Introduction
Lung cancer is the most common type of cancer and
the leading cause of cancer-associated mortality worldwide
(1). Non-small cell lung cancer
(NSCLC) is the most common form of lung cancer (2). Recent advances in the diagnosis and
treatment of cancer have been achieved; however, the 5-year overall
survival rate of NSCLC is still only 11% (3). Consequently, an in-depth analysis of
the mechanisms underlying NSCLC development and progression is
required.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs,
which negatively regulate the expression of target genes by binding
to the 3′ untranslated region (3′UTR) (4). It has previously been demonstrated
that miRNAs serve key roles in the development and progression of
numerous types of cancer, including lung cancer (5). A previous study suggested that
miR-140-5p is involved in cancer progression (6). Furthermore, numerous studies
regarding miR-140-5p have been performed; for example, miR-140-5p
has been demonstrated to inhibit the growth of ovarian cancer by
suppressing the expression of platelet-derived growth factor
receptor A (7). miR-140-5p has
also been reported to inhibit the invasion and angiogenesis of
breast cancer by targeting vascular endothelial growth factor-A
(VEGFA) (8). The present study
aimed to investigate the precise molecular mechanism of miR-140-5p
in NSCLC.
VEGF, which serves a major role in angiogenesis, is
a dimeric glycoprotein secreted from numerous cell types, including
cancer cells (9). VEGF is a member
of the platelet-derived growth factor family, which includes VEGFA,
VEGFB, VEGFC, VEGFD and VEGFE (10). Upregulation of the VEGFA gene has
been identified as a poor prognostic element for tumor-free
survival in osteosarcoma (11).
Therefore, VEGFA may be considered a potential target for cancer
therapy.
The results of the present study demonstrated that
downregulation of miR-140-5p was associated with clinical grading
and metastasis in lung cancer tissues. In addition, VEGFA was
verified to be a direct target of miR-140-5p. These findings
suggested that miR-140-5p may function as a tumor suppressor, which
affects lung cancer cell migration and invasion by inhibiting the
expression of VEGFA.
Materials and methods
Cell culture and transfection
The lung cancer cell line A549 was purchased from
The Cell Bank of Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China) and cultured in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100
U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in
a humidified atmosphere containing 5% CO2.
For cell transfection, A549 cells were seeded into
6-well plates at a density of 2×105 cells/well. Briefly,
a miR-140-5p mimic (5′-CAGUGGUUUUACCCUAUGGUAG-3′; 100 nM; Guangzhou
RiboBio Co., Ltd., Guangzhou, China) or a negative control (NC)
mimic (5′-CUCACCAAAAACCCUAUGGUAG-3′; 100 nM; Guangzhou RiboBio Co.,
Ltd.) and Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) were diluted in DMEM. Following this, the mixture
was added to the 6-well plates to obtain a final concentration of
20 nmol/l of hsa-miR-140-5p mimics or miR-NC mimics, and
subsequently incubated at 37°C for 48 h.
Luciferase reporter assay
The potential targets of miR-140-5p were analyzed
using TargetScan (http://www.targetscan.org/vert_71/). The 3′-UTR of
VEGFA was amplified by polymerase chain reaction (PCR), which was
inserted into a pGL3-basic (Promega Corporation, Madison, WI, USA)
using PrimeSTAR Max DNA Polymerase (Takara Biotechnology Co., Ltd.,
Dalian, China) to obtain a VEGFA-3′-UTR reporter construct. The
primer sequences used for the amplification of VEGFA were as
follows: Forward, 5′-GCTCTAGAGAGCCTCCCTCAGGGTTTC-3′ and reverse,
5′-GCTCTAGAAAGGAATGTGTGCTGGGGAG-3′. The thermocycling conditions
used for PCR were are follows: Denaturation for 10 sec at 98°C;
followed by 35 cycles of annealing for 10 sec at 55°C and
elongation for 10 sec at 72°C. A VEGFA-3′-UTR-mutant (mut) reporter
construct was also obtained by mutating the complementary seed
sequences in the miR-140-5p-binding region using the QuikChange
Site-Directed Mutagenesis kit (Agilent Technologies, Inc., Santa
Clara, CA, USA). A549 cells (2.5×104) were transfected
with 0.1 µg reporter construct (mut or wild type), 100 nM
miR-140-5p mimic or NC, and 5 ng Renilla luciferase vector
(phRL-TK; Promega Corporation) by Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. A total of 24 h post-transfection,
luciferase activity was measured by Luminoskan Ascent (Thermo
Labsystems, Helsinki, Finland) and a Dual-Luciferase Reporter Assay
kit (Promega Corporation), according to the manufacturers'
protocols. Luciferase activity was normalized to Renilla
luciferase activity.
Tissue samples
Paired NSCLC and adjacent non-tumor lung tissues
were obtained from 30 patients who were treated at Linyi People's
Hospital (Linyi, China) during tumor resection between January 2013
and February 2015. None of patients had previously undergone
treatment prior to tissue collection. Prior written informed
consent was obtained from each subject. The present study was
approved by the Ethics Committee of Linyi People's Hospital.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total miRNA was extracted using the mirVana miRNA
Isolation kit (Ambion; Thermo Fisher Scientific, Inc.), according
to the manufacturer's protocol. Total RNA was prepared by
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. cDNA was prepared using
the PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. Subsequently, cDNA was
used to detect mRNA expression levels by qPCR using the
SYBR® Premix Ex Taq™ II Perfect Real time kit
(Takara Biotechnology Co., Ltd.) and an ABI 7500 Real-time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). mRNA
and miRNA expression levels were determined using the
2−ΔΔCq method (12).
For the detection of miRNAs, miRNA purification miRNeasy Mini kit
(Qiagen GmbH, Hilden, Germany) was used for the extraction of total
miRNA.TransScript First-Strand cDNA Synthesis SuperMix (Beijing
Transgen Biotech Co., Ltd., Beijing, China) was used to perform
reverse transcription, SYBR-Green 2× Master Mix (Thermo Fisher
Scientific, Inc.) and RT-PCR primer sets were used. qPCR was
performed using a CFX96™ real-time PCR detection system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). U6 was used as an internal
control for miR-140-5p. β-actin was used as an internal control for
VEGFA. The thermocycling conditions used for qPCR were as follows:
4 min at 94°C (initial activation); followed by 40 cycles of 30 sec
at 94°C (denaturation), 30 sec at 58°C (annealing) and 30 sec at
72°C (extension). The primer sequences used were as follows:
miR-140-5p forward, 5′-GAGTGTCAGTGGTTTTACCCT-3′ and reverse,
5′-GCAGGGTCCGAGGTATTC-3′; VEGFA forward,
5′-TTTCTGCTGTCTTGGGTGCATTGG-3′ and reverse,
5′-ACCACTTCGTGATGATTCTGCCCT-3′; β-actin forward,
5′-TCAAGATCATTGCTCCTCCTG-3′ and reverse,
5′-CTGCTTGCTGATCCACATCTG-3′; and U6 forward,
5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′.
Western blot analysis
A total of 48 h post-transfection, A549 cells were
washed with PBS and proteins were extracted using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Haimen, China). Protein concentration was determined
using the bicinchoninic acid protein assay (Pierce; Thermo Fisher
Scientific, Inc.). Samples (15 µg/lane) were separated by 12%
SDS-PAGE and transferred to polyvinylidene difluoride membranes.
Subsequently, the membranes were blocked using 5% non-fat milk at
room temperature for 1 h, and subsequently probed with the
following primary antibodies at 4°C overnight: Anti-AKT (cat. no.
ab81283; 1:1,000), anti-phosphorylated AKT (cat. no. ab38449;
1:1,000) and anti-GAPDH (cat. no. ab8245; 1:1,000; all Abcam,
Cambridge, UK). Following this, membranes were incubated with goat
anti-rabbit horseradish peroxidase-conjugated secondary antibodies
(cat. no. sc2004; 1:5,000; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) at 37°C for 1 h. Finally, the blots were visualized using
enhanced chemiluminescent reagents (Thermo Fisher Scientific,
Inc.).
Wound-healing assay
A549 cells transfected with miR-140-5p and NC mimics
were seeded into 6-well plates at 1×105 cells/l, and
were cultivated in DMEM supplemented with 1% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) for 6 h at 37°C to
allow adherent growth. Subsequently, a scratch was made in the cell
layer using a 10-µl pipette tip. After washing with serum-free
medium, A549 cells were cultured at 37°C in an atmosphere
containing 5% CO2 for 24 h. The wound-healing ability of
the cells was visualized under a light microscope and calculated by
measuring the distance between the scratches, as follows: Mobility
ratio=(initial scratch width-current scratch width)/initial scratch
width.
Transwell assay
Cell invasion was measured using Transwell cell
culture chambers (Corning Incorporated, Corning, NY, USA) coated
with 10 µl Matrigel (1:3; BD Biosciences, San Jose, CA, USA).
Briefly, A549 cells (5×105/200 µl) transfected with
miR-140-5p and NC mimics were added to the upper chambers with
complete culture medium, whereas serum-free medium with 5% FBS was
added into the lower chambers. Following 24 h of incubation at
37°C, cells that had invaded through the Matrigel were fixed with
4% paraformaldehyde for 30 min at room temperature and stained with
0.1% crystal violet at room temperature for 20 min. Finally, cells
within five fields of view were counted under a light
microscope.
Statistical analysis
The quantitative values are presented as the means ±
standard error of the mean. The two-tailed Student's t-test was
applied to compare between two groups. One-way analysis of variance
followed by Student-Newman-Keuls post hoc analysis was applied to
analyze the miR-140-5p expression among the T classification and N
classification groups, as well as the differences between multiple
groups. Statistical analyses were conducted using SPSS 18.0 (SPSS
software, Inc., Chicago, IL, USA) and GraphPad Prism 5 (GraphPad
Software, Inc., La Jolla, CA, USA). All experiments were repeated
in triplicate. P<0.05 was considered to indicate a statistically
significant difference.
Results
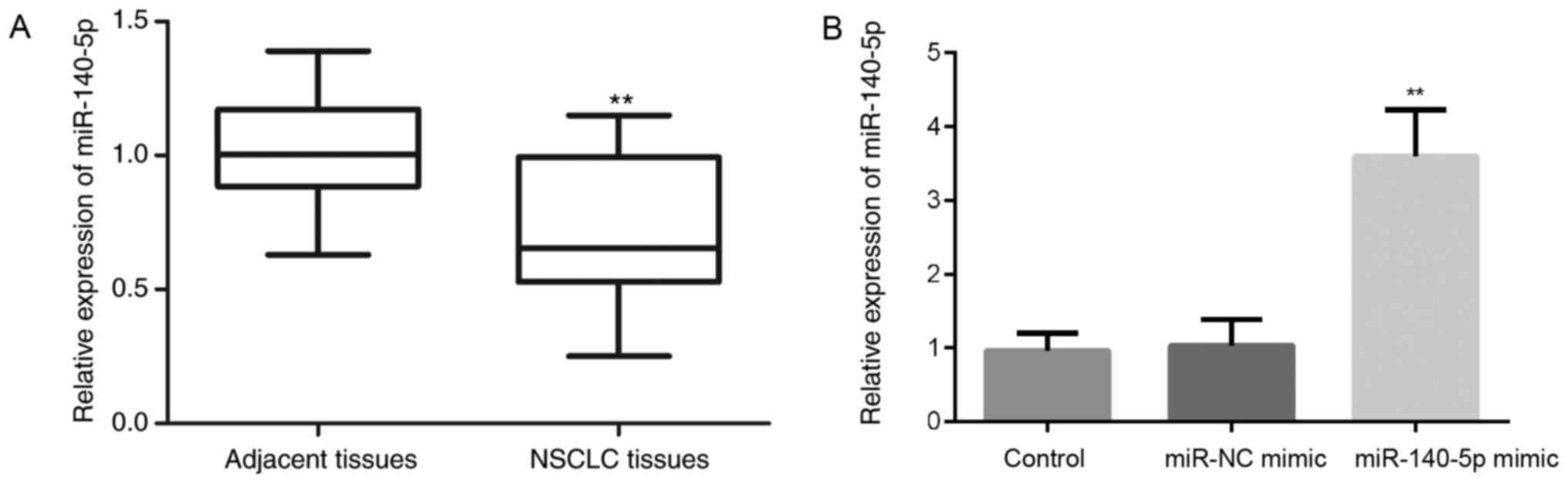
Loss of miR-140-5p in human NSCLC is
associated with patient N/M classification
To determine the expression levels of miR-140-5p in
NSCLC, the expression levels of miR-140-5p were measured in 30
pairs of NSCLC samples and matched normal lung tissues by RT-qPCR.
The results indicated that the miR-140-5p expression was
significantly reduced in NSCLC tissues compared with in the matched
normal tissues (P<0.01; Fig.
1A). These findings are consistent with those of a previous
study, which identified a decreased expression of miR-140 in NSCLC
samples (13).
Furthermore, the association between miR-140-5p
expression and the clinicopathological parameters of patients with
NSCLC were analyzed and exhibited in Table I. Statistical analysis demonstrated
that downregulation of miR-140-5p was significantly associated with
N classification (P<0.001, f=9.131) and M classification
(P=0.002, t=3.456); however, no significant association was
observed with regards to the other parameters, including sex
(P=0.432, t=3.223), age (P=0.064, t=1.932), smoking history
(P=0.108, t=1.663) or T classification (P=0.752, f=0.403).
 | Table I.Basic characteristics of patients with
non-small cell lung cancer. |
Table I.
Basic characteristics of patients with
non-small cell lung cancer.
| Characteristic | Case number | miR-140-5p
expression | t-value | f-value | P-value |
|---|
| Sex |
|
| 3.223 |
| 0.432 |
| Male | 20 | 0.741±0.066 |
|
|
|
|
Female | 10 | 0.655±0.071 |
|
|
|
| Age (years) |
|
| 1.932 |
| 0.064 |
| ≥60 | 19 | 0.700±0.068 |
|
|
|
|
<60 | 11 | 0.648±0.061 |
|
|
|
| Smoking history |
|
| 1.663 |
| 0.108 |
| No | 11 | 0.692±0.098 |
|
|
|
|
Yes | 19 | 0.647±0.051 |
|
|
|
| T
classification |
|
|
| 0.403 | 0.752 |
| T1 | 9 | 0.648±0.065 |
|
|
|
| T2 | 11 | 0.623±0.074 |
|
|
|
| T3 | 8 | 0.633±0.047 |
|
|
|
| T4 | 2 | 0.603±0.033 |
|
|
|
| N
classification |
|
|
| 9.131 | <0.001 |
| N0 | 11 | 0.776±0.087 |
|
|
|
| N1 | 10 | 0.657±0.071 |
|
|
|
| N2 | 6 | 0.601±0.065 |
|
|
|
| N3 | 3 | 0.596±0.058 |
|
|
|
| M
classification |
|
| 3.456 |
| 0.002 |
| M0 | 25 | 0.703±0.091 |
|
|
|
| M1 | 5 | 0.559±0.031 |
|
|
|
miR-140-5p has previously been demonstrated to be
dysregulated in human colorectal cancer tissues (14), ovarian cancer tissues (7) and gastric cancer (15). In addition, miR-140-5p has been
reported to inhibit cell invasion and migration (14,16).
Therefore, the following experiments were conducted to analyze the
effects of miR-140-5p on NSCLC cell behavior, including cell
invasion and migration.
The effects of the miR-140-5p mimic were initially
detected on miR-140-5p expression in A549 cells by RT-qPCR. The
results demonstrated that, compared with in cells in the control
and miR-NC mimic groups, the expression levels of miR-140-5p were
significantly increased post-transfection with the miR-140-5p mimic
(P<0.01; Fig. 1B).
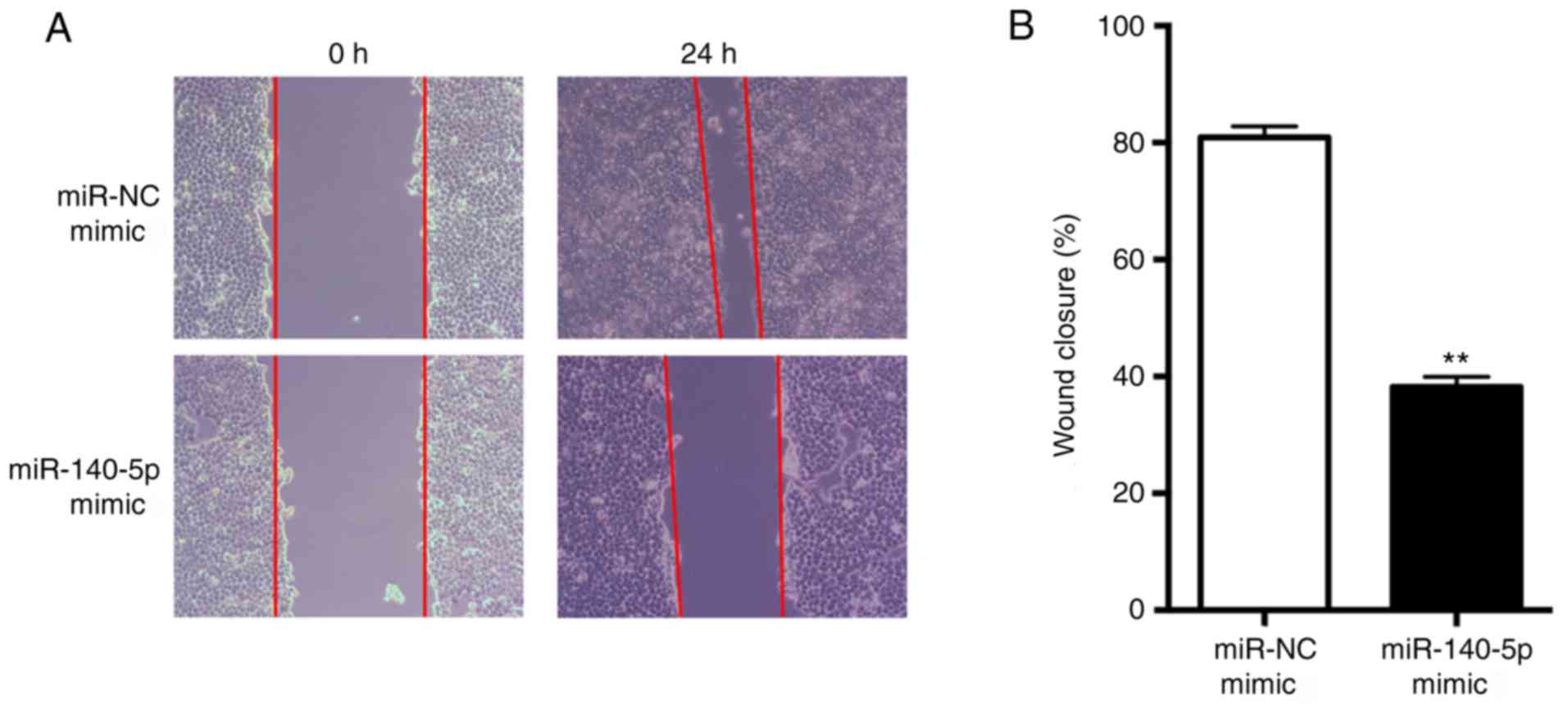
miR-140-5p suppresses NSCLC cell
migration in vitro
Using a wound-healing assay, it was demonstrated
that overexpression of miR-140-5p significantly suppressed tumor
cell migration in A549 cells compared with in the NC group
(P<0.01; Fig. 2). This result
suggested that miR-140-5p may suppress NSCLC cell migration in
vitro.
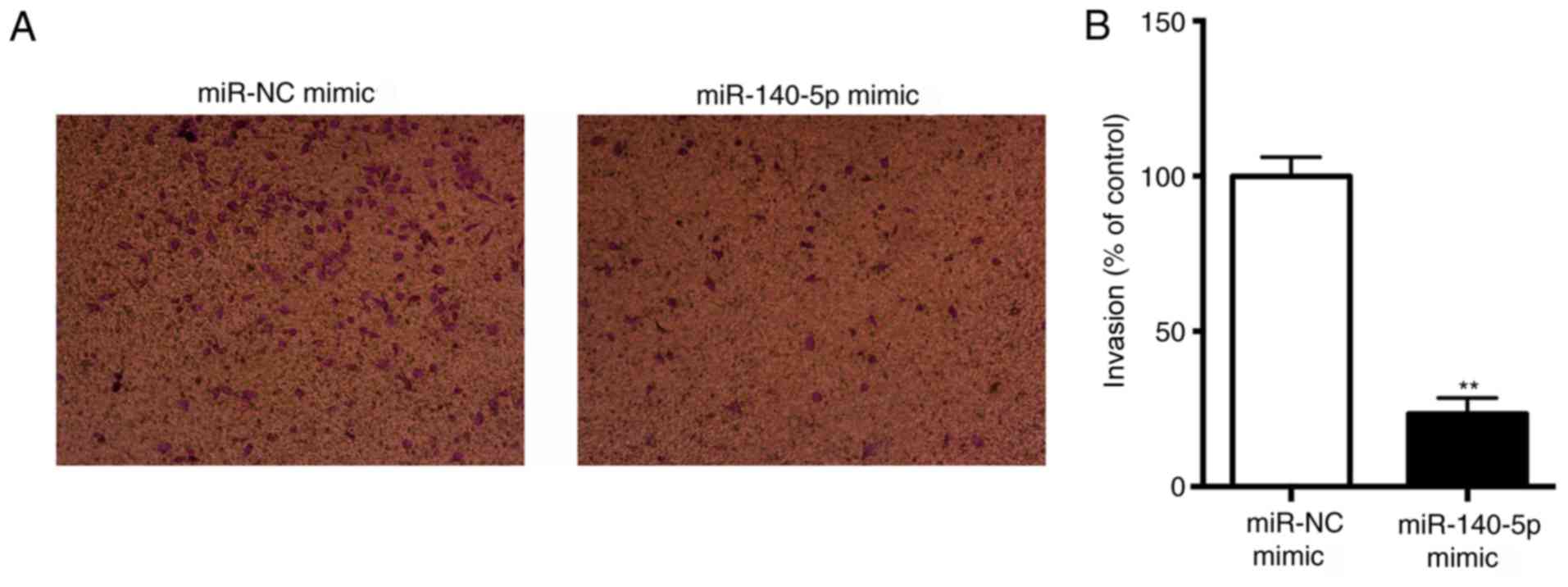
miR-140-5p suppresses NSCLC cell
invasion in vitro
Transwell assays with Matrigel revealed that
miR-140-5p significantly decreased the invasive capacity of A549
cells (P<0.01; Fig. 3). This
result suggested that miR-140-5p may suppress NSCLC cell invasion
in vitro.
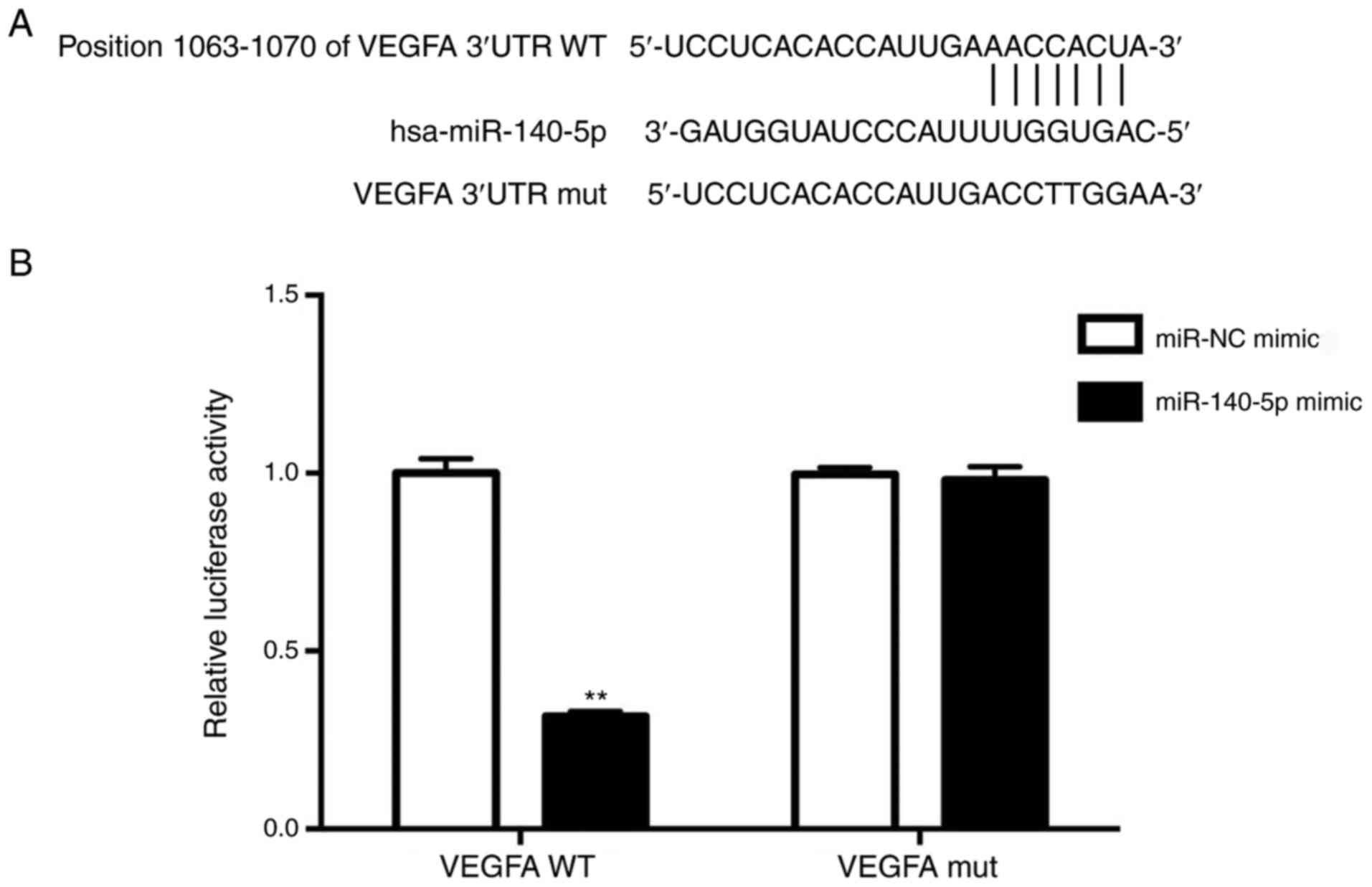
miR-140-5p directly targets the 3UTR
of VEGFA
To elucidate the molecular mechanisms by which
miR-140-5p performs its function, the potential targets of
miR-140-5p were analyzed using computational methods including
TargetScan. In particular, oncogenes that could be targeted by
miR-140-5p were focused on. VEGFA was demonstrated to be a
potential miR-140-5p target with a complementary sequence to
miR-140-5p being identified in the 3′UTR of VEGFA by TargetScan
analysis (Fig. 4A).
To confirm whether VEGFA is a direct downstream
target of miR-140-5p, a luciferase reporter assay was performed.
The results demonstrated that overexpression of miR-140-5p
significantly decreased the relative luciferase activity of
VEGFA-3′UTR in A549 cells (P<0.01; Fig. 4B), but had no significant effect on
the VEGFA-3′UTR mut. These results suggested that miR-140-5p may
downregulate VEGFA expression by directly targeting its 3′UTR.
miR-140-5p inactivates p-AKT signaling
by targeting and downregulating VEGFA
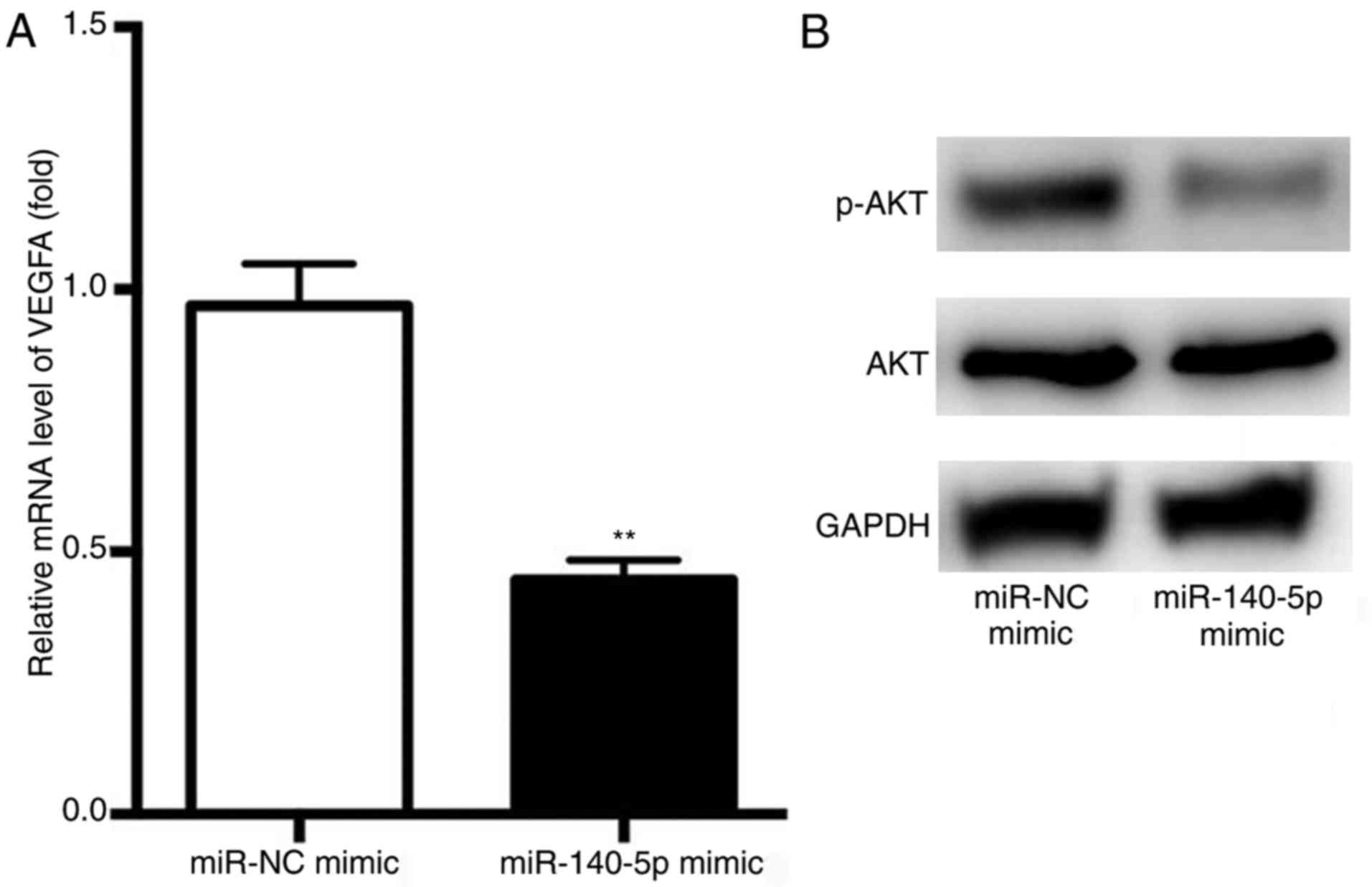
Finally, RT-qPCR analysis revealed that the average
expression levels of VEGFA were significantly decreased in the
miR-140-5p mimic group compared with in the NC group (Fig. 5A). These results indicated that
VEGFA may be targeted by miR-140-5p in NSCLC A549 cells.
Subsequently, the mechanisms by which the miR-140-5p/VEGFA axis
affected the migration and invasion of NSCLC cells were
investigated. The phosphatidylinositol 3-kinase/protein kinase B
(PI3K/AKT) signaling pathway has been reported to be involved in
angiogenesis and lung cancer progression (16). Therefore, the activation of p-AKT
in NSCLC cells was investigated. Overexpression of miR-140-5p was
observed to decrease the levels of p-AKT, with no obvious effects
on the levels of total AKT (Fig.
5B). These results suggested that the miR-140-5p/VEGFA axis may
negatively regulate the migration and invasion of NSCLC cells via
p-AKT signaling.
Discussion
NSCLC is the most common type of lung cancer, which
is associated with a high morbidity and mortality rate worldwide
(17). The prognosis of patients
with lung cancer is poor and treatment efficacy is not satisfactory
(18). It is therefore necessary
to develop novel therapeutic strategies for patients with NSCLC,
and to perform an in-depth investigation into the development and
progression of NSCLC.
It has previously been demonstrated that miRNAs
serve key roles in the development and progression of lung cancer
(5). The association between
miR-140-5p and VEGFA has not been comprehensively investigated;
however, miR-140-5p (13) and
VEGFA (19,20) have been implicated in NSCLC.
The present study demonstrated that the expression
levels of miR-140-5p were decreased in NSCLC tissues compared with
in matched normal tissues, which was consistent with a previous
study (13). Furthermore, it was
demonstrated that downregulation of miR-140-5p was significantly
associated with the N classification and M classification of
patients. These results indicated that miR-140-5p served a tumor
suppressive role in NSCLC; however, the effects of miR-140-5p on
cell behavior remain to be investigated. Therefore, a miR-140-5p
mimic was used to upregulate the expression levels of miR-140-5p in
A549 cells, as verified by RT-qPCR.
NSCLC cell migration and invasion were demonstrated
to be inhibited by overexpression of miR-140-5p, which is in
agreement with the role of miR-140-5p in gastric cancer, as
reported by Fang et al (15), who demonstrated that it
significantly inhibited cell migration and invasion of AGS and
BGC823 cells. In addition, the function of miR-140-5p in breast
cancer was demonstrated by Lu et al (8); miR-140-5p markedly suppressed the
invasion of MCF-7 and MDA-MB-231 cells. However, to the best of our
knowledge, it remains unknown how miR-140-5p executes its function
in NSCLC.
The present study also aimed to determine the target
mRNAs of miR-140-5p that functioned as oncogenes. VEGFA is
essential for migration, invasion and angiogenesis in
hepatocellular carcinoma (21), as
well as tumor progression of NSCLC (22). Notably, VEGFA was identified as a
candidate gene of miR-140-5p by TargetScan; this finding was
validated by luciferase reporter assays in A549 cells.
Finally, the mechanisms by which the
miR-140-5p/VEGFA axis affected the migration and invasion of NSCLC
cells were investigated. The PI3K/AKT signaling pathway has been
reported to serve a role in angiogenesis and lung cancer
progression (16). As a crucial
component of the PI3K/AKT signaling pathway, AKT mediates a large
number of cellular responses through activating the expression of
VEGFA (23,24). A link between activation of the
PI3K/AKT signaling pathway and increased expression of VEGF has
been demonstrated in numerous studies (25,26).
Furthermore, VEGFA has been reported to induce AKT phosphorylation
in NSCLC (22). Therefore, in the
present study, the expression levels of VEGFA and p-AKT were
analyzed by RT-qPCR and western blotting, respectively. Results
revealed that the expression levels of VEGFA and p-AKT level were
decreased in the miR-140-5p mimic group compared with in the NC
group. Conversely, there were no obvious alterations in the protein
expression levels of AKT between the miR-140-5p mimic and NC
groups. These results suggested that the miR-140-5p/VEGFA axis may
inhibit the migration and invasion of NSCLC cells via inactivation
of p-AKT signaling. In conclusion, targeting miR-140-5p or VEGFA
may serve as an appealing strategy for lung cancer therapy.
Activation of PI3K/AKT has previously been reported
to stimulate VEGFA expression (27). In the present study, the results
suggested that the miR-140-5p/VEGFA axis may inactivate p-AKT;
however, several issues require further investigation: i) The
association between VEGFA and p-AKT; ii) the association between
p-AKT and miR-140-5p; and iii) the effects of VEGF on the PI3K/AKT
signaling pathway, these issues will be investigated in future
studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ designed the study, performed the data analyses
and wrote the manuscript. PY, JX and LZ performed the experiments
and analyzed the data. KL was responsible for patient enrollment,
analyzed patient data and wrote the first draft of the manuscript
prior to the re-editing of the manuscript by HZ.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Linyi People's Hospital. Prior written informed
consent was obtained from each subject.
Patient consent for publication
Prior written informed consent was obtained from
each subject.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Verdecchia A, Francisci S, Brenner H,
Gatta G, Micheli A, Mangone L and Kunkler I: EUROCARE-4 Working
Group: Recent cancer survival in Europe: A 2000–02 period analysis
of EUROCARE-4 data. Lancet Oncol. 8:784–796. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Del Vescovo V, Grasso M, Barbareschi M and
Denti MA: MicroRNAs as lung cancer biomarkers. World J Clin Oncol.
5:604–620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ruan K, Fang X and Ouyang G: MicroRNAs:
Novel regulators in the hallmarks of human cancer. Cancer Lett.
285:116–126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lan H, Chen W, He G and Yang S: miR-140-5p
inhibits ovarian cancer growth partially by repression of PDGFRA.
Biomed Pharmacother. 75:117–122. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu Y, Qin T, Li J, Wang L, Zhang Q, Jiang
Z and Mao J: MicroRNA-140-5p inhibits invasion and angiogenesis
through targeting VEGF-A in breast cancer. Cancer Gene Ther.
24:386–392. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Petrovic N: Targeting angiogenesis in
cancer treatments: Where do we stand? J Pharm Pharm Sci.
19:226–238. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shinkaruk S, Bayle M, Laïn G and Déléris
G: Vascular endothelial cell growth factor (VEGF), an emerging
target for cancer chemotherapy. Curr Med Chem Anticancer Agents.
3:95–117. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang J, Yang D, Sun Y, Sun B, Wang G,
Trent JC, Araujo DM, Chen K and Zhang W: Genetic amplification of
the vascular endothelial growth factor (VEGF) pathway genes,
including VEGFA, in human osteosarcoma. Cancer. 117:4925–4938.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan Y, Shen Y, Xue L and Fan H: miR-140
suppresses tumor growth and metastasis of non-small cell lung
cancer by targeting insulin-like growth factor 1 receptor. PLoS
One. 8:e736042013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhai H, Fesler A, Ba Y, Wu S and Ju J:
Inhibition of colorectal cancer stem cell survival and invasive
potential by hsa-miR-140-5p mediated suppression of Smad2 and
autophagy. Oncotarget. 6:19735–19746. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y,
Zhang T, Khaliq J and Li Y: miR-140-5p suppresses the
proliferation, migration and invasion of gastric cancer by
regulating YES1. Mol Cancer. 16:1392017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jeannot V, Busser B, Brambilla E, Wislez
M, Robin B, Cadranel J, Coll JL and Hurbin A: The PI3K/AKT pathway
promotes gefitinib resistance in mutant KRAS lung adenocarcinoma by
a deacetylase-dependent mechanism. Int J Cancer. 134:2560–2571.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen HY, Yu SL, Chen CH, Chang GC, Chen
CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, et al: A five-gene
signature and clinical outcome in non-small-cell lung cancer. N
Engl J Med. 356:11–20. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao D, Pan C, Sun J, Gilbert C,
Drews-Elger K, Azzam DJ, Picon-Ruiz M, Kim M, Ullmer W, El-Ashry D,
et al: VEGF drives cancer-initiating stem cells through
VEGFR-2/Stat3 signaling to upregulate Myc and Sox2. Oncogene.
34:3107–3119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gu A, Lu J, Wang W, Shi C, Han B and Yao
M: Role of miR-497 in VEGF-A-mediated cancer cell growth and
invasion in non-small cell lung cancer. Int J Biochem Cell Biol.
70:118–125. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ghosh A, Dasgupta D, Ghosh A, Roychoudhury
S, Kumar D, Gorain M, Butti R, Datta S, Agarwal S, Gupta S, et al:
MiRNA199a-3p suppresses tumor growth, migration, invasion and
angiogenesis in hepatocellular carcinoma by targeting VEGFA,
VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 8:e27062017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Geng J, Li X, Zhou Z, Wu CL, Dai M and Bai
X: EZH2 promotes tumor progression via regulating VEGF-A/AKT
signaling in non-small cell lung cancer. Cancer Lett. 359:275–287.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Osaki M, Oshimura M and Ito H: PI3K-Akt
pathway: Its functions and alterations in human cancer. Apoptosis.
9:667–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shiojima I and Walsh K: Role of Akt
signaling in vascular homeostasis and angiogenesis. Circ Res.
90:1243–1250. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hudson C, Liu M, Chiang G, Otterness D,
Loomis D, Kaper F, Giaccia A and Abraham R: Regulation of
hypoxia-inducible factor 1alpha expression and function by the
mammalian target of rapamycin. Mol Cell Biol. 22:7004–7014. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu GT, Chen HT, Tsou HK, Tan TW, Fong YC,
Chen PC, Yang WH, Wang SW, Chen JC and Tang CH: CCL5 promotes
VEGF-dependent angiogenesis by down-regulating miR-200b through
PI3K/Akt signaling pathway in human chondrosarcoma cells.
Oncotarget. 5:10718–10731. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qi L, Zhu F, Li SH, Si LB, Hu LK and Tian
H: Retinoblastoma binding protein 2 (RBP2) promotes
HIF-1α-VEGF-induced angiogenesis of non-small cell lung cancer via
the Akt pathway. PLoS One. 9:e1060322014. View Article : Google Scholar : PubMed/NCBI
|