Introduction
Epithelial ovarian cancer (EOC) is a major cause for
malignancy-associated female mortality (1). The mortality rate is frequently
increased by a delay in diagnosis and drug resistance (2). Investigating the mechanisms behind
EOC initiation and progression can help us to find a therapeutic
target.
MicroRNAs (miRs), non-coding RNAs with a length of
20–24 nucleotides, regulate gene expression by inhibiting
translation or degrading messenger RNA (3). Increasing evidence reveals that miRs
regulate cellular processes, including proliferation,
differentiation and apoptosis (3–5). A
number of miRs are associated with ovarian cancer, and may regulate
tumor progression, function as potential prognostic markers and
contribute to drug resistance (6–9). The
functions of miRs in carcinogenesis have been illustrated in
multiple studies. For example, miR-224, as an oncogene in EOC,
improves cancer cell proliferation by downregulating KILLIN
expression (10). By targeting
pyruvate dehydrogenase E1 β subunit, miR-203 promotes the
proliferation and migration of EOC cells (11). Other miRs function as tumor
suppressors expressed (12,13).
For example, by targeting cyclin dependent kinase (CDK1),
miR-490-3p inhibits the proliferation, migration and invasion of
EOC cells (12). miR-101
suppresses the expression of the suppressor of cytokine signaling 2
gene and inhibits the proliferation and invasion of EOC cells
(13). Among them, miR-655 was the
focus of the present study as it has been demonstrated to play a
role in certain malignancies, including esophageal squamous cell
carcinoma (14), bladder
urothelial carcinoma (15) and
gastric signet ring cell carcinoma (16). Currently, to the best of our
knowledge, no studies have been conducted to investigate the
association between miR-665 and EOC.
Homeobox A10 (HOXA10), one member of the homeobox
gene family, acts as a transcription factor in embryonic
development (17). The aberrant
expression of HOXA10 was first observed in leukemia (18). The association between HOXA10 and
EOC can be demonstrated by the downregulation and upregulation of
HOXA10 in cancer cellular processes, including cell proliferation,
epithelial-mesenchymal transition, apoptosis and drug resistance
(6,18–22).
HOXA10 was demonstrated to participate in G1 phase arrest of
endometrial cancer that may be caused by P21 expression
(23). It was hypothesized that
miR-665 can regulate HOXA10 expression, based on the data from
PicTar, TargetScan and miRBase. However, the role of miR-665 in the
development of EOC and its association with HOXA10 remains
uninvestigated.
In the present study, the expression of miR-665 in
human EOC and normal ovary tissues were compared, and the impact of
miR-665 expression on cell proliferation and migration in
vitro was investigated. The results of the present study
suggested that miR-665 serves a suppressive role in human EOC
pathogenesis.
Materials and methods
Tissue samples and cell lines
Informed consent was obtained from each patient. The
present study was approved by the Institutional Ethic Committee of
Nanjing Medical University (Nanjing, China). Tissues from EOC
patients were frozen immediately and stored at −80°C. EOC tissue
specimens (n=28) and normal ovarian tissue specimens (n=15) were
collected from patients (age range, 24–73) who underwent surgery at
the Department of Gynecology at the First Affiliated Hospital of
Nanjing Medical University (Nanjing, China) between December 2014
and December 2015. EOC cell lines (HO8910 and OVCAR-3) and 293T
cells were purchased from American Type Culture Collection
(Manassas, VA, USA). Cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin, and 100 mg/ml (Beijing
Solarbio Science and Technology Co., Ltd., Beijing, China) at 37°C
with 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from ovarian tissues and
ovarian cancer cell lines using TRIzol reagent (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) following the manufacturer's
protocol. Following isolation, the integrity of RNA was determined
using Agilent Bioanalyzer 2100 and RNA 6000 Nano kit (Agilent
Technologies, Inc., Santa Clara, CA, USA). According to
manufacturer's protocol, single-stranded complementary DNA (cDNA)
was synthesized from 1 µg RNA in a 20 µl reaction volume with the
High-Capacity cDNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.), at 25°C for 10 min, 37°C for 120 min and 85°C
for 5 sec, followed by a 4°C hold. Quantification of miR and mRNA
was carried out using a SYBR Green PCR kit (Thermo Fisher
Scientific, Inc.), and the cycle quantification (Cq) of each gene
was recorded. The relative expression of miR-665 mRNA and HOXA10
was normalized to U6, and calculated using the 2−∆∆Cq
method (ΔCq=Cqtarget gene-Cqinternal control)
(24). The qPCR was performed
using the following parameters: 95°C, 10 min; 40 cycles, 95°C, 15
sec; 67°C, 30 sec; 72°C, 30 sec; and 72°C, 5 min. The primers used
are illustrated in Table I.
 | Table I.Primers used in reverse
transcription-quantitative polymerase chain reaction and miR
sequences. |
Table I.
Primers used in reverse
transcription-quantitative polymerase chain reaction and miR
sequences.
| Name | Direction | Sequence
(5′-3′) |
|---|
| U6 | Forward |
CTCGCTTCGGCAGCACA |
|
| Reverse |
AACGCTTCACGAATTTGCGT |
| Hsa-miR-665
mimic | Forward |
ACCAGGAGGCUGAGGCCCCUTT |
|
| Reverse |
AGGGGCCUCAGCCUCCUGGUTT |
| miR-665 mimic
negative control | Forward |
UUCUCCGAACGUGUCACGUTT |
|
| Reverse |
ACGUACACGUUCGGAGAATT |
| Hsa-miR-665
inhibitor |
|
AGGGGCCUCAGCCUCCUGGU |
| miR inhibitor
negative control |
|
CAGUACUUUUGUGUAGUACAA |
| HOXA10 | Forward |
GGGTAAGCGGAATAAACT |
|
| Reverse |
GCACAGCAGCAATACAATA |
Protein extraction and western
blotting
At 48 h following transfection, the cells were
harvested, washed twice with phosphate-buffered saline, lysed using
M-PER™ Mammalian Protein Extraction reagent (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with 0.01% protease and
phosphatase inhibitor, and incubated on ice for 30 min. The cell
lysate was centrifuged at 4°C and 12,000 × g for 15 min. Protein
concentrations were determined by a bicinchoninic acid protein
assay (Beyotime Institute of Biotechnology, Haimen, China). The
supernatant (50 µg) of total protein was run on 10% SDS-PAGE and
transferred electrophoretically to a polyvinylidene fluoride
membrane (EMD Millipore, Billerica, MA, USA). Following blocking in
Western Blocking Reagent (10%; Hoffman; Roche Diagnostics, Basel,
Switzerland) for 15 min at room temperature, the membrane was
incubated overnight at 4°C with polyclonal rabbit anti-human HOXA10
(1:1,000; Abcam, Cambridge, MA, USA; cat. no. ab90641) and mouse
anti-human β-actin (1:10,000; cat. no. ab49900; Abcam),
respectively. The membranes were washed with 1X Tris-buffered
saline containing 0.1% Tween-20 (TBST), incubated with the
horseradish peroxidase (HRP)-conjugated anti-rabbit IgG secondary
antibody (1:1,000; cat. no. 7074; Cell Signaling Technology, Inc.)
at room temperature for 1 h. Membranes were washed again with TBST
three times for 10 min each, prior to visualization and analysis
using the Odyssey IR imaging system (LI-COR Biosciences, Lincoln,
NE, USA).
Bioinformatics analysis
To investigate the target genes of miR-665,
TargetScan version 7.1 (http://www.targetscan.org), PicTar 5 (https://pictar.mdc-berlin.de/) and miRBase release 22
(http://www.mirbase.org/) were used to predict the
potential target gene of miR-665.
Transwell migration assay
Following being placed into a 24-well plate, the
migration assays of OVCAR-3 and HO8910 cells were carried out using
Transwell chambers (EMD Millipore). For the migration assay, a
total of 1×105 cells were resuspended in 200 µl
serum-free medium and placed in the top chambers. RPMI-1640 medium
(600 µl) containing 10% FBS was added into the bottom chambers.
Cells were incubated for another 20 h at 37°C with 5%
CO2. The cells were fixed with 4% polyoxymethylene
following the incubation for 20 min at room temperature, stained
with 0.1% crystal violet for 20 min at room temperature and
observed using a light microscope (magnification, ×100; Olympus
Corporation, Tokyo, Japan). The numbers of migrated cells were
calculated from five randomly selected fields.
Colony formation assay
Cells were transfected with NC, miR-665, as
described above. Then 24 h later, transfected cells were
trypsinized, counted and replated at a concentration of 500
cells/well. Following another 10 days, colonies formed by the
surviving cells were fixed with 3.7% methanol for 20 min at room
temperature, stained with 0.1% crystal violet for 20 min at room
temperature and counted. Colonies containing ≥50 cells were scored.
Each assay was performed in triplicate.
CCK-8 assay
The cell proliferation reagent WST-8 (Roche
Diagnostics GmbH, Mannheim, Germany) was used to measure cell
growth. Cells were seeded into 96-well microtiter plates (Corning
Inc., Corning, NY, USA) at a density of 1.0×103
cells/well. CCK-8 was added to each well according to the
manufacturer's protocol. Following incubation at 37°C with 5%
CO2 for 2 h, the absorbance of the converted dye was
detected at 450 nm to determine the cellular viability.
Dual-luciferase reporter assay
The 3′-untranslated regions (3′-UTRs) of human
HOXA10 cDNA with the potential target sites for miR-665 were
synthesized and inserted at the XbaI site downstream of the
luciferase gene in the pGL3-control (Promega Corporation, Madison,
WI, USA) vector by Integrated Biotech Solutions Co., Ltd.,
(Shanghai, China).
At 24 h prior to transfection, cells were seeded in
24-well plates (1.5×105 wells/well). Then, 200 ng of
pGL3-HOXA10-3′-UTR and 80 ng of pRL-TK (Promega Corporation) were
co-transfected with 60 pmol of miR-665 mimic or NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. At 24 h
following the transfection, the Dual-luciferase assay system
(Promega Corporation) was used to determine luciferase activity as
previously described (6). The
firefly luciferase activity in each well was normalized to that of
the Renilla luciferase. Three independent experiments were
performed in duplicate. The sequence of 3′ UTR of human HOXA10 cDNA
containing the putative target site for the miR-665; the
underlined, italicized area indicates the putative target site for
miR-665:
5′-TGAATCTCCAGGCGACGCGGTTTTTTCACTTCCCGAGCGCTGGTCCCCTCCCTCTGTCTTCAGGCTCTGCCCAGGAACTCGCACCTGTGCTGGAGCCCTGTTCCTCCCTCCCACACTCGCCATCTCCTGGGCCGTTACATCTGTGCAGGGCTGGTTTGTTCTGACTTTTTGTTTCTTTGTGTTTGCTTGGTGCTGGTTTATTTGTTGTTTTCTGGGGGAAAAAGCCATATCATGCTAAAATTCTATAGAGATA-3′.
Immunohistochemistry
The tissues were fixed in 10% formalin for 24 h at
room temperature, embedded in paraffin and 4 µm thick sections were
prepared. Microwave irradiation in 10 mol/l citrate buffer (pH 6.0)
was used for antigen retrieval for tissue slides incubated with a
HOXA10 antibody (1:500; cat. no. ab90641; Abcam) at 4°C overnight,
followed by incubation with a HRP-conjugated secondary antibody
(1:1,000; cat. no. ab6721; Abcam) for 1 h at room temperature. The
staining was repeated if the result was uncertain. Immunostaining
of the slides was objectively evaluated by two pathologists under a
light microscope (magnification, ×200; Olympus Corporation, Tokyo,
Japan). Discordant scores were reevaluated until consensus was
reached. The level of positive HOXA10 expression in cancer cells
was analyzed by HMIAS-2000 automatic medical color image analysis
system (Qianping Image Technology Co., Ltd., Wuhan, China). HOXA10
staining was determined semi-quantitatively according to the
staining intensity observed (0, no staining; 1,weak staining; 2,
moderate staining; and 3, strong staining) and the percentage of
positive cells (0, none or <10; 1, 11–25; 2, 26–50; and 3,
51–75%; 4, >75%). Scores of 0–3 were considered to indicate
negative expression, and scores of 3–12 were considered to indicate
positive expression. Cells were counted in at least three randomly
selected fields (magnification, ×200) in the tumor areas.
Statistical analysis
All experiments were repeated three times
independently. The results were summarized as the mean ± standard
error. Independent sample t-tests were performed to compare
differences between two groups with SPSS 19.0 software (IBM Corp.,
Armonk, NY, USA). Spearman's correlation test was used to analyze
the correlation between miR-655 and HOXA10 expression. P<0.05
was considered to indicate a statistically significant
difference.
Results
Expression level of HOXA10 increases
in EOC tissues
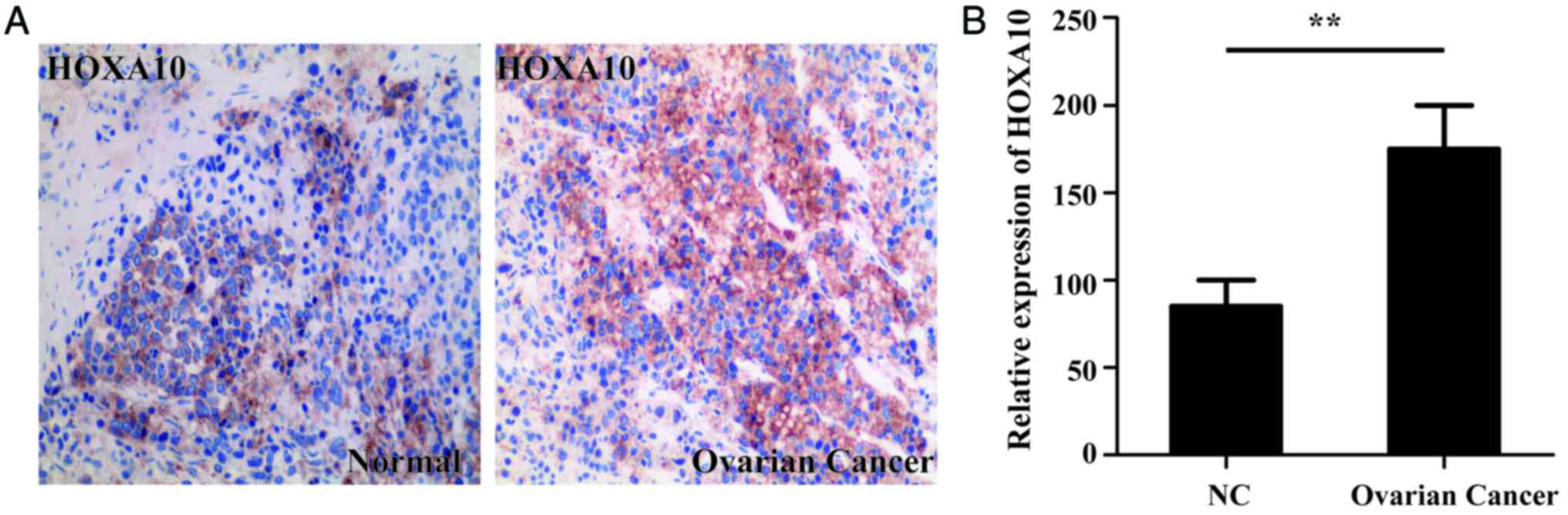
Immunohistochemical staining was used to determine
the level of HOXA10 expression in both normal and ovarian cancer
tissues. HOXA10 expression of 15 EOC specimens and 8 normal
specimens were analyzed in the present study. Immunohistochemical
staining identified that the HOXA10 protein levels were
significantly increased in the ovarian cancer tissues compared with
the normal tissues (P<0.01; Fig.
1).
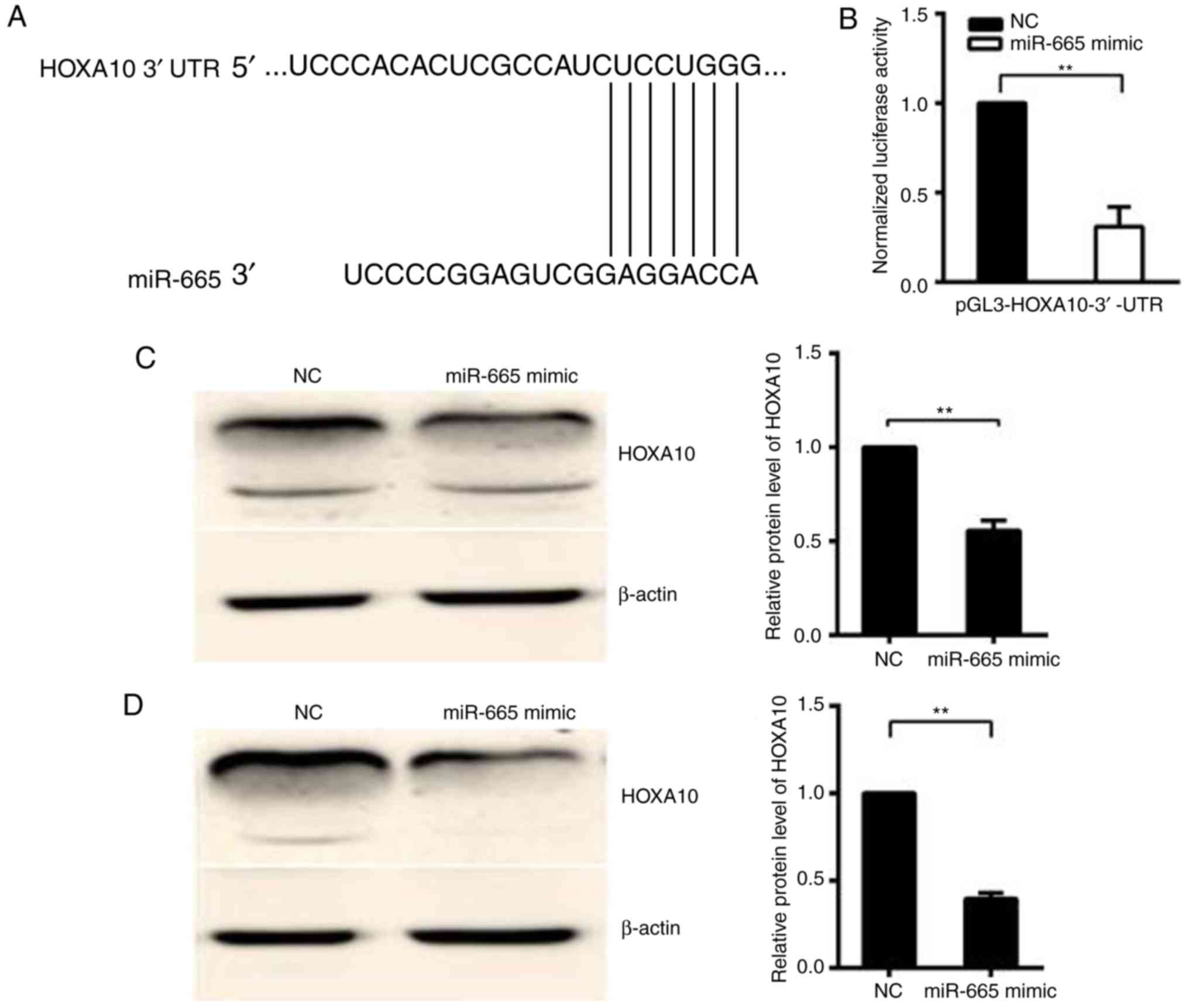
miR-665 targets the 3′-UTR of
HOXA10
TargetScan7.1 (http://www.targetscan.org) was used to predict whether
HOXA10 is targeted by miR-665 (Fig.
2A). In the 293T cell line, relative activity of luciferase was
significantly decreased (P<0.01) following the co-transfection
of the pGL3-HOXA10-3′-UTR vector with the miR-665 mimic, but not
following the co-transfection of pGL3-HOXA10-3′-UTR vector with
mimic NC, suggesting that HOXA10 is the target gene of miR-665
(Fig. 2B). Western blotting was
performed to confirm the downregulation of the HOXA10 protein
following the miR-665 transfection in HO8910 and OVCAR-3 EOC cells.
The protein expression level of HOXA10 significantly decreased in
miR-665-transfected cells, compared with cells transfected with
miR-NC (P<0.01; Fig. 2C and D).
Then whether HOXA10 is the target of miR-665 was investigated. A
luciferase reporter vector was constructed with the target sites of
putative HOXA10 3′-UTR (luciferase gene) located in the downstream
of miR-665. The luciferase reporter vector, miR-665 mimic and mimic
NC was then transfected into 293T cells.
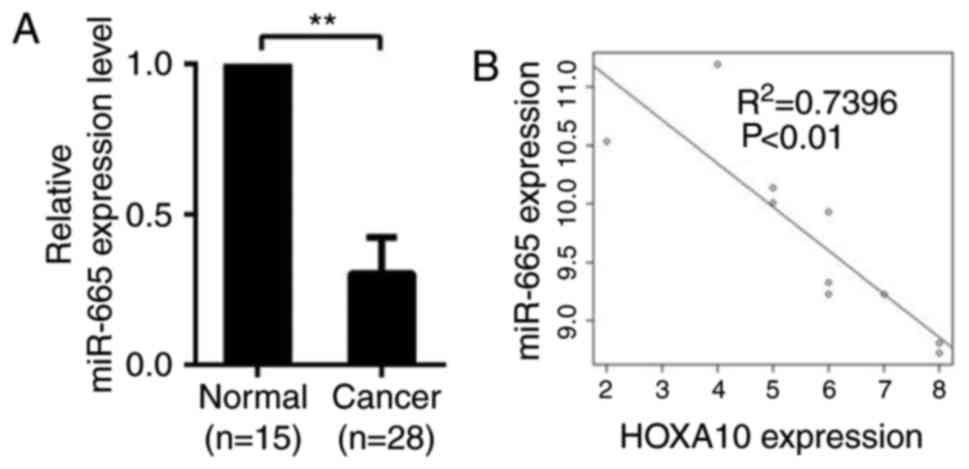
miR-665 is negatively associated with
HOXA10
qPCR was used to determine the expression of miR-665
in 28 EOC specimens and 15 normal specimens. Compared with normal
tissues, expression level of miR-665 significantly dropped in
ovarian cancer tissues, suggesting that miR-665 is downregulated in
ovarian cancer tissues and may be involved in the development of
ovarian cancer (P<0.01; Fig.
3A). A significant inverse correlation (R2=0.7496;
P<0.01) was observed between miR-665 and HOXA10 using Spearman's
correlation analysis (Fig.
3B).
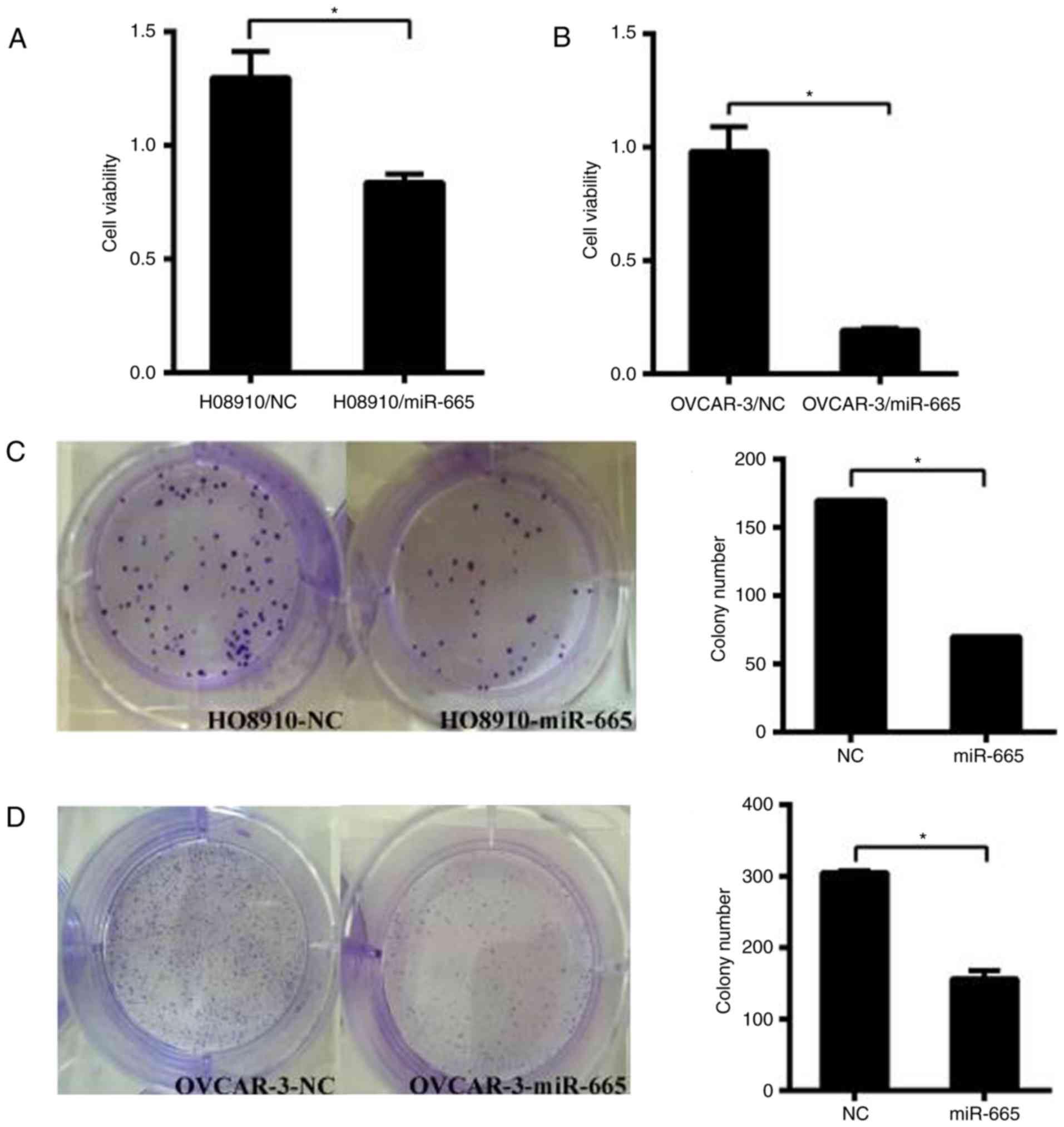
miR-665 suppresses cell growth in
vitro
HO8910 and OVCAR-3 cells were transfected with the
miR-665 mimic. Subsequently, CCK-8 assays were performed to
determine the impact of miR-665 on the proliferation of ovarian
cancer cells. The results identified that increased expression of
miR-665 significantly suppressed the proliferation of ovarian
cancer cells in both cell lines (P<0.05; Fig. 4A and B). The colony formation
capacity of HO8910 and OVCAR-3 cells transfected with miR-665 mimic
was significantly inhibited compared with the miR-NC group
(P<0.05; Fig. 4C and D). These
results demonstrated that miR-665 inhibits the proliferative
ability of HO8910 and OVCAR-3 cells.
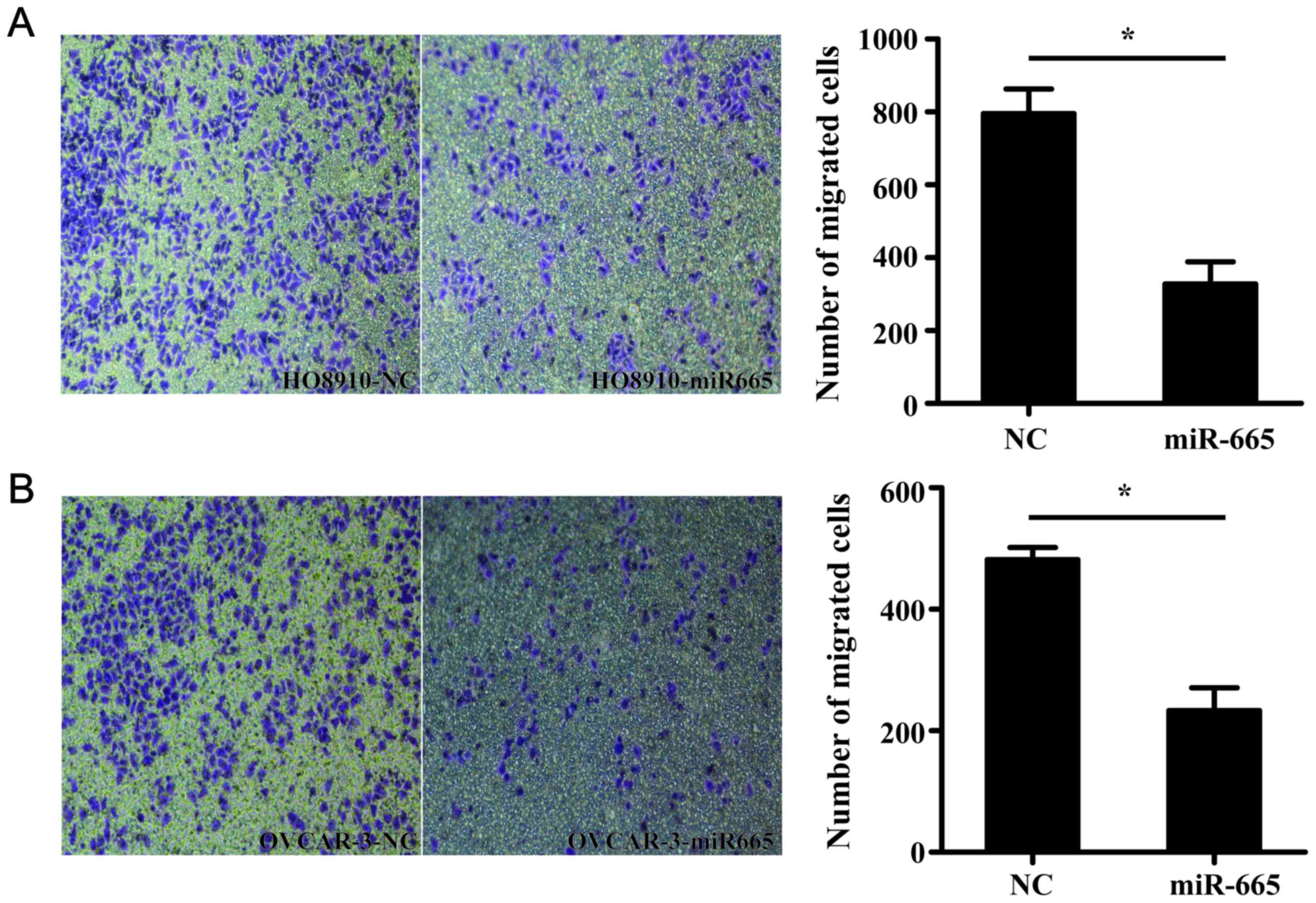
miR-665 suppresses the migration of
ovarian cancer cells
Transwell migration assay demonstrated that
miR-665-overexpressed HO8910 and OVCAR-3 cells exhibited a
significantly decreased ability to migrate compared with the
control cells (P<0.05; Fig.
5).
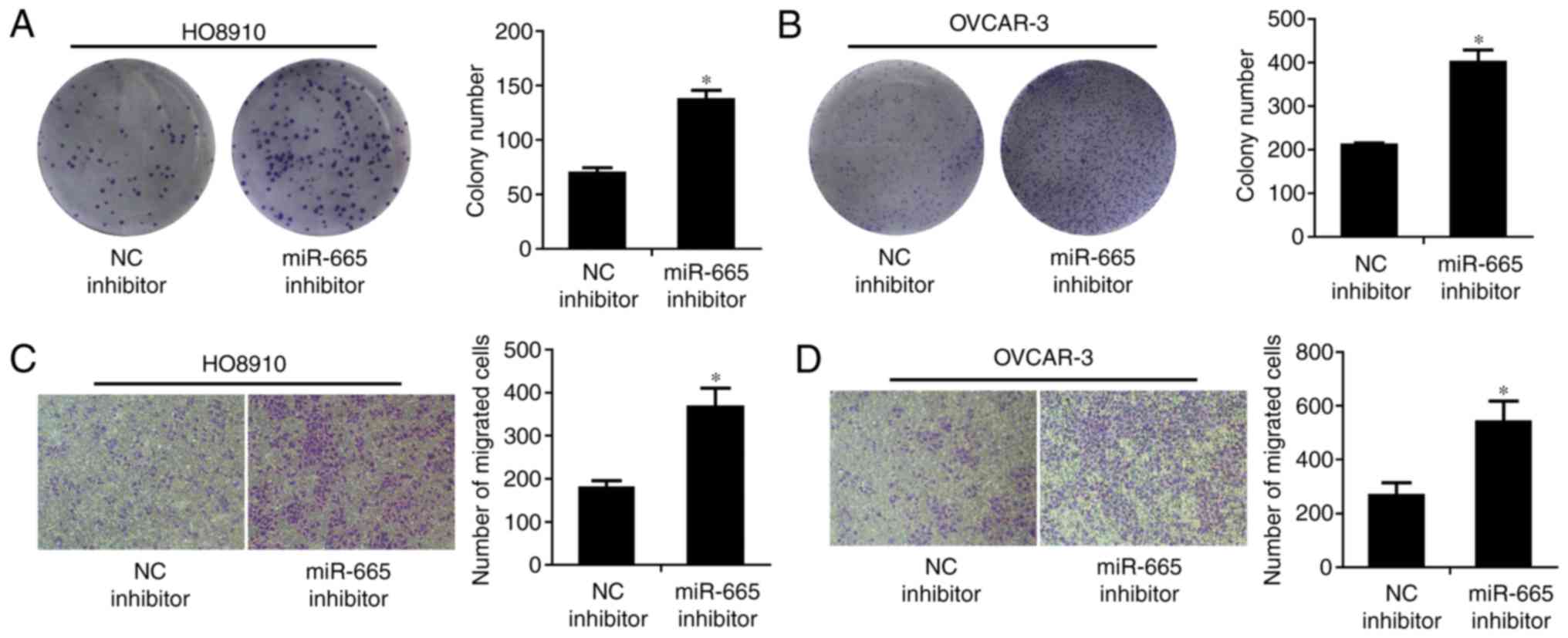
Inhibition of miR-665 promotes the
growth and migration of ovarian cancer cell lines
The colony formation rate of HO8910 and OVCAR-3
cells transfected with miR-665 inhibitor, was significantly
increased, compared to cells transfected with the miR-NC inhibitor
(P<0.05; Fig. 6A and B). The
migratory abilities of miR-665-downregulated HO8910 and OVCAR-3
cells were significantly enhanced compared with the miR-NC
inhibitor (P<0.05; Fig. 6C and
D).
Discussion
Ovarian cancer is the most lethal cancer for women,
with an overall survival rate of ~35% (25). Although modified chemotherapy can
improve the prognosis, its effectiveness has reached its limit.
Consequently, novel therapies, such as targeted therapy combined
with standard treatment, are being investigated in clinical trials
(26). In these trials, predictive
markers can be used to personalize and optimize the therapeutic
strategy for ovarian cancer (27).
miRs modulate gene expression in a
post-transcriptional manner either by inhibiting translation or
destroying the target mRNA (28).
miRs are aberrantly expressed in ovarian cancer. For example, the
expression of miR-145 is significantly reduced in human ovarian
cancer tissues, leading to relapse and poorer outcomes of ovarian
cancer (29). miR-490-3p
overexpression inhibits the proliferation, migration and invasion
of tumor cells by directly targeting CDK1 (12). In addition, miR-497 downregulation
triggers chemotherapy resistance in ovarian cancer cells (30). The level of miR-125b expression in
ovarian cancer tissue is significantly lower compared with normal
ovarian tissues; the increased expression of miR-125b induces cell
cycle arrest and inhibits the proliferation and clonal formation of
ovarian cancer cells by targeting BCL-3 (31).
In carcinogenesis, miR-665 demonstrates diverse
functions (14–16) that are co-regulated by its targets
(32). miR-665, located at
14q32.2, with a length of 20 amino acids, can inhibit B7-H3
expression. However, the underlying mechanism remains
uninvestigated. In the present study, the analysis of RNA
expression revealed that the expression of hsa-miR-665 was
decreased in EOC, which is consistent with the results of a
previous study on breast cancer (33). Therefore, it is hypothesized that
miR-665 may function as a tumor-suppressor gene in ovarian cancer.
Certain studies demonstrated that miR-665 expression was
dysregulated in a number of types of cancer, including esophageal
squamous cell carcinoma (14),
bladder urothelial carcinoma (15)
and gastric signet ring cell carcinoma (16).
HOXA10, from the homeobox gene family, functions as
a transcription factor in embryonic development (17). It has been suggested that HOXA10
acts as a key factor in endometrial receptivity and embryo
implantation, and is expressed in endometrial glandular epithelium
and mesenchymal cells in normal humans (34). The HOXA10 expression level in the
middle and late stages of secretion is increased compared with the
period of endometrial proliferation and the early stage of
secretion (35). As the
progesterone concentration rises during implantation and embryonic
circulation, the level of HOXA10 gradually reaches a peak,
indicating that HOX genes regulate endometrial development and
embryonic planting (35,36). In addition, HOXA10 is highly
expressed in endometroid, clear or mucinous cells, but not in
serous epithelial ovary cancer cells (37,38).
Aberrant HOX gene expression has been reported in several types of
cancer, including glioblastoma (39), oral cancer and gastric cancer
(21,22,40).
Increased expression of HOXA10 promotes the proliferation,
migration and invasion of clear cell adenocarcinoma of the ovary,
reducing the survival of patients (41). The present study identified that
miR-665 was downregulated and negatively correlated with the
expression of HOXA10 in ovarian tumor tissues, indicating that
miR-665 may be a tumor suppressor gene in the development of
ovarian cancer and a potential therapeutic target for ovarian
cancer.
In the present study, the RT-qPCR results
demonstrated that miR-665 was downregulated in ovarian cancer
tissues compared with normal tissues. Immunohistochemistry revealed
that HOXA10 was overexpressed in ovarian cancer tissues and this
expression was negatively correlated with the expression of
miR-665. It was also demonstrated that miR-665 suppressed the
proliferation and migration of cell lines, whereas the
downregulation of miR-665 led to the opposite effect, as it bound
to the 3′-UTR of HOXA10, and downregulated HOX10 by reducing HOXA10
protein levels. Further studies are needed to elucidate the
mechanism of HOXA10 silencing.
The present study investigated the expression and
biological function of miR-665 in ovarian cancer. miR-665
downregulated the expression of HOXA10 and weakened the ability of
ovarian cancer cells to proliferate and migrate. miR-665, through
targeting HOXA10, serves as a suppressor gene in ovarian cancer.
Therefore, therapeutic miR that mimics miR-665 could possibly be
developed to treat ovarian cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Nature Science Foundation of China (grant nos. 81472442 and
81272871) and the Postgraduate Research & Practice Innovation
Program of Jiangsu Province (grant no. JX22013366)
Availability of data and materials
Not applicable.
Authors' contributions
Conceived and designed the experiments: WJC.
Performed the experiments: JHL, YJ. Analyzed the data and wrote the
manuscript: JHL, YJ, YCW. Performed part of the experiments and
bioinformatics analyses: SLZ, ST. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Nanjing Medical University and samples were obtained with
informed consent from all patients.
Patient consent for publication
Informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EOC
|
epithelial ovarian cancer
|
|
miR
|
microRNA
|
|
PCR
|
polymerase chain reaction
|
|
3′-UTRs
|
3′-untranslated regions
|
References
|
1
|
Xian H, Xian Y, Liu L, Wang Y, He J and
Huang J: Expression of β-nerve growth factor and homeobox A10 in
experimental cryptorchidism treated with exogenous nerve growth
factor. Mol Med Rep. 11:2875–2881. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo R, Sherman-Baust C and Abdelmohsen K:
miRNA-based ovarian cancer diagnosis and therapySarkar F: microRNA
Targeted Cancer Therapy. Springer; Cham: pp. 115–127. 2014,
View Article : Google Scholar
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hornstein E, Mansfield JH, Yekta S, Hu JK,
Harfe BD, McManus MT, Baskerville S, Bartel DP and Tabin CJ: The
microRNA miR-196 acts upstream of Hoxb8 and Shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boehm M and Slack FJ: microRNA control of
lifespan and metabolism. Cell Cycle. 5:837–840. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang W, Jiang Y, Mu X, Xu L, Cheng W and
Wang X: MiR-135a functions as a tumor suppressor in epithelial
ovarian cancer and regulates HOXA10 expression. Cell Signal.
26:1420–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang F, Chang TH, Kao CJ and Huang RS:
High expression of miR-532-5p, a tumor suppressor, leads to better
prognosis in ovarian cancer both in vivo and in vitro. Mol Cancer
Ther. 15:1123–1131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao H, Yu X, Ding Y, Zhao J, Wang G, Wu
X, Jiang J, Peng C, Guo GZ and Cui S: MiR-770-5p inhibits cisplatin
chemoresistance in human ovarian cancer by targeting ERCC2.
Oncotarget. 7:53254–53268. 2016.PubMed/NCBI
|
|
9
|
Zhao X, Zhou Y, Chen YU and Yu F: miR-494
inhibits ovarian cancer cell proliferation and promotes apoptosis
by targeting FGFR2. Oncol Lett. 11:4245–4251. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu K and Liang M: Upregulated microRNA-224
promotes ovarian cancer cell proliferation by targeting KLLN. In
Vitro Cell Dev Biol Anim. 53:149–156. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xiaohong Z, Lichun F, Na X, Kejian Z,
Xiaolan X and Shaosheng W: MiR-203 promotes the growth and
migration of ovarian cancer cells by enhancing glycolytic pathway.
Tumour Biol. 37:14989–14997. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen S, Chen X, Xiu YL, Sun KX and Zhao Y:
MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial
carcinoma tumorigenesis and progression. Cancer Lett. 362:122–130.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng HB, Zheng XG and Liu BP: miRNA-101
inhibits ovarian cancer cells proliferation and invasion by
down-regulating expression of SOCS-2. Int J Clin Exp Med.
8:20263–20270. 2015.PubMed/NCBI
|
|
14
|
Zang W, Wang Y, Du Y, Xuan X, Wang T, Li
M, Ma Y, Li P, Chen X, Dong Z and Zhao G: Differential expression
profiling of microRNAs and their potential involvement in
esophageal squamous cell carcinoma. Tumor Biol. 35:3295–3304. 2014.
View Article : Google Scholar
|
|
15
|
Cheng W, Gao JP, Zhang ZG, Jing-Ping GE,
Feng XU and Wei ZF: Study on microRNAs in urothelial carcinoma(II
grade) of the bladder. J Med Postgrad. 23:48–52. 2010.
|
|
16
|
Chen J, Sun D, Chu H, Gong Z, Zhang C,
Gong B, Li Y, Li N and Jiang L: Screening of differential microRNA
expression in gastric signet ring cell carcinoma and gastric
adenocarcinoma and target gene prediction. Oncol Rep. 33:2963–2971.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hatanaka Y, De Velasco MA, Kura Y,
Yamamoto Y, Kodama M, Nozawa M, Shimizu N, Yoshimura K, Yoshikawa
K, Nishio K and Uemura H: Abstract 3629: HOXA10 expression profiles
in prostate cancer. Cancer Res. 72 8 Suppl:S36292012. View Article : Google Scholar
|
|
18
|
Thorsteinsdottir U, Sauvageau G, Hough MR,
Dragowska W, Lansdorp PM, Lawrence HJ, Largman C and Humphries RK:
Overexpression of HOXA10 in murine hematopoietic cells perturbs
both myeloid and lymphoid differentiation and leads to acute
myeloid leukemia. Mol Cell Biol. 17:495–505. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chu MC, Selam FB and Taylor HS: HOXA10
regulates p53 expression and matrigel invasion in human breast
cancer cells. Cancer Biol Ther. 3:568–572. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Y, Zhang J, Wang H, Zhao J, Xu C, Du
Y, Luo X, Zheng F, Liu R, Zhang H and Ma D: miRNA-135a promotes
breast cancer cell migration and invasion by targeting HOXA10. BMC
Cancer. 12:1112012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carrera M, Bitu CC, de Oliveira CE,
Cervigne NK, Graner E, Manninen A, Salo T and Coletta RD: HOXA10
controls proliferation, migration and invasion in oral squamous
cell carcinoma. Int J Clin Exp Pathol. 8:3613–3623. 2015.PubMed/NCBI
|
|
22
|
Kim JW, Kim JY, Kim JE, Kim SK, Chung HT
and Park CK: HOXA10 is associated with temozolomide resistance
through regulation of the homologous recombinant DNA repair pathway
in glioblastoma cell lines. Genes Cancer. 5:165–174.
2014.PubMed/NCBI
|
|
23
|
Zhang L, Wan Y, Jiang Y, Ma J, Liu J, Tang
W, Wang X and Cheng W: Upregulation HOXA10 homeobox gene in
endometrial cancer: Role in cell cycle regulation. Med Oncol.
31:522014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jougla E: Survival of cancer patients in
Europe (EUROCARE study). Rev Epidemiol Sante Publique. 44:473–475.
1996.(In French). PubMed/NCBI
|
|
26
|
Coward JI, Middleton K and Murphy F: New
perspectives on targeted therapy in ovarian cancer. Int J Womens
Health. 7:189–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zaman MS, Maher DM, Khan S, Jaggi M and
Chauhan SC: Current status and implications of microRNAs in ovarian
cancer diagnosis and therapy. J Ovarian Res. 5:442012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nikitina EG, Urazova LN and Stegny VN:
MicroRNAs and human cancer. Exp Oncol. 34:2–8. 2012.PubMed/NCBI
|
|
29
|
Kim TH, Song JY, Park H, Jeong JY, Kwon
AY, Heo JH, Kang H, Kim G and An HJ: miR-145, targeting
high-mobility group A2, is a powerful predictor of patient outcome
in ovarian carcinoma. Cancer Lett. 356:937–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu S, Fu GB, Tao Z, OuYang J, Kong F,
Jiang BH, Wan X and Chen K: MiR-497 decreases cisplatin resistance
in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget.
6:26457–26471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guan Y, Yao H, Zheng Z, Qiu G and Sun K:
MiR-125b targets BCL3 and suppresses ovarian cancer proliferation.
Int J Cancer. 128:2274–2283. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nygren MK, Tekle C, Ingebrigtsen VA,
Mäkelä R, Krohn M, Aure MR, Nunes-Xavier CE, Perälä M, Tramm T,
Alsner J, et al: Identifying microRNAs regulating B7-H3 in breast
cancer: The clinical impact of microRNA-29c. Br J Cancer.
110:2072–2080. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim JJ, Taylor HS, Lu Z, Ladhani O,
Hastings JM, Jackson KS, Wu Y, Guo SW and Fazleabas AT: Altered
expression of HOXA10 in endometriosis: Potential role in
decidualization. Mol Hum Reprod. 13:323–332. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kelly M, Daftary G and Taylor HS: An
autoregulatory element maintains HOXA10 expression in endometrial
epithelial cells. Am J Obstet Gynecol. 194:1100–1109. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Daftary GS, Troy PJ, Bagot CN, Young SL
and Taylor HS: Direct regulation of beta3-integrin subunit gene
expression by HOXA10 in endometrial cells. Mol Endocrinol.
16:571–579. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng W, Liu J, Yoshida H, Rosen D and
Naora H: Lineage infidelity of epithelial ovarian cancers is
controlled by HOX genes that specify regional identity in the
reproductive tract. Nat Med. 11:531–537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shih IeM, Chen L, Wang CC, Gu J, Davidson
B, Cope L, Kurman RJ, Xuan J and Wang TL: Distinct DNA methylation
profiles in ovarian serous neoplasms and their implications in
ovarian carcinogenesis. Am J Obstet Gynecol. 203(584): e1–22.
2010.
|
|
39
|
Pan BL, Tong ZW, Wu L, Pan L, Li JE, Huang
YG, Li SD, Du SX and Li XD: Effects of MicroRNA-206 on osteosarcoma
cell proliferation, apoptosis, migration and invasion by targeting
ANXA2 through the AKT signaling pathway. Cell Physiol Biochem.
45:1410–1422. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Libório-Kimura TN, Jung HM and Chan EK:
miR-494 represses HOXA10 expression and inhibits cell proliferation
in oral cancer. Oral Oncol. 51:151–157. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li B, Jin H, Yu Y, Gu C, Zhou X, Zhao N
and Feng Y: HOXA10 is overexpressed in human ovarian clear cell
adenocarcinoma and correlates with poor survival. Int J Gynecol
Cancer. 19:1347–1352. 2009. View Article : Google Scholar : PubMed/NCBI
|