Introduction
Increasing attention has been focused on the
function and significance of mRNA in sperm in light of the role it
serves in sperm development and maintenance (1,2).
Thus, mRNAs that aid in detection of sperm abnormalities are
potential biomarkers to evaluate the quality of sperm in the
diagnosis and treatment of male infertility (3–5).
Teratozoospermia is a condition characterized by a
large number of spermatozoa with abnormal morphology and is
considered to be a factor that may result in male infertility
(6). There are two manifestations
of teratozoospermia: Monomorphic morphological defects and
flagellum morphological defects (6,7).
Previous studies have demonstrated that abnormal expression of mRNA
is a primary cause of abnormal sperm morphology (6–9).
Kang-Decker et al (8)
reported that male mice with ArfGAP with FG repeats 1 depletion
were infertile due to a lack of acrosome formation. Casein kinase 2
α 2 knock-out male mice were also infertile due to abnormal
morphology of the spermatid nucleus (9). Sptrx-2, expressed exclusively in
human testis, was reported to be associated with flagellar
anomalies (10). Allegrucci et
al (11) also reported
specific epigenetic signatures of flagellar anomalies. However, the
specific mechanism underlying male infertility remains to be
elucidated, as it is a complex process involving a large number of
genes (12).
The rapid development of high-throughput
technologies, including microarrays and RNA-sequencing has resulted
in successful profiling of RNA expression, which enhances
understanding of various diseases and helps further exploration of
their underlying molecular mechanisms (13). HM et al (14) detected the gene expression of
crossbred cattle sperm by microarray assessment and identified 305
genes that were significantly and differentially expressed. Hu
et al (15) profiled long
non-coding (lnc)RNA expression in male mice germ cells and revealed
that a variety of lncRNAs may regulate male reproduction by serving
as competing-endogenous RNAs to modulate the function of germ
cells.
Exome sequencing analysis of two brothers with
azoospermia demonstrated that the deficiency of homozygous serine
peptidase inhibitor, Kazal type 2 is a factor in the development of
azoospermia (16). However, to the
best of our knowledge, there are limited studies that have
systematically analyzed the gene expression profiles in patients
with teratozoospermia using integration bioinformatics analysis.
The present study obtained a gene expression dataset for
teratozoospermia from the Gene Expression Omnibus (4) and performed systemic bioinformatics
analysis, including identification of differentially expression
genes (DEGs), functional enrichment analysis, co-expression network
analysis and identified several significantly and differentially
expressed biomarkers for teratozoospermia. The results of the
present study may be beneficial in understanding the mechanism
underlying teratozoospermia.
Materials and methods
Microarray data
The microarray dataset GSE6872 was downloaded from
the GEO website (ncbi.nlm.nih.gov/gds/) which was based on the GPL570
platform. This dataset was submitted by Platts et al
(17) and included 13 semen
samples, collected from healthy fertile males. A total of 8 semen
samples were collected from infertile patients with
teratozoospermia without any other abnormal semen parameters.
Data preprocessing
The present study imported original CEL data into R
(version 3.2.4, http://www.r-project.org/) and used an Affy R-package
(Bioconductor version 3.6) to correct data background and data
normalization. The mas5calls method for AffyBatch returns an
ExpressionSet by multi probes which correspond to specific
genes.
Differentially expressed gene
selection
DEGs were identified between 13 healthy semen and 8
infertile semen samples, using the limma package (version 3.6,
http://bioinf.wehi.edu.au/limma). False
discovery rate (FDR)-value <0.01 and |log2 fold change| >2
were selected as the cutoff values.
Functional annotation and pathway
analysis of DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID V6.8; http://david.ncifcrf.gov/) (18) was used to annotate and conclude
gene lists or protein identifiers via comprehensive categorical
data for Gene Ontology (GO) (19).
In order to extensively evaluate connected pathways and biological
processes associated with teratozoospermia, pathway enrichment
analyses of DEGs were performed with the DAVID analysis system,
with a threshold of P≤0.05.
Protein interaction network and module
analyses
The STRING database (http://string-db.org) was used to construct a
protein-protein interaction (19)
network of upregulated and downregulated DEGs, with a cutoff score
of >0.4. The significant modules from the constructed PPI
network of downregulated DEGs were selected using the ClusterONE
plugin of the Cytoscape software v3.6.1 (cytoscape.org/plugins.html) with P<0.01 considered
to indicate a statistical significance.
Results
Analysis of DEGs
The expression profile data were pre-processed and
then analyzed with the Affy package in R language. The whole gene
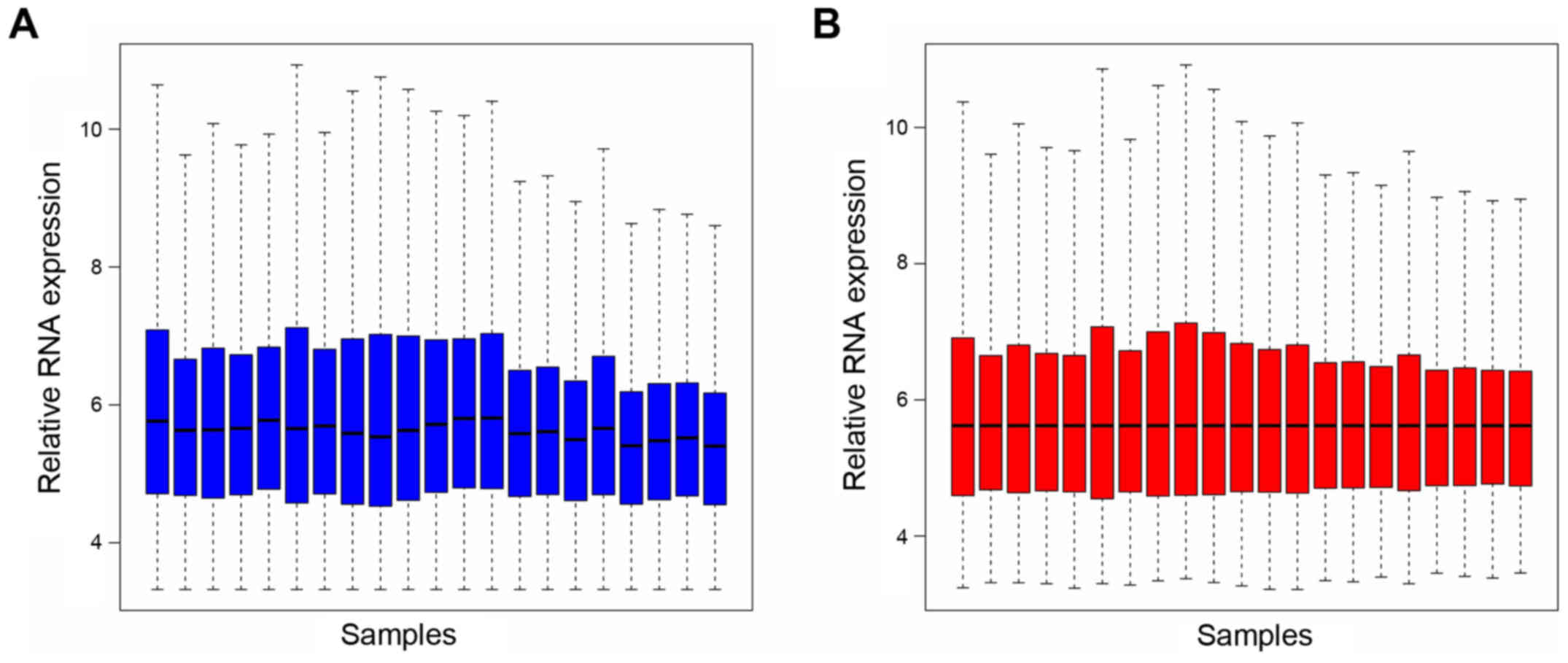
expression was screened. Box plots following data standardization
are presented in Fig. 1A and B.
Median values in Fig. 1 are
similar, which suggests a good degree of standardization. All RNA
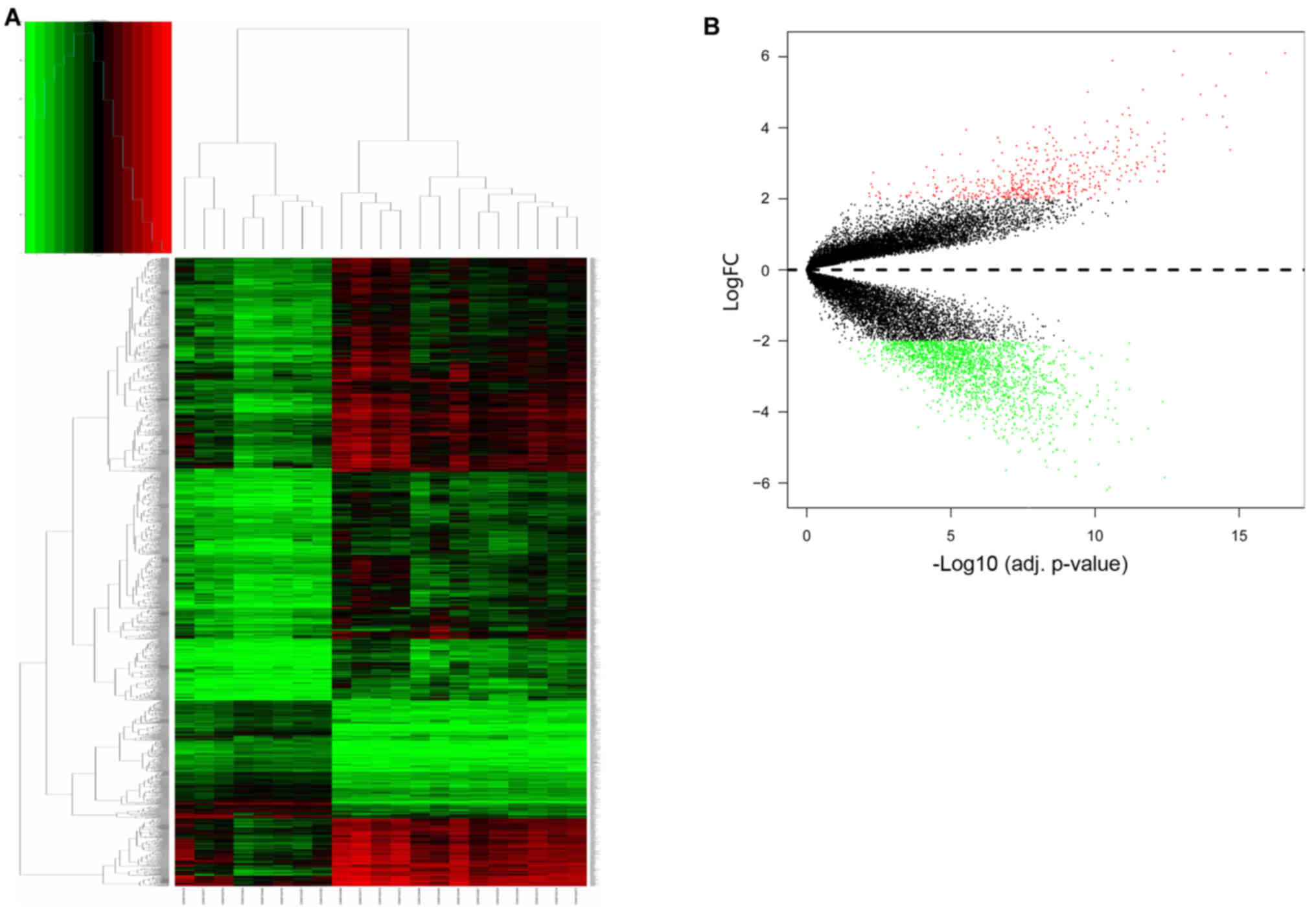
expression levels are presented in Fig. 2A. Hierarchical cluster analysis
indicated that the 8 samples from patients with teratozoospermia
and the 13 normal samples exhibited differing distributions. The
results revealed that grouping was reasonable, and the data
successfully underwent further analysis. Microarray data from the
normal semen samples were compared with those from the
teratozoospermia semen samples and a total of 2,392 DEGs were
identified. There were 450 upregulated genes and 1,942
downregulated genes (Fig. 2B). The
top 10 upregulated genes were heparan sulfate-glucosamine
3-sulfotransferase 3A1 (HS3ST3A1), XK related 4, armadillo-like
helical domain containing 4, hydatidiform mole associated and
imprinted (non-protein coding), WNT inhibitory factor 1, SLIT-ROBO
Rho GTPase activating protein 2C, SLIT-ROBO Rho GTPase activating
protein 2, DQ592442 (GenBank, http://www.ncbi.nlm.nih.gov/genbank/),
monoacylglycerol O-acyltransferase 1 and LOC101928622. The most
unregulated gene HS3ST3A1 is a component in heparan sulfate
generation pathway that few studies reported to be associated with
spermatogenesis (20,21). The top 10 downregulated genes were
zona pellucida binding protein (ZPBP), pancreatic progenitor cell
differentiation and proliferation factor, microseminoprotein β,
TSSK6 activating cochaperone, prolactin induced protein,
transcription elongation factor A like 4, ribosomal protein
(16) S5, ribosomal protein L7a
pseudogene 12, semenogelin 1 and semenogelin 2 (Table I). The protein produced from the
most downregulated gene, ZPBP, is usually located in the acrosome
of spermatozoa (22). Abnormal
morphogenesis is a major performance if patient lace of ZPBP
expression (22).
 | Table I.Top 10 upregulated and downregulated
DEGs. |
Table I.
Top 10 upregulated and downregulated
DEGs.
| A, Upregulated
DEGs |
|---|
| Gene | logFC | AveExpr | t | P-value | Adjusted P-value | B |
|---|
| HS3ST3A1 | 6.15829 | 6.264543 | 22.41724 |
1.07×10−16 |
1.78×10−13 | 28.22125 |
| XKR4 | 6.108402 | 6.904738 | 38.15492 |
1.13×10−21 |
2.44×10−17 | 38.46529 |
| C14orf37 | 6.089262 | 5.604961 | 29.23487 |
3.63×10−19 |
1.97×10−15 | 33.48849 |
| HYMAI | 5.891272 | 7.247713 | 16.23585 |
9.01×10−14 |
2.38×10−11 | 21.67427 |
| WIF1 | 5.55279 | 5.831665 | 34.46781 |
1.03×10−20 |
1.11×10−16 | 36.61418 |
| SRGAP2C | 5.491472 | 6.150009 | 23.34658 |
4.51×10−17 |
8.89×10−14 | 29.03932 |
| SRGAP2 | 5.182356 | 5.577265 | 26.86696 |
2.24×10−18 |
6.06×10−15 | 31.83706 |
| DQ592442 | 5.070846 | 7.77992 | 19.09086 |
3.15×10−15 |
2.07×10−12 | 24.96463 |
| MOGAT1 | 5.004399 | 5.407998 | 14.35237 |
1.10×10−12 |
1.71×10−10 | 19.18967 |
| LOC101928622 | 4.938621 | 5.265134 | 25.09824 |
9.64×10−18 |
2.09×10−14 | 30.48735 |
|
| B, downregulated
degs |
|
| Gene | logFC | AveExpr | t | P-value | Adjusted
P-value | B |
|
| ZPBP | −5.39989 | 9.272953 | −13.465 |
3.91×10−12 |
4.71×10−10 | 17.91695 |
| PPDPF | −5.48137 | 7.809478 | −15.1896 |
3.50×10−13 |
7.22×10−11 | 20.32858 |
| MSMB | −5.54533 | 8.057753 | −11.4953 |
8.49×10−11 |
4.52×10−09 | 14.82184 |
| TSACC | −5.62232 | 9.854213 | −12.3248 |
2.22×10−11 |
1.74×10−09 | 16.17356 |
| PIP | −5.63595 | 8.930515 | −9.11178 |
6.10×10−09 |
1.17×10−07 | 10.49482 |
| TCEAL4 | −5.81103 | 7.351813 | −13.5001 |
3.72×10−12 |
4.57×10−10 | 17.96871 |
| RPS5 | −5.83978 | 7.284755 | −21.5724 |
2.41×10−16 |
3.71×10−13 | 27.44499 |
| RPL7AL2 | −6.12204 | 7.736477 | −16.0315 |
1.17×10−13 |
3.01×10−11 | 21.41787 |
| SEMG1 | −6.1932 | 10.05513 | −15.8135 |
1.54×10−13 |
3.67×10−11 | 21.14096 |
| SEMG2 | −6.69875 | 7.822177 | −10.4268 |
5.32×10−10 |
1.81×10−08 | 12.96753 |
Functional and pathway enrichment
analysis
A total of 450 upregulated and 1,942 downregulated
genes were uploaded to DAVID and GO analysis was conducted, with
P≤0.05 used to determine statistical significance. The top 10 GO
terms enriched by up and downregulated genes are presented in
Table II. The upregulated genes
were primarily enriched in ‘nervous system development’,
‘developmental processes’, ‘anatomical structural development’,
‘synapse’, ‘regulation of developmental processes’, ‘regulation of
multicellular organismal development’, ‘synaptic membranes’,
‘positive regulation of developmental processes’, ‘regulation of
multicellular organismal process’ and ‘postsynaptic membranes’. The
top downregulated genes were primarily associated with ‘protein
targeting to the endoplasmic reticulum’, ‘membrane-enclosed lumen’,
‘nuclear part’, ‘SRP-dependent co-translational protein targeting
to the membrane’, ‘translational initiation’, ‘RNA binding’,
‘cytoplasm’, ‘macromolecular complex’, ‘intracellular organelle
part’ and ‘organelle part’. The KEGG (http://www.genome.jp/kegg/) pathways of up and
downregulated genes are presented in Table III. The upregulated genes were
primarily enriched in ‘neuroactive ligand-receptor interaction’,
‘retrograde endocannabinoid signaling’, ‘morphine addiction’,
‘GABAergic synapses’, ‘nicotine addiction’, ‘Rap1 signaling’, ‘Ras
signaling’, ‘PI3K-Akt signaling’, and ‘glutamatergic and
cholinergic synapses’. Down-regulated genes were associated with
‘ribosomes’, ‘Huntington's disease’, ‘oxidative phosphorylation’,
‘Parkinson's and Alzheimer's diseases’, ‘proteasomes’,
‘non-alcoholic fatty liver disease’, ‘metabolic pathways’, ‘protein
processing in the endoplasmic reticulum’ and ‘RNA transport’.
 | Table II.Gene Ontology terms enriched in the
teratozoospermia-related module. |
Table II.
Gene Ontology terms enriched in the
teratozoospermia-related module.
| A, Upregulated
genes |
|---|
| ID | Term | Count | FDR |
|---|
| GO.0007399 | Nervous system
development | 37 |
4.12×10−07 |
| GO.0032502 | Developmental
process | 60 |
3.97×10−07 |
| GO.0048856 | Anatomical
structure development | 56 |
3.52×10−07 |
| GO.0098794 | Postsynapse | 16 |
3.04×10−07 |
| GO.0050793 | Regulation of
developmental process | 39 |
1.73×10−07 |
| GO.2000026 | Regulation of
multicellular organismal development | 34 |
6.97×10−08 |
| GO.0097060 | Synaptic
membrane | 15 |
6.89×10−08 |
| GO.0051094 | Positive regulation
of developmental process | 30 |
1.85×10−08 |
| GO.0051239 | Regulation of
multicellular organismal process | 44 |
1.85×10−08 |
| GO.0045211 | Postsynaptic
membrane | 16 |
4.26×10−10 |
|
| B, Downregulated
genes |
|
| ID | Term | Count | FDR |
|
| GO.0045047 | Protein targeting
to ER | 62 |
1.05×10−37 |
| GO.0031974 | Membrane-enclosed
lumen | 497 |
3.44×10−38 |
| GO.0044428 | Nuclear part | 456 |
3.40×10−38 |
| GO.0006614 | SRP-dependent
cotranslational protein targeting to membrane | 62 |
2.24×10−38 |
| GO.0006413 | Translational
initiation | 89 |
6.39×10−39 |
| GO.0003723 | RNA binding | 262 |
8.87×10−40 |
| GO.0005737 | Cytoplasm | 941 |
2.62×10−43 |
| GO.0032991 | Macromolecular
complex | 552 |
4.40×10−48 |
| GO.0044446 | Intracellular
organelle part | 807 |
1.03×10−54 |
| GO.0044422 | Organelle part | 821 |
8.62×10−55 |
 | Table III.KEGG pathways enriched in the
teratozoospermia-related module. |
Table III.
KEGG pathways enriched in the
teratozoospermia-related module.
| A, Up-regulated
genes |
|---|
| Term | Count | FDR |
|---|
| Neuroactive
ligand-receptor interaction | 15 |
7.99×10−08 |
| Retrograde
endocannabinoid signaling | 9 |
1.95×10−06 |
| Morphine
addiction | 8 |
9.94×10−06 |
| GABAergic
synapse | 7 |
8.70×10−05 |
| Nicotine
addiction | 5 |
3.09×10−04 |
| Rap1 signaling
pathway | 9 |
3.89×10−04 |
| Ras signaling
pathway | 9 |
5.16×10−04 |
| PI3K-Akt signaling
pathway | 11 |
5.16×10−04 |
| Glutamatergic
synapse | 6 |
2.62×10−03 |
| Cholinergic
synapse | 5 |
1.98×10−02 |
|
| B,
Down-regulated genes |
|
| Term | Count | FDR |
|
| Ribosome | 63 |
1.12×10−34 |
| Huntington s
disease | 56 |
8.43×10−19 |
| Oxidative
phosphorylation | 46 |
3.97×10−18 |
| Parkinson's
disease | 45 |
4.14×10−16 |
| Alzheimer's
disease | 46 |
7.12×10−14 |
| Proteasome | 20 |
1.94×10−10 |
| Non-alcoholic fatty
liver disease | 38 |
2.39×10−10 |
| Metabolic
pathways | 147 |
2.17×10−09 |
| Protein processing
in endoplasmic reticulum | 35 |
2.29×10−07 |
| RNA transport | 30 |
1.65×10−05 |
PPI network construction and module
analysis
In order to extract PPI data, the present study
uploaded 450 upregulated genes and 1,942 downregulated genes to the
STRING website. Subsequently, the samples with PPI data >0.4
were selected to assemble PPI networks. The PPI networks of
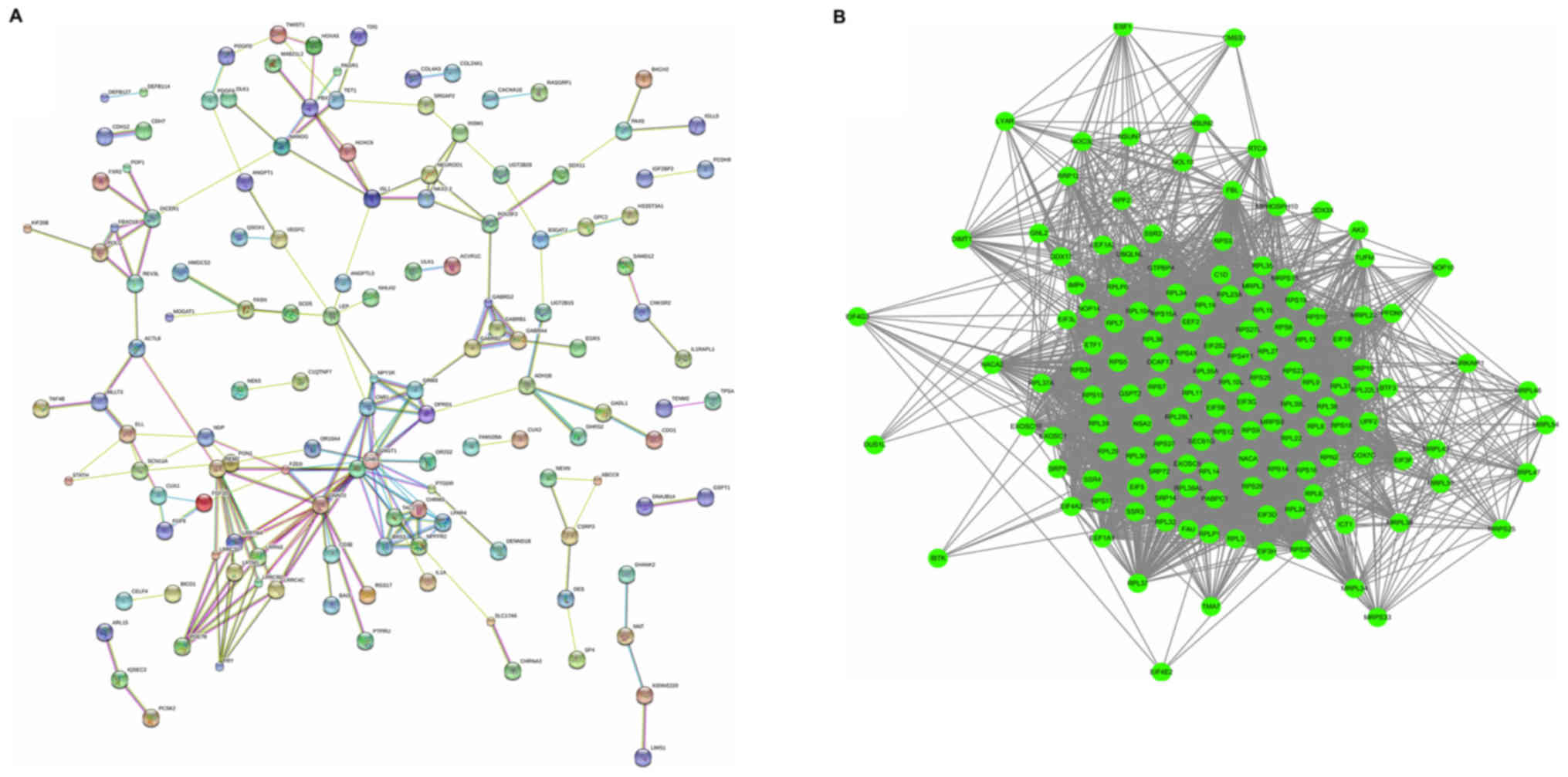
upregulated genes are displayed in Fig. 3A. The upregulated network was
constructed with 134 nodes and 199 edges. The G protein subunit β 3
(GNB3; degree=20), G protein subunit α o1 (GNAO1; degree=16) and G
protein subunit γ transducin 1 (GNGT1; degree=15), were hub nodes
in this network, which had almost twice the degree compared with
other nodes in the network. The downregulated PPI network was
subsequently constructed. The most significant modules were
selected, with 160 nodes and 1,024 edges, as presented in Fig. 3B. Ribosomal protein S3 (RPS3;
degree=32), RPS5 (degree=30), RPS16 (degree=29), RPS6 (degree=25)
and RPS23 (degree=24) were hub nodes in this network.
Discussion
Over the last decade, the molecular mechanism
underlying teratozoospermia has been of great research interest,
with studies conducted in animal, human and cell models (6,7).
With the development of high-throughput technology, an increased
number of genes/proteins have been demonstrated to be associated
with male infertility (23,24).
However, a comprehensive understanding of how the biological
processes at the molecular level are associated with the
pathogenesis of teratozoospermia remains to be elucidated.
Therefore, it is necessary to elucidate the latent pathogenesis of
teratozoospermia at the systems biology level. The present study
identified the disease module associated with teratozoospermia,
systematically investigated the interaction of module genes through
pathway and network analyses and PPI data, and constructed a
comprehensive and systematic framework to trace relevant genes.
There are several upregulated module genes that were
observed to be involved in the pathogenesis of teratozoospermia,
including GNB3, GNAO1 and GNGT1, all of them belong to the G
proteins family, also known as guanine nucleotide-binding proteins.
It has been reported that G proteins are present in human
spermatozoa, transmit various stimulation signals from outside the
cell to its interior and are associated with propagation (25–27).
The aforementioned studies indicate that G proteins serve a role in
the maintenance of fertilization capacity in human and mouse sperm
(28). The aforementioned three G
protein genes have not yet been associated with teratozoospermia;
however, other members of the same class have been demonstrated to
be necessary during spermatogenesis. Decreased expression of G
protein subunit α i2 (Gαi2) was detected in low-motility
spermatozoa with midpieces that were bent on themselves (29). Similarly, the activation of
Gαi2 may affect the volume of ejaculated spermatozoa
(11). Defective expression of
GNA13 was observed in macrocephalic and global nucleus spermatozoa
(30). The axonemal-associated
localization within the midpiece and principal piece of various
mammalian mature spermatozoa indicates that the G protein α-subunit
gustducin likely affects sperm motility via intracellular signal
transduction (31).
A comparative study of epigenetic research between
fertile and infertile boars indicated significantly increased DNA
methylation levels in the GNAS complex locus of infertile boars
(32). These data suggest that G
proteins may be downregulated in abnormal spermatogenesis. However,
the results of the present study suggested that one of the G
protein clusters that have never been proposed to have a function
during spermatogenesis was enriched. GNB3, GNAO1 and GNGT1 are
upregulated in sperm of patients with teratozoospermia, which may
indicate a more comprehensive function of the G protein during
spermatogenesis.
In addition, various ribosomal genes, including
RPS3, RPS5, RPS6, RPS16 and RPS23, were observed to be
downregulated in abnormal sperm, in the present study. Prior to the
present study, RPS3 had not been reported to be associated with
spermatogenesis. A previous study suggested that RPS6 may regulate
the viability of sertoli cells in blood-testis barrier dynamics in
rats (33). Furthermore, it has
also been reported that RPS6 levels are downregulated via the
serine/threonine-protein kinase mTOR signaling pathway in rats with
sperm defects (34).
The function of the RPS23 gene, which is reported to
be expressed in bovine sperm, remains to be fully elucidated
(35). A previous study
demonstrated that the downregulation of RPS16 and RPS5 in infertile
patients is purportedly associated with asthenozoospermia (36). The consistency between previous
studies and the results of the present study suggest that the
methods used in the present study were effective in the study of
teratozoospermia.
In conclusion, the present study used a systems
biology framework for a comprehensive and systematic biological
function- and network-based analysis of teratozoospermia. By
integrating the information from GO, KEGG pathway and pathway
crosstalk, it was revealed that three upregulated genes and five
downregulated genes are enriched in the teratozoospermia-associated
module. This systematic and comprehensive investigation of the
teratozoospermia-associated module genes may improve the
understanding of the contribution of genetic factors and their
interactions with the pathogenesis of teratozoospermia, and may aid
in identification of potential biomarkers for further
investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Family
Planning Project, Shanghai Municipal Commission of 415 Health and
Family Planning (grant no. 201440002); Youth Foundation of Shanghai
Institute of Planned Parenthood Research; Youth Foundation of
Shanghai municipal commission of health and family planning (grant
no. 20164Y0267).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TZ and CL conducted data acquisition and analysis,
and drafted the manuscript. JW and ZN contributed to the analysis
of the results and revised the manuscript critically for important
intellectual content. JZ and FY made substantial contributions to
the design of the present study and critically revised the
manuscript for important intellectual content.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Browne RK, Kaurova SA, Uteshev VK,
Shishova NV, McGinnity D, Figiel CR, Mansour N, Agney D, Wu M,
Gakhova EN, et al: Sperm motility of externally fertilizing fish
and amphibians. Theriogenology. 83:1–13. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Soulavie F, Piepenbrock D, Thomas J,
Vieillard J, Duteyrat JL, Cortier E, Laurencon A, Gopfert MC and
Durand B: Hemingway is required for sperm flagella assembly and
ciliary motility in Drosophila. Mol Biol Cell. 25:1276–1286. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sendler E, Johnson GD, Mao S, Goodrich RJ,
Diamond MP, Hauser R and Krawetz SA: Stability, delivery and
functions of human sperm RNAs at fertilization. Nucleic Acids Res.
41:4104–4117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Georgiadis AP, Kishore A, Zorrilla M,
Jaffe TM, Sanfilippo JS, Volk E, Rajkovic A and Yatsenko AN: High
quality RNA in semen and sperm: isolation, analysis and potential
application in clinical testing. J Urol. 193:352–359. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ostermeier GC, Miller D, Huntriss JD,
Diamond MP and Krawetz SA: Reproductive biology: Delivering
spermatozoan RNA to the oocyte. Nature. 429:1542004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Coutton C, Escoffier J, Martinez G,
Arnoult C and Ray PF: Teratozoospermia: Spotlight on the main
genetic actors in the human. Hum Reprod Update. 21:455–485. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Braekeleer M, Nguyen MH, Morel F and
Perrin A: Genetic aspects of monomorphic teratozoospermia: A
review. J Assist Reprod Genet. 32:615–623. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang-Decker N, Mantchev GT, Juneja SC,
McNiven MA and van Deursen JM: Lack of acrosome formation in
Hrb-deficient mice. Science. 294:1531–1533. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu X, Toselli PA, Russell LD and Seldin
DC: Globozoospermia in mice lacking the casein kinase II alpha'
catalytic subunit. Nat Genet. 23:118–121. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sadek CM, Damdimopoulos AE, Pelto-Huikko
M, Gustafsson JA, Spyrou G and Miranda-Vizuete A: Sptrx-2, a fusion
protein composed of one thioredoxin and three tandemly repeated
NDP-kinase domains is expressed in human testis germ cells. Genes
Cells. 6:1077–1090. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Allegrucci C, Liguori L and Minelli A:
Stimulation by n6-cyclopentyladenosine of A1 adenosine receptors,
coupled to galphai2 protein subunit, has a capacitative effect on
human spermatozoa. Biol Reprod. 64:1653–1659. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matzuk MM and Lamb DJ: Genetic dissection
of mammalian fertility pathways. Nat Cell Biol. 4 Suppl:S41–S49.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
O'Doherty AM and McGettigan PA: Epigenetic
processes in the male germline. Reprod Fertil Dev. 27:725–738.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
H MY, Kumar S, Dubey PP, Modi RP,
Chaudhary R, A SK, Ghosh SK, Sarkar M and B S: Profiling of sperm
gene transcripts in crossbred (Bos taurus × Bos indicus) bulls.
Anim Reprod Sci. 177:25–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu K, Zhang J and Liang M: LncRNA AK015322
promotes proliferation of spermatogonial stem cell C18-4 by acting
as a decoy for microRNA-19b-3p. In Vitro Cell Dev Biol Anim.
53:277–284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kherraf ZE, Christou-Kent M, Karaouzene T,
Amiri-Yekta A, Martinez G, Vargas AS, Lambert E, Borel C, Dorphin
B, Aknin-Seifer I, et al: SPINK2 deficiency causes infertility by
inducing sperm defects in heterozygotes and azoospermia in
homozygotes. EMBO Mol Med. 9:1132–1149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Platts AE, Dix DJ, Chemes HE, Thompson KE,
Goodrich R, Rockett JC, Rawe VY, Quintana S, Diamond MP, Strader LF
and Krawetz SA: Success and failure in human spermatogenesis as
revealed by teratozoospermic RNAs. Hum Mol Genet. 16:763–773. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sanchez MC, Sedo CA, Julianelli VL,
Romanato M, Calvo L, Calvo JC and Fontana VA: Dermatan sulfate
synergizes with heparin in murine sperm chromatin decondensation.
Syst Biol Reprod Med. 59:82–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Langsdorf A, Schumacher V, Shi X, Tran T,
Zaia J, Jain S, Taglienti M, Kreidberg JA, Fine A and Ai X:
Expression regulation and function of heparan sulfate
6-O-endosulfatases in the spermatogonial stem cell niche.
Glycobiology. 21:152–161. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin YN, Roy A, Yan W, Burns KH and Matzuk
MM: Loss of zona pellucida binding proteins in the acrosomal matrix
disrupts acrosome biogenesis and sperm morphogenesis. Mol Cell
Biol. 27:6794–6805. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu Y, Ma M, Wan L, Zhang D, Zhao L, Wei L
and Li L: Analysis of DAZL SNP260 and SNP386 in infertile Chinese
males using multi-analyte suspension array. Mol Med Rep.
10:2949–2954. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li YZ, Chen ZY, Wang H, Huang H, Song QX
and Zhou GH: Establishment of a hydrogel chip for high-throughput
detection of Y chromosome microdeletions. Zhonghua Nan Ke Xue.
18:109–114. 2012.(In Chinese). PubMed/NCBI
|
|
25
|
Hinsch KD, Schwerdel C, Habermann B,
Schill WB, Muller-Schlosser F and Hinsch E: Identification of
heterotrimeric G proteins in human sperm tail membranes. Mol Reprod
Dev. 40:345–354. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Merlet F, Weinstein LS, Goldsmith PK,
Rarick T, Hall JL, Bisson JP and de Mazancourt P: Identification
and localization of G protein subunits in human spermatozoa. Mol
Hum Reprod. 5:38–45. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Modarressi MH, Taylor KE and Wolfe J:
Cloning, characterization, and mapping of the gene encoding the
human G protein gamma 2 subunit. Biochem Biophys Res Commun.
272:610–615. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Baxendale RW and Fraser LR:
Immunolocalization of multiple galpha subunits in mammalian
spermatozoa and additional evidence for galphas. Mol Reprod Dev.
65:104–113. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu Y, Lu Y, Zhou Z, Du Y, Xing J, Wang L,
Lin M and Sha J: Defective expression of Galpha12 in the testes of
azoospermia patients and in the spermatozoa with low motility. J
Mol Med (Berl). 84:416–424. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu Y, Xing J, Chen L, Guo X, Du Y, Zhao C,
Zhu Y, Lin M, Zhou Z and Sha J: RGS22, a novel testis-specific
regulator of G-protein signaling involved in human and mouse
spermiogenesis along with GNA12/13 subunits. Biol Reprod.
79:1021–1029. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fehr J, Meyer D, Widmayer P, Borth HC,
Ackermann F, Wilhelm B, Gudermann T and Boekhoff I: Expression of
the G-protein alpha-subunit gustducin in mammalian spermatozoa. J
Comp Phys A Neuroethol Sens Neural Behav Physiol. 193:21–34. 2007.
View Article : Google Scholar
|
|
32
|
Congras A, Yerle-Bouissou M, Pinton A,
Vignoles F, Liaubet L, Ferchaud S and Acloque H: Sperm DNA
methylation analysis in swine reveals conserved and
species-specific methylation patterns and highlights an altered
methylation at the GNAS locus in infertile boars. Biol Reprod.
91:1372014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mok KW, Chen H, Lee WM and Cheng CY: rpS6
regulates blood-testis barrier dynamics through Arp3-mediated actin
microfilament organization in rat sertoli cells. An in vitro study.
Endocrinology. 156:1900–1913. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu H, Shen L, Chen X, Ding Y, He J, Zhu J,
Wang Y and Liu X: mTOR/P70S6K promotes spermatogonia proliferation
and spermatogenesis in Sprague Dawley rats. Reprod Biomed Online.
32:207–217. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han CM, Chen R, Li T, Chen XL, Zheng YF,
Ma MT and Gao QH: Evaluation of the semen swim-up method for bovine
sperm RNA extraction. Genet Mol Res. 15:gmr.15027713. 2016.
View Article : Google Scholar
|
|
36
|
Bansal SK, Gupta N, Sankhwar SN and
Rajender S: Differential genes expression between fertile and
infertile spermatozoa revealed by transcriptome analysis. PloS One.
10:e01270072015. View Article : Google Scholar : PubMed/NCBI
|