposing findings indicated
that miR-141 might play different roles in different cancer types
as an oncogene or a tumor suppressor gene. In the present study, we
found that exogenous miR-141 overexpression results in a dramatic
suppression of glioma-derived cell proliferation, migration,
invasion and VM formation, and an increase in cell cycle arrest.
Additional in vivo analyses indicated that exogenous miR-141
overexpression resulted in profound decreases in glioma tumor
weight and concomitant increases in apoptosis. Therefore, we
conclude that miR-141 is involved in regulating glioma tumor
progression, and may act as an anti-oncogenic factor.
Undoubtedly, one tumor usually requires a blood
supply to sustain growth. The tumor microcirculation has central
and key effect on the tumor's hematogenous dissemination. The
mechanisms by which tumors obtain their blood supply have attracted
considerable attention in these years. And two distinctive VM types
have been reported in tumors: the patterned matrix type in VM and
the tubular type in VM, and human glioblastoma tissues could formed
VM of the tubular type (34). In
glioma, Liu X pointed out that VM channels correlated with the
increasing malignancy and higher aggressiveness, which may provide
a complementation to the tumor's blood supply, especially in less
vascularized regions (35). The
cancer cells with highly metastatic and aggressive are capable of
forming highly patterned vascular channels with basement membrane
stained PAS-positive (36). Thus
it may provide a potential method for GBM treatment by exploring a
new way to inhibit VM. In GBM cells, we found the typical VM
channels structure (PAS-positive channels) in vivo, and
overexpression of miR-141 could inhibit VM formation. The findings
in our study suggest that miR-141 and EphA2 are potential targets
against VM treatment.
In conclusion, the experimental results from us
indicate that miR-141 plays a key and important role in progression
of glioma tumor by modulating cell proliferation, apoptosis,
migration, invasion and VM formation through controlling EphA2
expression. As such, miR-141 and its target EphA2 may serve as a
specific biomarker for glioma diagnosis/prognosis and as a
potential anti-VM therapeutic target. In this study, we elucidate
the regulatory relationship between miR-141 and EphA2, and their
effects on the proliferation, migration, invasion and VM in glioma,
which provides a laboratory basis for the treatment of glioma. The
other specific target genes that are involved in tumorigenesis and
regulated by miR-141, which could be developed as drug-oriented
targets in the future. Since individual miRNAs may regulate several
different target genes (miRNAs) that affect carcinogenic processes
in different ways, studies aimed at gathering more detailed
information on the molecular and cellular mechanisms underlying the
action of miR-141 and its targets are required.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Innovation of Science and Technology Committee of Shenzhen City
(grant no. JCYJ20150402155418386), the Natural Science Foundation
of China (grant nos. 81302177 and 81272806), the Natural Science
Foundation of Guangdong Province, China (grant no. S2012010009088)
and Medical Scientific Research Foundation of Guangdong Province,
China (grant no. B2013246).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GL and YK conceived and designed the experiments. GL
and MH performed the experiments, and YC and YY collected and
processed the clinical samples. XS analyzed the data, and GL wrote
the paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The Institutional Review Boards of Southern Medical
University and Zhujiang Hospital approved the protocol used in the
present study, and all procedures were performed in accordance with
the ethical standards established in the Declaration of Helsinki.
All patients provided written informed consent prior to the
study.
Consent for publication
All patients provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Demuth T and Berens ME: Molecular
mechanisms of glioma cell migration and invasion. J Neurooncol.
70:217–228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu N, Zhao XZ, Liu M, Liu H, Yao W, Zhang
Y, Cao S and Lin X: Role of microRNA-26b in glioma development and
its mediated regulation on EphA2. PLoS One. 6:e162642011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tanase C, Albulescu R, Codrici E, Popescu
ID, Mihai S, Enciu AM, Cruceru ML, Popa AC, Neagu AI, Necula LG, et
al: Circulating biomarker panels for targeted therapy in brain
tumors. Future Oncol. 11:511–524. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cruceru ML, Enciu AM, Popa AC, Albulescu
R, Neagu M, Tanase CP and Constantinescu SN: Signal transduction
molecule patterns indicating potential glioblastoma therapy
approaches. Onco Targets Ther. 6:1737–1749. 2013.PubMed/NCBI
|
|
6
|
Song Y, Mu L, Han X, Li Q, Dong B, Li H
and Liu X: MicroRNA-9 inhibits vasculogenic mimicry of glioma cell
lines by suppressing Stathmin expression. J Neurooncol.
115:381–390. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang M, Ke Y, Sun X, Yu L, Yang Z, Zhang
Y, Du M, Wang J, Liu X and Huang S: Mammalian target of rapamycin
signaling is involved in the vasculogenic mimicry of glioma via
hypoxia-inducible factor-1α. Oncol Rep. 32:1973–1980. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mosch B, Pietzsch D and Pietzsch J:
Irradiation affects cellular properties and Eph receptor expression
in human melanoma cells. Cell Adh Migr. 6:113–125. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lytle JR, Yario TA and Steitz JA: Target
mRNAs are repressed as efficiently by microRNA-binding sites in the
5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 104:9667–9672.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bouyssou JM, Manier S, Huynh D, Issa S,
Roccaro AM and Ghobrial IM: Regulation of microRNAs in cancer
metastasis. Biochim Biophys Acta. 1845:255–265. 2014.PubMed/NCBI
|
|
11
|
Bandres E, Agirre X, Bitarte N, Ramirez N,
Zarate R, Roman-Gomez J, Prosper F and Garcia-Foncillas J:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Wang X, Ruan A, Han W, Zhao Y, Lu
X, Xiao P, Shi H, Wang R, Chen L, et al: miR-141 is a key regulator
of renal cell carcinoma proliferation and metastasis by controlling
EphA2 expression. Clin Cancer Res. 20:2617–2630. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shimono Y, Zabala M, Cho RW, Lobo N,
Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mateescu B, Batista L, Cardon M, Gruosso
T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P,
Sastre-Garau X and Mechta-Grigoriou F: miR-141 and miR-200a act on
ovarian tumorigenesis by controlling oxidative stress response. Nat
Med. 17:1627–1635. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schickel R, Park SM, Murmann AE and Peter
ME: miR-200c regulates induction of apoptosis through CD95 by
targeting FAP-1. Mol Cell. 38:908–915. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hatano M, Eguchi J, Tatsumi T, Kuwashima
N, Dusak JE, Kinch MS, Pollack IF, Hamilton RL, Storkus WJ and
Okada H: EphA2 as a glioma-associated antigen: A novel target for
glioma vaccines. Neoplasi. 7:717–722. 2005. View Article : Google Scholar
|
|
17
|
Yang L, Zhao J, Zhou G, Wang Y, Li L, Yuan
H, Nan X, Guan L and Pei X: The 9L(LUC)/Wistar rat glioma model is
not suitable for immunotherapy. Neural Regen Res. 7:1406–1411.
2012.PubMed/NCBI
|
|
18
|
Li XY, Zhao Y, Sun MG, Shi JF, Ju RJ,
Zhang CX, Li XT, Zhao WY, Mu LM, Zeng F, et al: Multifunctional
liposomes loaded with paclitaxel and artemether for treatment of
invasive brain glioma. Biomaterials. 35:5591–5604. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:(Database Issue). D105–D110.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ogawa K, Pasqualini R, Lindberg RA, Kain
R, Freeman AL and Pasquale EB: The ephrin-A1 ligand and its
receptor, EphA2, are expressed during tumor neovascularization.
Oncogene. 19:6043–6052. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jin Q, Li X and Cao P: EphA2 modulates
radiosensitive of hepatocellular carcinoma cells via
p38/mitogen-activated protein kinase-mediated signal pathways.
Kaohsiung J Med Sci. 31:510–517. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zelinski DP, Zantek ND, Stewart JC,
Irizarry AR and Kinch MS: EphA2 overexpression causes tumorigenesis
of mammary epithelial cells. Cancer Res. 61:2301–2306.
2001.PubMed/NCBI
|
|
23
|
Shao Z, Zhang WF, Chen XM and Shang ZJ:
Expression of EphA2 and VEGF in squamous cell carcinoma of the
tongue: Correlation with the angiogenesis and clinical outcome.
Oral Oncol. 44:1110–1117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brantley-Sieders DM, Fang WB, Hicks DJ,
Zhuang G, Shyr Y and Chen J: Impaired tumor microenvironment in
EphA2-deficient mice inhibits tumor angiogenesis and metastatic
progression. FASEB J. 19:1884–1886. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nat.
435:834–838. 2005. View Article : Google Scholar
|
|
26
|
Chen B, Huang T, Jiang J, Lv L, Li H and
Xia S: miR-141 suppresses proliferation and motility of gastric
cancer cells by targeting HDGF. Mol Cell Biochem. 388:211–218.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou X, Xia Y, Su J and Zhang G:
Down-regulation of miR-141 induced by helicobacter pylori promotes
the invasion of gastric cancer by targeting STAT4. Cell Physiol
Biochem. 33:1003–1012. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu L, Li Q, Xu D, Wang Q, An Y, Du Q,
Zhang J, Zhu Y and Miao Y: hsa-miR-141 downregulates TM4SF1 to
inhibit pancreatic cancer cell invasion and migration. Int J Oncol.
44:459–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Neves R, Scheel C, Weinhold S, Honisch E,
Iwaniuk KM, Trompeter HI, Niederacher D, Wernet P, Santourlidis S
and Uhrberg M: Role of DNA methylation in miR-200c/141 cluster
silencing in invasive breast cancer cells. BMC Res Notes.
3:2192010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Y, Ding Y, Huang J, Wang S, Ni W, Guan
J, Li Q, Zhang Y, Ding Y, Chen B and Chen L: MiR-141 suppresses the
migration and invasion of HCC cells by targeting Tiam1. PLoS One.
9:e883932014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peng T, Zhang S, Li W, Fu S, Luan Y and
Zuo L: MicroRNA-141 inhibits glioma cells growth and metastasis by
targeting TGF-β2. Am J Transl Res. 8:3513–3521. 2016.PubMed/NCBI
|
|
32
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
El Hallani S, Boisselier B, Peglion F,
Rousseau A, Colin C, Idbaih A, Marie Y, Mokhtari K, Thomas JL,
Eichmann A, et al: A new alternative mechanism in glioblastoma
vascularization: Tubular vasculogenic mimicry. Brain. 133:973–982.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu XM, Zhang QP, Mu YG, Zhang XH, Sai K,
Pang JC, Ng HK and Chen ZP: Clinical significance of vasculogenic
mimicry in human gliomas. J Neurooncol. 105:173–179. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Folberg R, Hendrix MJ and Maniotis AJ:
Vasculogenic mimicry and tumor angiogenesis. Am J Pathol.
156:361–381. 2000. View Article : Google Scholar : PubMed/NCBI
|