Introduction
Pancreatic cancer, the most malignant type of
digestive system tumour, ranks as the seventh-most common cause of
cancer-related deaths globally (1). Pancreatic ductal adenocarcinoma
(PDAC), the main subtype of pancreatic cancer, accounts for
approximately 90% of all pancreatic cancer cases (2). Despite a remarkable development in
treatments and perioperative management, the prognosis of patients
with PDAC remains poor. The median survival period and 5-year
overall survival rate of patients with PDAC are approximately 6
months and less than 5%, respectively (3). The poor therapeutic outcomes of PDAC
patients are due to the late onset of presentation, metastasis and
unresponsiveness to chemotherapy and radiation therapy (4). The pathogenesis of PDAC is influenced
by various factors, including poor dietary habits, smoking,
excessive drinking, long-term exposure to chemical carcinogens,
diabetes mellitus and chronic pancreatitis (5,6).
However, the detailed mechanisms of the formation and progression
of PDAC remain largely unknown. Therefore, the molecular mechanism
of PDAC onset and development should be understood, and new
therapeutic targets for the treatment of patients with this highly
aggressive malignancy should be explored.
MicroRNAs (miRNAs) are noncoding and highly
conserved short RNA molecules with 19 to 24 nucleotides implicated
in gene regulation (7). miRNAs are
involved in the regulation of their target genes by base pairing
with the 3′-untranslated regions (3′-UTRs) of their target genes,
thereby degrading mRNA and suppressing translation; thus, the
expression levels of associated proteins are inhibited (8). miRNA dysregulation frequently occurs
in numerous human malignancies, such as PDAC (9), melanoma (10), gastric cancer (11), colorectal cancer (12) and bladder cancer (13). Aberrantly expressed miRNAs play key
roles in the initiation and progression of PDAC by improving
oncogene expression or by downregulating the level of tumour
suppressor genes (14–16). Hence, an in-depth understanding of
miRNAs expression patterns and biological roles in PDAC may be
advantageous to the development of miRNA-based targeted therapy,
which may enhance the diagnosis, treatment and prognosis of
patients with this fatal disease.
miR-539 has been studied in multiple types of human
cancer (17–19). However, its expression and
potential biological function in PDAC remain unclear. In our
current study, we detected the expression level, clinical
significance, roles and underlying molecular mechanism of miR-539
in PDAC.
Materials and methods
Tissue samples
Forty-five pairs of PDAC tissues and adjacent normal
pancreatic tissues were collected from patients who received
surgical resection at Yidu Central Hospital of Weifang between
February 2014 and August 2016. All of the patients did not receive
other treatments before surgery. This research was approved by the
Ethics Committee of Yidu Central Hospital of Weifang (no. 2014036).
The use of these tissue samples was approved by all of the patients
before they participated in this project, and written informed
consent was obtained from all of the patients.
Cell lines
Normal human pancreatic cell line (HPDE6c7) was
acquired from American Type Culture Collection (Manassas, VA, USA).
Four PDAC cell lines (Sw1990, Panc-1, Bxpc-3 and Aspc-1) from
Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai,
China) were grown in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin/streptomycin mixture (all from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and then maintained in a
humidified atmosphere with 5% CO2 at 37°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA of tissue samples or cells was isolated
with a TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
in accordance with the manufacturer's instructions. Afterwards, the
concentration of total RNA was detected using a NanoDrop 2000
(NanoDrop Technologies; Thermo Fisher Scientific, Inc., Pittsburgh,
PA, USA). To quantify miR-539 level, total RNA was reversed
transcription into complementary DNA (cDNA) with a TaqMan MicroRNA
reverse transcription kit, and then quantitative PCR was conducted
with a TaqMan MicroRNA PCR kit (all from Applied Biosystems; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) under Applied
Biosystems® 7900HT Real-Time PCR system (Thermo Fisher
Scientific, Inc.). To analyse IGF-1R mRNA expression, cDNA was
synthesized from total RNA using a Primescript™ RT reagent kit.
Subsequently, the cDNA was subjected into amplification with a SYBR
Premix Ex Taq™ II kit (all from Takara Biotechnology Co., Ltd.,
Dalian, China). Relative miR-539 and IGF-1R mRNA expression was
normalized to U6 snRNA and β-action, respectively. Data were
analyzed using the 2−ΔΔCq method (20).
Cell transfection
miR-539 mimics and negative control miRNA mimics
(miR-NC) were obtained from GenePharma (Shanghai, China).
Insulin-like growth factor 1 receptor (IGF-1R) overexpression
vector (pCMV-IGF-1R) and empty pCMV vector were generated from
Amspring Biological Technology Co., Ltd. (Changsha, China). For
functional experiments, the cells were plated in 6-well plates 1
day prior to transfection. Cell transfection or cotransfection was
carried out using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The transfected cells were then cultured at 37°C in a
humidified atmosphere with 5% CO2, and the culture
medium was replaced with fresh DMEM containing 10% FBS at 6 h post
transfection.
Cell Counting Kit (CCK)-8 and colony
formation assays
CCK-8 assay was utilised to determine cell
proliferative ability. For CCK-8 assay, the transfected cells were
collected and seeded into 96-well plates at a density of
3×103 cells per well with 100 µl of the culture medium.
The extent of proliferation was evaluated at 0, 24, 48 and 72 h
after incubation at 37°C in a humidified atmosphere with 5%
CO2. At each time point, 10 µl of CCK-8 solution
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added,
and the cells were incubated for another 2 h. Absorbance was
detected at a wavelength of 450 nm by using an enzyme-linked
immunosorbent assay plate reader (BioTek Instruments, Inc.,
Winooski, VT, USA).
Colony formation assay was carried out to examine
the cell colony formation ability. The transfected cells were
harvested and plated into 6-well plates at a density of
1×103 cells per well. The plates were shaken to disperse
the cells equally, and the cells were cultured in an incubator at
37°C for 7 days. On day 8, the colonies were fixed with 100%
methanol, stained with 0.5% crystal violet and rinsed in
phosphate-buffered saline (PBS). The colonies were observed using a
microscope, and the colonies containing >50 cells were
counted.
Cell invasion assay
Transwell chambers with 8 µm pores (BD Biosciences,
San Jose, CA, USA) were applied to assess the cell invasive
ability. The upper chambers were precoated with 100 µl of diluted
Matrigel (1 mg/ml; BD Biosciences) and then incubated at 37°C for
additional 1 h. Afterwards, 1×105 transfected cells in
FBS-free DMEM were plated into the upper chambers, and 500 µl of
DMEM containing 10% FBS was added into the lower chambers. The
cells were cultured at 37°C for 24 h, and the cells remaining on
the upper chambers were scraped off gently with cotton swabs. The
invading cells were fixed with 100% methanol, stained with 0.5%
crystal violet and washed with PBS. The invading cells were
photographed and counted under an inverted microscope (CKX41;
Olympus, Tokyo, Japan) in five randomly selected fields.
Bioinformatics prediction
TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdcberlin.de/) were employed to predict
the potential targets of miR-539. IGF-1R, a well-known oncogene,
was predicted as a major highly conserved target of miR-539.
Luciferase reporter assay
Luciferase reporter plasmids, pMIR-IGF-1R-3′-UTR
wild-type (Wt) and pMIR-IGF-1R-3′-UTR mutant (Mut), were designed,
synthesised and confirmed by GenePharma. The cells were seeded into
24-well plates at a density of 6×104 cells per well 1
day prior to transfection. These cells were then transfected with
miR-539 mimics or miR-NC and cotransfected with pMIR-IGF-1R-3′-UTR
Wt or pMIR-IGF-1R-3′-UTR Mut by using Lipofectamine®
2000, according to the manufacturer's instructions. Relative
luciferase activities were measured at 48 h posttransfection with a
dual-luciferase reporter assay kit (Promega Corporation, Madison,
WI, USA) and normalised to that of Renilla activities.
Western blot analysis
Total protein of tissues or cells was lysed in a
radioimmunoprecipitation assay lysis buffer, and the concentration
of the total protein was detected with a bicinchoninic acid assay
kit (all from Beyotime Institute of Biotechnology, Haimen, China).
Equal amounts of protein were separated through 10% sodium dodecyl
sulfate polyacrylamide gel electrophoresis and transferred to
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). These membranes were blocked with 5% non-fat powdered milk in
TBS-Tween-20 (TBST), incubated with primary antibodies overnight at
4°C, washed with TBST thrice and further incubated with horseradish
peroxidase-conjugated secondary antibody at room temperature for 1
h. The protein signals were visualised using an enhanced
chemiluminescence reagents (Pierce; Thermo Fisher Scientific,
Inc.). Densitometric analysis was performed using Quantity One
software version 4.62 (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The primary antibodies used in this study include mouse
anti-human IGF-1R monoclonal antibody (1:1,000 dilution; cat no.
sc-462; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and mouse
anti-human GAPDH monoclonal antibody (1:1,000 dilution; cat no.
sc-365062; Santa Cruz Biotechnology, Inc.).
Statistical analysis
Data were expressed as mean ± standard deviation
from at least three separate experiments. SPSS 19.0 (SPSS, Inc.,
Chicago, IL, USA) was used to perform statistical analysis.
Qualitative data were analysed with chi-square test. Independent
Student's t-test and one-way ANOVA with Student-Newman-Keuls post
hoc test were performed to compare the differences between groups.
Spearman correlation analysis was used to examine the correlation
between miR-539 and IGF-1R mRNA in PDAC tissues. P<0.05 was
considered statistically significant.
Results
miR-539 is significantly downregulated
in PDAC tissues and cell lines
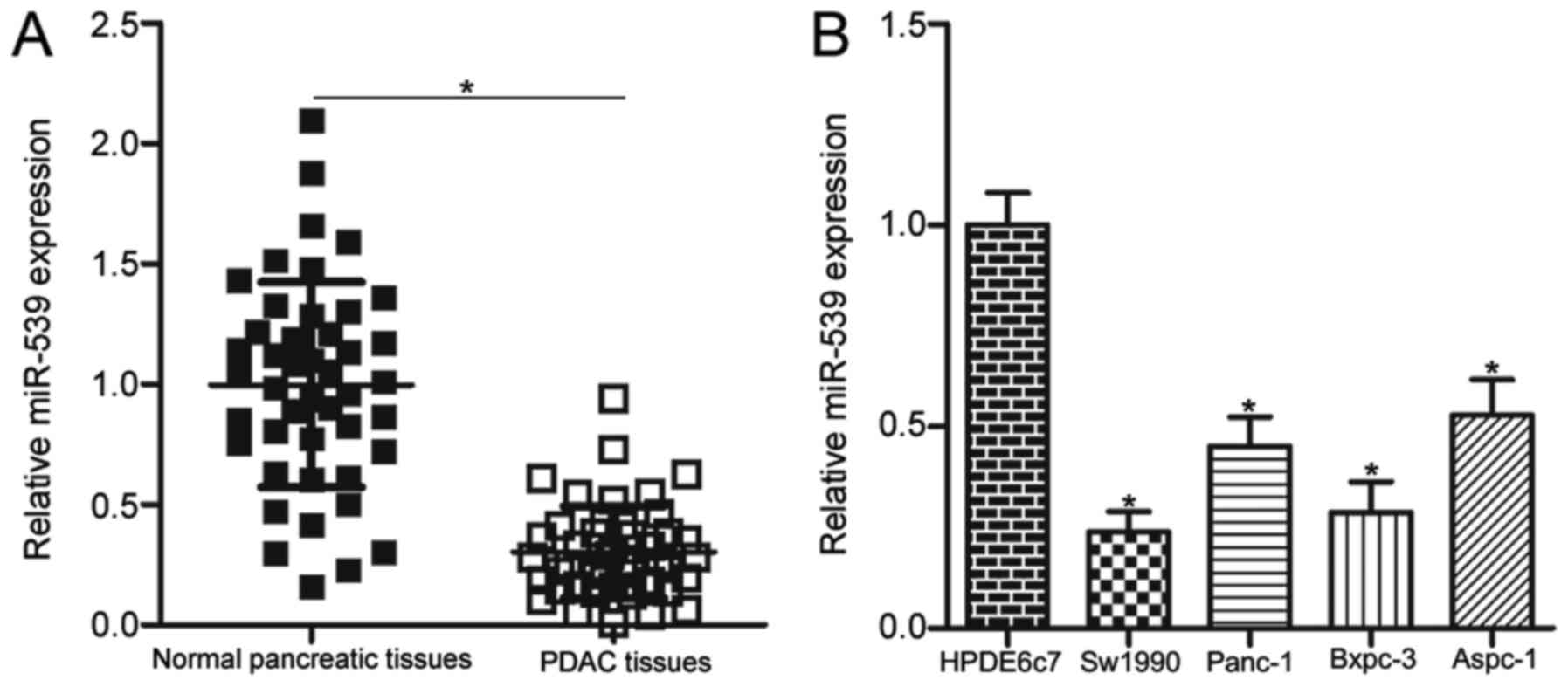
To investigate the expression pattern of miR-539 in
PDAC, we measured the miR-539 expression in 45 pairs of PDAC
tissues and adjacent normal pancreatic tissues using RT-qPCR. Our
data showed that the miR-539 expression obviously decreased in PDAC
tissues compared with that in adjacent normal pancreatic tissues
(Fig. 1A, P<0.05). The
association between this expression and the clinicopathological
characteristics of PDAC was evaluated to determine the clinical
value of miR-539 in PDAC. Table I
shows that the expression level of miR-539 was significantly
associated with TNM stage (P=0.011) and lymph node metastasis
(P=0.026). However, no correlation was observed between miR-539 and
other clinicopathological features, including age, sex, tumour site
and tumour differentiation (all P>0.05). RT-qPCR analysis was
performed to quantify the miR-539 expression in four PDAC cell
lines, including Sw1990, Panc-1, Bxpc-3 and Aspc-1. The expression
level of miR-539 was underexpressed in all four PDAC cell lines
compared with that of the normal human pancreatic cell line HPDE6c7
(P<0.05; Fig. 1B). These
results suggested that miR-539 downregulation might be correlated
with PDAC progression.
 | Table I.Association between miR-539
expression and clinicopathological characteristics of patients with
pancreatic ductal adenocarcinoma. |
Table I.
Association between miR-539
expression and clinicopathological characteristics of patients with
pancreatic ductal adenocarcinoma.
|
| miR-539
expression |
|
|---|
|
|
|
|
|---|
| Clinicopathological
characteristics | Low | High | P-value |
|---|
| Age |
|
| 0.273 |
| <60
years | 11 | 7 |
|
| ≥60
years | 12 | 15 |
|
| Sex |
|
| 0.465 |
|
Male | 15 | 12 |
|
|
Female | 8 | 10 |
|
| Tumour size |
|
| 0.661 |
| <2
cm | 10 | 11 |
|
| ≥2
cm | 13 | 11 |
|
| Tumour
differentiation |
|
| 0.449 |
|
Well | 11 | 13 |
|
|
Poor | 12 | 9 |
|
| TNM stage |
|
| 0.011a |
|
I–II | 5 | 13 |
|
|
III–IV | 18 | 9 |
|
| Lymph node
metastasis |
|
| 0.026a |
|
Negative | 7 | 14 |
|
|
Positive | 16 | 8 |
|
miR-539 plays a negative role in human
PDAC cell proliferation, colony formation and invasion
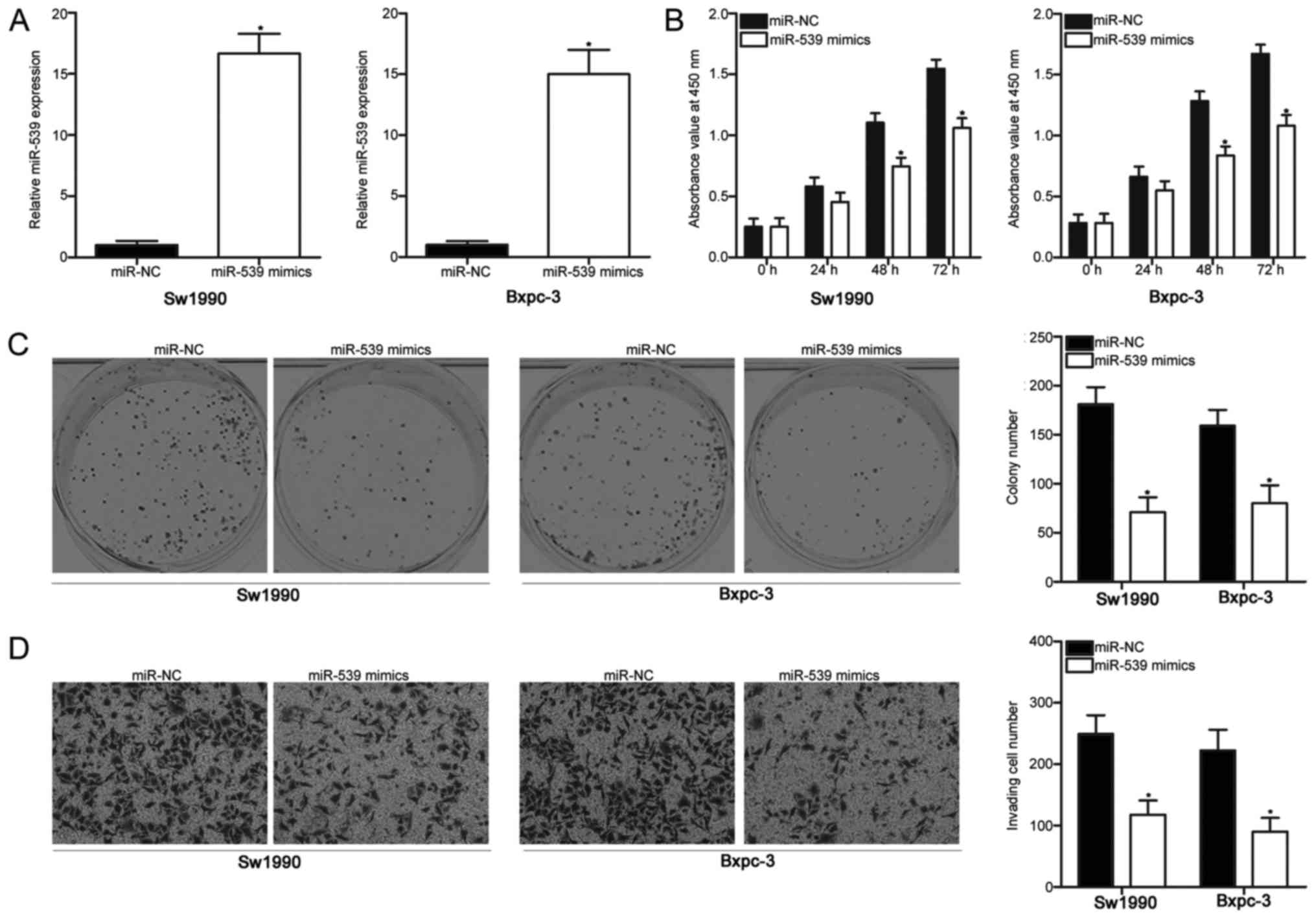
To evaluate the effect of miR-539 on the oncogenic
phenotype of PDAC, we upregulated the miR-539 expression in Sw1990
and Bxpc-3 cells, whose endogenous miR-539 expression was
relatively lower among that of the four PDAC cell lines (P<0.05;
Fig. 2A). The effect of miR-539
overexpression on PDAC cell proliferation was examined with a CCK-8
assay. We found that miR-539 upregulation reduced Sw1990 and Bxpc-3
cell proliferation (P<0.05; Fig.
2B). A colony formation assay was further conducted to confirm
the inhibitory effect of miR-539 on PDAC cell proliferation. In
Fig. 2C, the upregulation of
miR-539 caused a significant decrease in the colony formation of
Sw1990 and Bxpc-3 cells (P<0.05). We next explored the effect of
miR-539 on the cell invasion ability of PDAC. The results of cell
invasion assay revealed that miR-539 overexpression decreased the
invasion capacities of Sw1990 and Bxpc-3 cells (P<0.05; Fig. 2D). These results suggested that
miR-539 could perform tumour suppressive roles in PDAC.
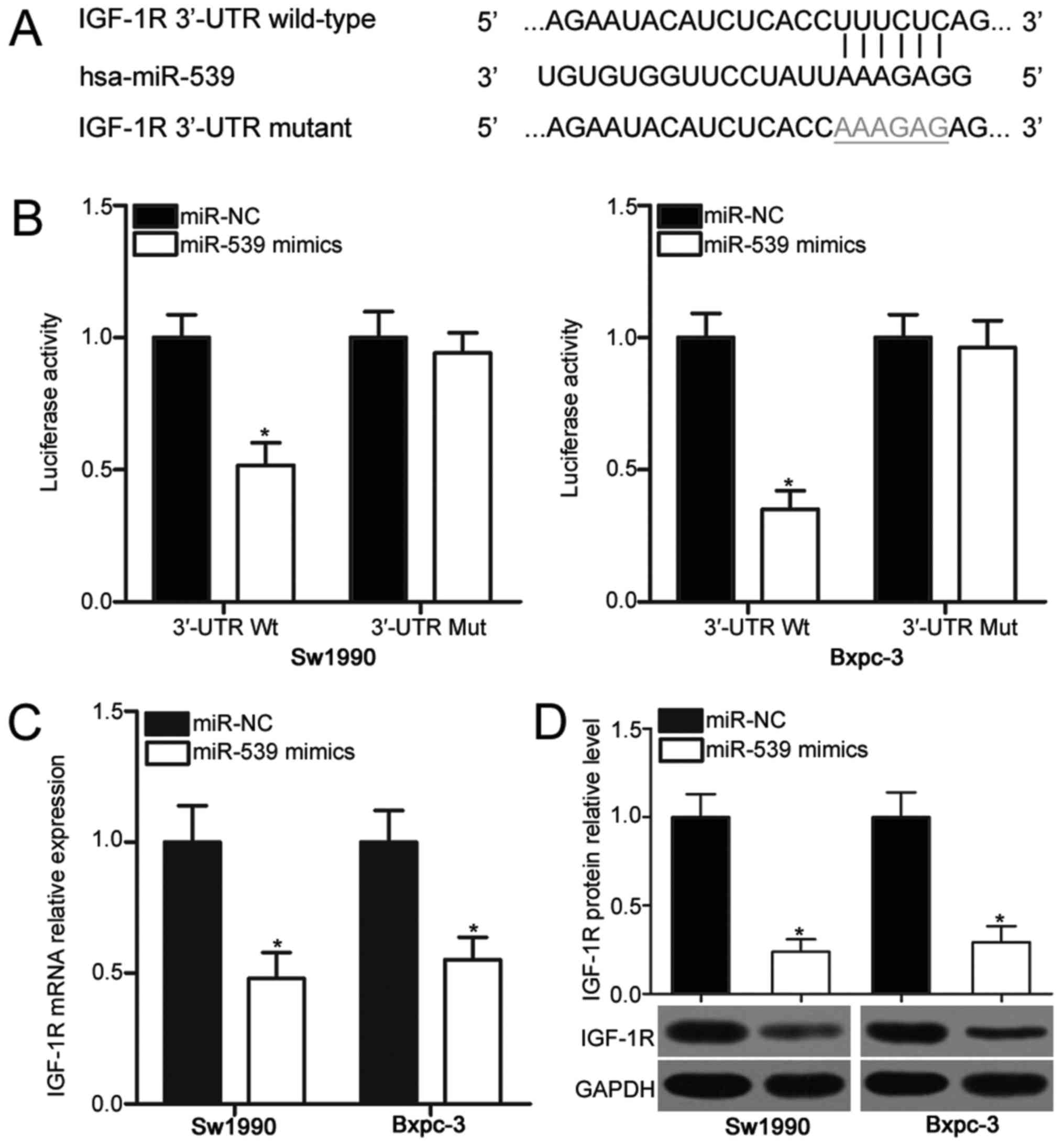
IGF-1R is a direct target of miR-539
in PDAC
To elucidate the mechanisms by which miR-539
executed its inhibitory effects on PDAC cells, we predicted the
putative targets of miR-539 through bioinformatics prediction.
IGF-1R (Fig. 3A), a well-known
oncogene in PDAC (21–27), was predicted as a major highly
conserved target of miR-539 and was selected for further analysis.
Luciferase reporter assay was performed to validate this
speculation and to investigate whether miR-539 could directly
interact with the 3′-UTR of IGF-1R. miR-539 mimics or miR-NC was
transfected into Sw1990 and Bxpc-3 cells in combination with
pMIR-Report vector containing wild-type (Wt) IGF-1R 3′-UTR or
mutant (Mut) IGF-1R 3′-UTR. The ectopic of miR-539 expression
resulted in a significant decrease in the luciferase activities of
pMIR-IGF-1R-3′-UTR Wt (P<0.05). However, the mutation of the
binding sequences of miR-539 in the 3′-UTR of IGF-1R abolished the
suppressive effect of miR-539 on luciferase activities (Fig. 3B). We also examined the effect of
miR-539 on the endogenous IGF-1R expression in PDAC. RT-qPCR and
Western blot analysis demonstrated that the enforced expression of
miR-539 reduced the IGF-1R expression in Sw1990 and Bxpc-3 cells at
mRNA (P<0.05; Fig. 3C) and
protein (P<0.05; Fig. 3D)
levels. These results evidently suggested that IGF-1R is a direct
target of miR-539 in PDAC.
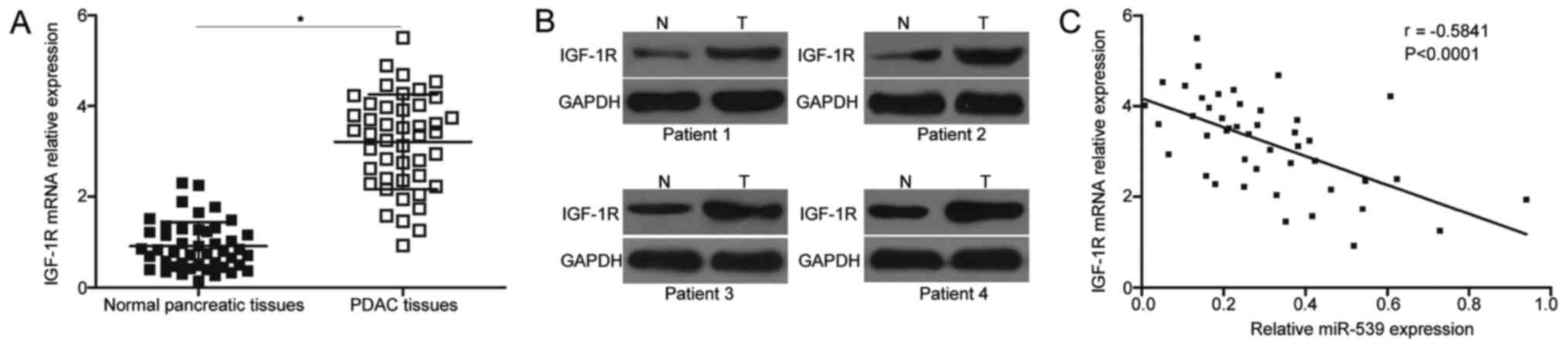
IGF-1R upregulation in PDAC tissues is
inversely correlated with miR-539 level
RT-qPCR analysis was carried out on 45 pairs of PDAC
tissues and adjacent normal pancreatic tissues to further examine
the association between miR-539 and IGF-1R in PDAC. The mRNA
expression level of IGF-1R was remarkably overexpressed in the PDAC
tissues compared with that in the adjacent normal pancreatic
tissues (P<0.05; Fig. 4A).
Western blot analysis also revealed that PDAC tissues exhibited a
significantly upregulated protein level of IGF-1R compared with
that in the adjacent normal pancreatic tissues (Fig. 4B). Furthermore, the association
between the mRNA expression of IGF-1R and miR-539 levels in the
PDAC tissues was examined through Spearman correlation analysis. In
Fig. 4C, the mRNA expression of
IGF-1R was inversely associated with miR-539 expression levels in
the PDAC tissues (r=−0.5841, P<0.0001). These results suggested
that the miR-539 downregulation might at least partly increase the
IGF-1R expression in PDAC tissues.
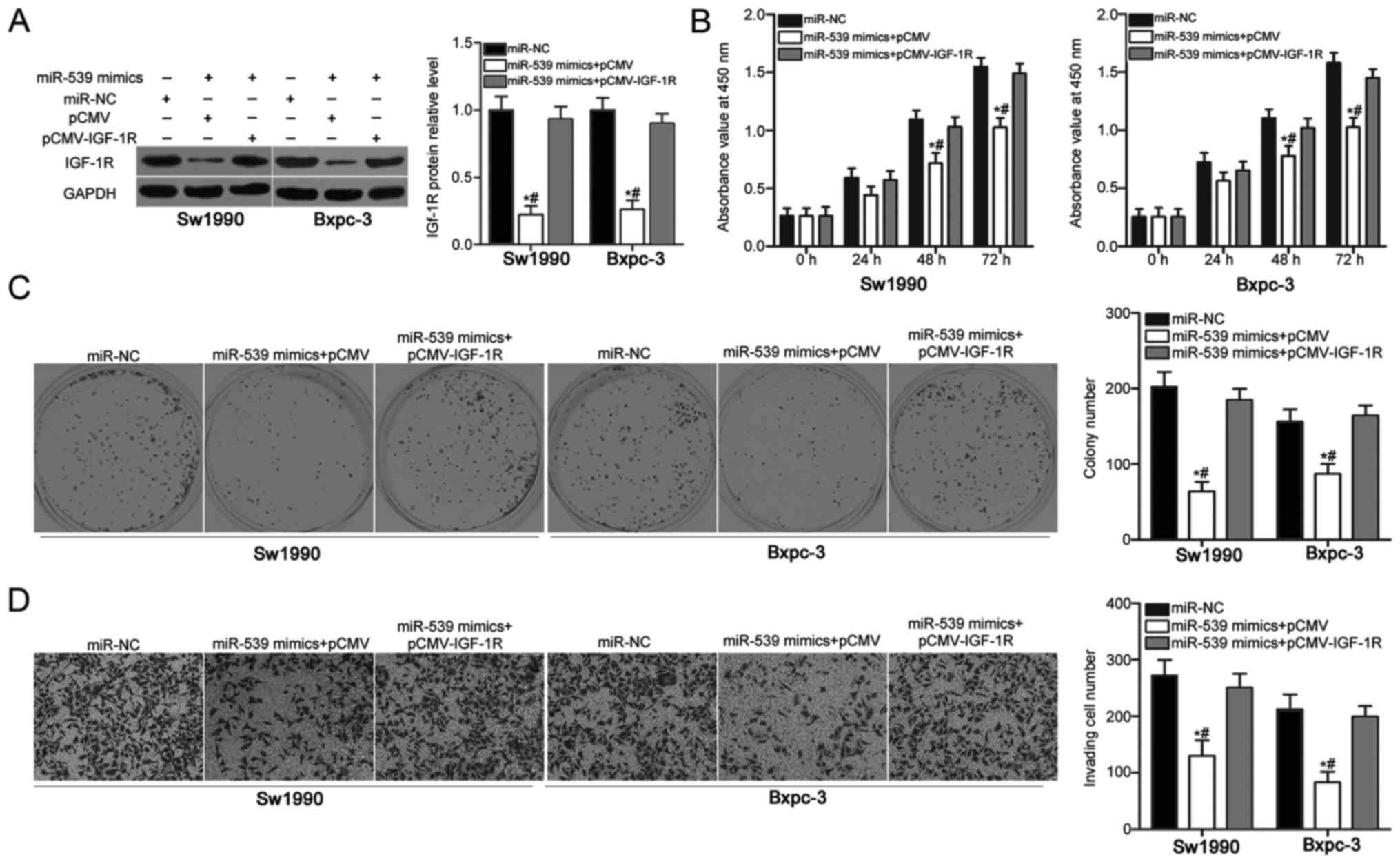
Recovered IGF-1R expression partially
counteracts the suppressive effects of miR-539 overexpression on
PDAC cells
A series of rescue experiments were applied to
further address that the tumor suppressive roles of miR-539 on PDAC
cells were mediated by the inhibition of IGF-1R. Sw1990 and Bxpc-3
cells were cotransfected with miR-539 mimics and empty pCMV vector
or IGF-1R overexpression vector (pCMV-IGF-1R). After transfection
for 72 h, western blot analysis confirmed that the IGF-1R protein
levels were restored in Sw1990 and Bxpc-3 cells cotransfected with
miR-539 mimics and pCMV-IGF-1R compared with those in the cells
cotransfected with miR-539 mimics and pCMV (P<0.05; Fig. 5A). CCK-8, colony formation and cell
invasion assays indicated that the IGF-1R overexpression partially
rescued the inhibitory effects of the miR-539 overexpression on
Sw1990 and Bxpc-3 cell proliferation (P<0.05; Fig. 5B), colony formation (P<0.05;
Fig. 5C) and invasion (P<0.05;
Fig. 5D). Overall, the
tumour-suppressing effects of miR-539 overexpression on PDAC cells
are partly achieved by downregulating the IGF-1R expression.
Discussion
miRNAs possess the oncogenic and tumour-suppressive
roles in the carcinogenesis and progression of PDAC by regulating
the expression of numerous cancer-related genes (28–30).
Thus, the investigation on the expression and roles of miRNAs in
PDAC may promote the identification of novel and effective targets
for the clinical diagnosis and treatment of patients with this
cancer. This study is the first to explore the expression pattern,
biological roles and underlying mechanisms of miR-539 in PDAC.
Here, we found that miR-539 was obviously downregulated in the PDAC
tissues and cell lines. The decreased miR-539 level was strongly
correlated with TNM stage and lymph node metastasis. Functional
experiments indicated that the upregulation of miR-539 restricted
the cell proliferation, colony formation and invasion of PDAC.
Furthermore, IGF-1R was confirmed as a direct target of miR-539 in
PDAC. IGF-1R upregulation in the PDAC tissues was inversely
correlated with the miR-539 level. IGF-1R overexpression partially
rescued the suppressive roles in the PDAC cells induced by miR-539
overexpression. These results suggested that miR-539 might be a
suitable therapeutic target for the treatment of patients with this
fatal malignancy.
miR-539 dysregulation has been observed in multiple
types of human cancer. For example, miR-539 is downregulated in
colorectal cancer tissues, and this phenomenon is associated with
clinical stage and lymph node metastasis (17). The expression pattern of miR-539 is
also decreased in glioma (18),
oesophageal cancer (19),
hepatocellular carcinoma (31,32),
prostate cancer (33),
nasopharyngeal carcinoma (34),
osteosarcoma (35) and thyroid
cancer (36). These findings
suggested that miR-539 downregulation is a common event in human
cancer and may represent useful markers for cancer diagnosis.
miR-539 performs important functional roles in
tumour occurrence and development. For instance, miR-539
re-expression suppresses the growth and metastasis of colorectal
cancer cells in vitro and impairs tumour growth in
vivo (17). Quan et al
(18) found that ectopic miR-539
expression causes an evident reduction in the cell proliferation
and invasion of glioma. Li et al (19) reported that the ectopic of miR-539
expression represses the epithelial-to-mesenchymal transition of
cells in oesophageal cancer. Zhu (31) and Liu et al (32) demonstrated that miR-539
upregulation restricts the growth and metastasis of hepatocellular
carcinoma cells, induces apoptosis in vitro, decreases
tumour growth and tumourigenesis in vivo and increases the
chemosensitivity of trioxide-resistant cells to arsenic trioxide.
Zhang et al (33) revealed
that miR-539 overexpression inhibits the proliferation, migration
and invasion of prostate cancer cells in vitro and in
vivo. Lv et al indicated that the induced miR-539
expression attenuates the proliferation of nasopharyngeal carcinoma
cells, triggers cell cycle arrest in vitro and reduces cell
growth in vivo (34). Jin
and Wang (35) and Gu and Sun
(36) showed that the resumption
of miR-539 expression prevents the migration and invasion of
osteosarcoma and thyroid cancer cells. These findings suggested
that exogenous miR-539 may have a therapeutic value for patients
with cancer.
miRNAs can affect carcinogenesis and cancer
progression by directly regulating target genes expression. Several
targets identified for miR-539 thus far include RUNX2 (17) in colorectal cancer, DIXDC1
(18) in glioma, TWIST1 (19) in esophageal cancer, FSCN1 (32) in hepatocellular carcinoma, SPAG5
(33) in prostate cancer, CDK4
(34) in nasopharyngeal carcinoma,
MMP8 (35) in osteosarcoma and
CARMA1 (36) in thyroid cancer. In
our study, IGF-1R, a transmembrane tyrosine kinase receptor, was
validated as a novel target of miR-539 in PDAC. IGF-1R protein is
composed of two extracellular α subunits with a ligand-binding site
and two transmembrane β subunits with intracellular tyrosine kinase
activity (37). IGF-1R is
overexpressed in numerous kinds of human malignancies, such as
ovarian cancer (38),
hepatocellular carcinoma (39),
renal cell carcinoma (40), lung
cancer (41), gastric cancer
(42) and bladder cancer (43). IGF-1R is also highly expressed in
PDAC tissues, and this upregulation is strongly associated with
tumour location, histological grade and TNM stage (21,22).
The prognosis of patients with PDAC with a high IGF-1R expression
is poorer than that of patients with a low IGF-1R expression
(22). IGF-1R dysregulation is
implicated in the aggressiveness of PDAC by regulating various
pathological processes, such as cell proliferation, apoptosis,
migration, invasion, epithelial-to-mesenchymal transition,
angiogenesis and chemoresistance (23–27).
Thus, the inhibition of IGF-1R is a promising therapeutic strategy
for patients with PDAC.
In conclusion, this study demonstrated that the
miR-539 expression was downregulated in PDAC tissues and cell
lines. Decreased miR-539 levels were significantly correlated with
TNM stage and lymph node metastasis. miR-539 upregulation
restricted the proliferation, colony foramtion and invasion of PDAC
cells by directly targeting and inhibiting IGF-1R. These findings
may provide novel insights into the mechanisms associated with the
rapid growth and early metastasis of PDAC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SL and YL designed the research. YL, LR, JZ, and RL
performed functional experiments. All authors read and approved the
final draft.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Yidu Central Hospital of Weifang, and was performed in
accordance with the Declaration of Helsinki and the guidelines of
the Ethics Committee of Yidu Central Hospital of Weifang. Written
informed consent was obtained from all patients for the use of
their clinical tissues.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Modolell I, Guarner L and Malagelada JR:
Vagaries of clinical presentation of pancreatic and biliary tract
cancer. Ann Oncol. 10 Suppl 4:82–84. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jiao F, Hu H, Yuan C and Wang L, Jiang W,
Jin Z, Guo Z and Wang L: Elevated expression level of long
noncoding RNA MALAT-1 facilitates cell growth, migration and
invasion in pancreatic cancer. Oncol Rep. 32:2485–2492. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maitra A and Hruban RH: Pancreatic cancer.
Annu Rev Pathol. 3:157–188. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moir J, White SA, French JJ, Littler P and
Manas DM: Systematic review of irreversible electroporation in the
treatment of advanced pancreatic cancer. Eur J Surg Oncol.
40:1598–1604. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Burkey MD, Feirman S, Wang H, Choudhury
SR, Grover S and Johnston FM: The association between smokeless
tobacco use and pancreatic adenocarcinoma: A systematic review.
Cancer Epidemiol. 38:647–653. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan Y, Shi C, Li T and Kuang T:
MicroRNA-454 shows anti-angiogenic and anti-metastatic activity in
pancreatic ductal adenocarcinoma by targeting LRP6. Am J Cancer
Res. 7:139–147. 2017.PubMed/NCBI
|
|
10
|
Zhang G, Ai D, Yang X, Ji S, Wang Z and
Feng S: MicroRNA-610 inhibits tumor growth of melanoma by targeting
LRP6. Oncotarget. 8:97361–97370. 2017.PubMed/NCBI
|
|
11
|
Jin Y, Tao LP, Yao SC, Huang QK, Chen ZF,
Sun YJ and Jin SQ: MicroRNA-582-5p suppressed gastric cancer cell
proliferation via targeting AKT3. Eur Rev Med Pharmacol Sci.
21:5112–5120. 2017.PubMed/NCBI
|
|
12
|
Wang X and Wu X: The Role of
MicroRNA-1207-5p in colorectal cancer. Clin Lab. 63:1875–1882.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang D, Du G, Xu A, Xi X and Li D:
Expression of miR-149-3p inhibits proliferation, migration, and
invasion of bladder cancer by targeting S100A4. Am J Cancer Res.
7:2209–2219. 2017.PubMed/NCBI
|
|
14
|
Liu S, Liu K, Zhang W, Wang Y, Jin Z, Jia
B and Liu Y: miR-449a inhibits proliferation and invasion by
regulating ADAM10 in hepatocellular carcinoma. Am J Transl Res.
8:2609–2619. 2016.PubMed/NCBI
|
|
15
|
Cheng RF, Wang J, Zhang JY, Sun L, Zhao
YR, Qiu ZQ, Sun BC and Sun Y: MicroRNA-506 is up-regulated in the
development of pancreatic ductal adenocarcinoma and is associated
with attenuated disease progression. Chin J Cancer. 35:642016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tu MJ, Pan YZ, Qiu JX, Kim EJ and Yu AM:
MicroRNA-1291 targets the FOXA2-AGR2 pathway to suppress pancreatic
cancer cell proliferation and tumorigenesis. Oncotarget.
7:45547–45561. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wen D, Li S, Jiang W, Zhu J, Liu J and
Zhao S: miR-539 inhibits human colorectal cancer progression by
targeting RUNX2. Biomed Pharmacother. 95:1314–1320. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Quan J, Qu J and Zhou L: MicroRNA-539
inhibits glioma cell proliferation and invasion by targeting
DIXDC1. Biomed Pharmacother. 93:746–753. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li S, Yang F, Wang M, Cao W and Yang Z:
miR-378 functions as an onco-miRNA by targeting the
ST7L/Wnt/β-catenin pathway in cervical cancer. Int J Mol Med.
40:1047–1056. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu JW, Wang TX, You L, Zheng LF, Shu H,
Zhang TP and Zhao YP: Insulin-like growth factor 1 receptor
(IGF-1R) as a target of MiR-497 and plasma IGF-1R levels associated
with TNM stage of pancreatic cancer. PLoS One. 9:e928472014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hirakawa T, Yashiro M, Murata A, Hirata K,
Kimura K, Amano R, Yamada N, Nakata B and Hirakawa K: IGF-1
receptor and IGF binding protein-3 might predict prognosis of
patients with resectable pancreatic cancer. BMC Cancer. 13:3922013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramani R, Lopez-Valdez R, Arumugam A,
Nandy S, Boopalan T and Lakshmanaswamy R: Targeting insulin-like
growth factor 1 receptor inhibits pancreatic cancer growth and
metastasis. PLoS One. 9:e970162014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng D, Golubovskaya V, Kurenova E, Wood
C, Massoll NA, Ostrov D, Cance WG and Hochwald SN: A novel strategy
to inhibit FAK and IGF-1R decreases growth of pancreatic cancer
xenografts. Mol Carcinog. 49:200–209. 2010.PubMed/NCBI
|
|
25
|
Farhana L, Dawson MI, Murshed F, Das JK,
Rishi AK and Fontana JA: Upregulation of miR-150* and miR-630
induces apoptosis in pancreatic cancer cells by targeting IGF-1R.
PLoS One. 8:e610152013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tian X, Hao K, Qin C, Xie K, Xie X and
Yang Y: Insulin-like growth factor 1 receptor promotes the growth
and chemoresistance of pancreatic cancer. Dig Dis Sci.
58:2705–2712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moser C, Schachtschneider P, Lang SA,
Gaumann A, Mori A, Zimmermann J, Schlitt HJ, Geissler EK and
Stoeltzing O: Inhibition of insulin-like growth factor-I receptor
(IGF-IR) using NVP-AEW541, a small molecule kinase inhibitor,
reduces orthotopic pancreatic cancer growth and angiogenesis. Eur J
Cancer. 44:1577–1586. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qadir MI and Faheem A: miRNA: A diagnostic
and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene
Expr. 27:197–204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang S, Gong X, Zhang G, Huang G, Lu Y
and Li Y: MicroRNA-140 regulates cell growth and invasion in
pancreatic duct adenocarcinoma by targeting iASPP. Acta Biochim
Biophys Sin (Shanghai). 48:174–181. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Keklikoglou I, Hosaka K, Bender C, Bott A,
Koerner C, Mitra D, Will R, Woerner A, Muenstermann E1, Wilhelm H,
et al: MicroRNA-206 functions as a pleiotropic modulator of cell
proliferation, invasion and lymphangiogenesis in pancreatic
adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene.
34:4867–4878. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu C, Zhou R, Zhou Q, Chang Y and Jiang
M: microRNA-539 suppresses tumor growth and tumorigenesis and
overcomes arsenic trioxide resistance in hepatocellular carcinoma.
Life Sci. 166:34–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Hong W, Zhou C, Jiang Z, Wang G,
Wei G and Li X: miR-539 inhibits FSCN1 expression and suppresses
hepatocellular carcinoma migration and invasion. Oncol Rep.
37:2593–2602. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang H, Li S, Yang X, Qiao B, Zhang Z and
Xu Y: miR-539 inhibits prostate cancer progression by directly
targeting SPAG5. J Exp Clin Cancer Res. 35:602016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lv LY, Wang YZ, Zhang Q, Zang HR and Wang
XJ: miR-539 induces cell cycle arrest in nasopharyngeal carcinoma
by targeting cyclin-dependent kinase 4. Cell Biochem Funct.
33:534–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jin H and Wang W: MicroRNA-539 suppresses
osteosarcoma cell invasion and migration in vitro and targeting
Matrix metallopeptidase-8. Int J Clin Exp Pathol. 8:8075–8082.
2015.PubMed/NCBI
|
|
36
|
Gu L and Sun W: MiR-539 inhibits thyroid
cancer cell migration and invasion by directly targeting CARMA1.
Biochem Biophys Res Commun. 464:1128–1133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu Q, Gong JP, Li J, Zhong SL, Chen WX,
Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH and Tang JH: Down-regulation
of miRNA-452 is associated with adriamycin-resistance in breast
cancer cells. Asian Pac J Cancer Prev. 15:5137–5142. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
An Y, Cai L, Wang Y, Zhu D, Guan Y and
Zheng J: Onkologie. Onkologie. 32:638–644. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
E C, Li J, Shao D, Zhang D, Pan Y, Chen L
and Zhang X: The insulin-like growth factor-I receptor inhibitor
picropodophyllin-induced selective apoptosis of hepatocellular
carcinoma cell through a caspase-dependent mitochondrial pathway.
Oncol Res. 21:103–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sichani MM, Yazdi FS, Moghaddam NA,
Chehrei A, Kabiri M, Naeimi A and Taheri D: Prognostic value of
insulin-like growth factor-I receptor expression in renal cell
carcinoma. Saudi J Kidney Dis Transpl. 21:69–74. 2010.PubMed/NCBI
|
|
41
|
Zhao S, Qiu Z, He J, Li L and Li W:
Insulin-like growth factor receptor 1 (IGF1R) expression and
survival in non-small cell lung cancer patients: A meta-analysis.
Int J Clin Exp Pathol. 7:6694–6704. 2014.PubMed/NCBI
|
|
42
|
Gryko M, Kiśluk J, Cepowicz D, Zińczuk J,
Kamocki Z, Guzińska-Ustymowicz K, Pryczynicz A, Czyżewska J, Kemona
A and Kędra B: Expression of insulin-like growth factor receptor
type 1 correlate with lymphatic metastases in human gastric cancer.
Pol J Pathol. 65:135–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xie QX, Lin XC, Zhang MF, Han CX and Guo
YH: Expression of IGF-I and IGF-IR in bladder cancer. Ai Zheng.
23:707–709. 2004.(In Chinese). PubMed/NCBI
|