Introduction
Bcl-2 associated athanogene (BAG) 3 is a member of
the human BAG family of co-chaperone proteins (1). The BAG domain of BAG3 protein binds
to heat shock protein (HSP) 70, a major chaperone protein involved
in anti-apoptosis through recovery of unfolded proteins as well as
interference with pro-apoptotic cytochrome c release from
mitochondria (2). BAG3 is also
known to bind to phospholipase C-γ and Bcl-2, which synergistically
inhibit cell death (3). In
addition, BAG3 was reported to interact with dual-specificity
phosphatase 6, which is involved in extracellular signal-regulated
kinase de-phosphorylation, resulting in the induction of cell
proliferation (4). BAG3 possesses
an N-terminal WW domain and a C-terminal PxxP domain that interact
with partner proteins other than HSP70, resulting in modulation of
various biological processes such as anti-apoptosis, proliferation,
cell adhesion, metastasis, invasion, and autophagy (3,5–7).
Under physiological conditions, constitutive
expression levels of BAG3 are low in normal cells other than muscle
cells. BAG3 expression is induced under stress conditions such as
heavy metals, heat, oxidative stress, ultrasound, and starvation
(8–13). The expression of BAG3 is reported
to be regulated partially by the activation of heat shock
transcription factor 1 as in the cases of HSPs (14). In addition, BAG3 and some HSP
family proteins are controlled by hypoxia-induced factor 1, which
is highly expressed under hypoxic conditions, such as in tumor
microenvironments (15). However,
many studies have revealed that BAG3 expression is also elevated
under normoxic conditions in numerous tumor cells including breast
cancer, prostate cancer, ovarian cancer, colorectal cancer,
melanoma, and osteosarcoma (3,6,16–19).
This is probably because BAG3 contributes to cell proliferation as
well as cell survival through interaction with anti-apoptotic
proteins such as Bcl-2, and myeloid leukemia cell differentiation
protein 1 that are overexpressed in cancer (20). Indeed, overexpression of BAG3
correlates with dismal prognosis in melanoma and several carcinomas
(6,16,18).
An impressive recent study revealed functional
categories of BAG3 partner proteins by using novel comprehensive
proteome analysis, called quantitative immunoprecipitation combined
with knockdown (QUICK) (21).
Protein analysis through evolutionary relationships classification
of the data obtained from QUICK demonstrated that the BAG3
interactome includes transcription factors, indicating that
BAG3-dependent cell proliferation and survival may be mediated by
gene transcription, at least in part. Despite accumulating data on
the functions of BAG3 and its protein-protein interactions, neither
the transcripts associated with BAG3 overexpression in cancer
cells, nor their functions, are fully elucidated. Here, we
performed DNA microarray-based comprehensive transcriptome analysis
and bioinformatics on two BAG3 knockout (KO) HeLa cell clones
established using the clustered regularly interspaced short
palindromic repeats (CRISPR)-Cas9 (CRISPR associated protein 9)
genome editing system. Finally, we identified genetic networks of
transcripts associated with proliferation and cell survival, which
may be dependent on BAG3 expression.
Materials and methods
Cell culture
Human cervical cancer HeLa cells were newly obtained
from the Human Science Research Resources Bank, Japan Health
Sciences Foundation (Tokyo, Japan) for this experiment. Cells were
cultured in E-MEM (Wako Pure Chemical Industries, Ltd., Osaka,
Japan) supplemented 10% fetal bovine serum (Equitech-Bio, Inc.,
Kerrville, TX, USA) and 1% penicillin/streptomycin (Nacali Tesque,
Inc., Kyoto, Japan).
Establishment of BAG3 KO HeLa cell
clones
In the present study, the BAG3 gene was deleted
using the CRISPR-Cas9 genome editing system as described below. For
the expression of Cas9 protein and guide RNA targeting the BAG3
gene, pX362 vector (Addgene, Cambridge, MA, USA) was used as
previously described (22). After
the digestion of pX362 with BbsI, the oligonucleotides
5′-caccGAGACTCCATCCTCTGCCAA-3′ and 5′-aaacTTGGCAGAGGATGGAGTCTC-3′
(upper and lower case letters are protospacer sequence and
additional sequence to clone into the BbsI site, respectively)
corresponding to the single guide RNA target sequence in exon 1 of
BAG3 were annealed and subcloned into the BbsI site of pX362. The
constructed plasmid was transfected into HeLa cells with Effectene
Transfection Reagent (Qiagen GmbH, Hilden, Germany) according to
the manufacture's procedure. The DNA-transfected HeLa cells were
selected by treatment with 1 µg/ml of puromycin (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) for 48 h, followed by limited
dilution to obtain the colonies. The grown colonies were picked up
and expanded. After initial screening by immunoblotting with
anti-BAG3 antibody, genomic DNA from the HeLa cell clones was
prepared and used for polymerase chain reaction to examine the
sequence of the DNA fragment containing the target site. The
forward and reverse primer sequences were
5′-CCAGCCTGTGTTTCTCCACTT-3′ and 5′-CTGTCTTTGCTGGGTGACCT-3′,
respectively.
SDS-PAGE and western blot
analysis
Cells were dissolved in a lysis buffer (150 mM NaCl,
1% Nonidet P-40 and 50 mM Tris-HCl, pH 8.0) containing a protease
inhibitor cocktail (Nacali Tesque, Inc.). SDS-PAGE and Western
blotting were carried out as described elsewhere (12). Proteins were detected using the
following primary antibodies: Anti-BAG3 rabbit monoclonal antibody
(GeneTex Inc., Irvine, CA, USA) and anti-glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) mouse monoclonal antibody (as a loading
reference; Proteintech, Rosemont, IL, USA). Secondary
fluorescent-conjugated anti-mouse and anti-rabbit IgGs (LI-COR
Bioscience, Lincoln, NE, USA) were also used. Protein expression
levels and images were acquired using an Odyssey Infrared Imager
(LI-COR Biosciences).
Cell counting assay
Ten thousand cells were seeded in 24-well culture
plates for cell counting. After trypsinization, the trypan blue dye
exclusion test was performed, by mixing cell suspension with an
equal amount of phosphate-buffered saline containing 0.4% trypan
blue. The number of cells excluding the dye was counted by using an
EVE™ automatic cell counter (NanoEnTek, Inc., Seoul,
Korea).
Cell cycle analysis
Fifty thousand cells were cultured in 60 mm culture
dishes two days before cell cycle analysis. For flow cytometry,
cells were fixed with 70% ice cold ethanol for at least 1 h, and
subsequently treated with 0.25 mg/ml RNase A (Nacali Tesque, Inc.)
and SYTOX AADvanced (Thermo Fisher Scientific, Inc.) as described
(23). The samples were finally
analyzed by flow cytometry using a Novocyte flow cytometer
(Novocyte, San Diego, CA, USA). A total of 10,000 cells per sample
were analyzed in each experiment. Distribution of cells in each
cell cycle phase was analyzed based on the Watson model (24).
RNA isolation
Total RNA was extracted from cells using a
NucleoSpin® RNA isolation kit (Macherey-Nagel GmbH &
Co., Düren, Germany) and treated with DNase I for 20 min at room
temperature to remove residual genomic DNA. The quality of the RNA
was analyzed using a Bioanalyzer 2100 and a RNA6000 Nano LabChip
kit (Agilent Technologies, Inc., Santa Clara, CA, USA). RNA samples
with RNA integrity number values above 9.5 were considered
acceptable.
Gene expression analysis
Microarray and computational gene expression
analyses were performed using a GeneChip® system with a
Human Genome U133-plus 2.0 array (Affymetrix, Inc., Santa Clara,
CA, USA), which was spotted with ~55,000 probe sets, as previously
described. Samples for array hybridization were prepared as
described in the Affymetrix GeneChip® Expression
Technical Manual. The scanned arrays were analyzed using the
GeneChip Analysis Suite Software (Affymetrix, Inc.). The microarray
data were deposited in the Gene Expression Omnibus: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103475.
For global normalization, microarray signals were
processed using a standard MAS5.0 algorithm. The obtained
hybridization intensity data and qualities were checked using the
GeneSpring® software (Agilent Technologies, Inc.).
Observed signals were normalized and genes that had no significant
signals were ignored to reduce noise. In addition, probe sets
targeting specific RefSeq transcripts, based on the RefDIC
database, were extracted (25).
Principal component analyses (PCA) were performed using R v3.2.1.
Venn diagrams and hierarchical clustering from the obtained
normalized intensity data were produced using
GeneSpring® software. Ward's linkage and squared
Euclidean distance were utilized in hierarchical clustering.
In order to examine the molecular functions of
differentially expressed genes and gene networks, data were
analyzed using Ingenuity Pathways Analysis (IPA) tools (Ingenuity
Systems, Mountain View, CA, USA), a web-delivered application that
enables the identification, visualization, and exploration of
molecular interaction networks in gene expression data. The top
five molecular functions were identified and the gene networks
containing the molecules were visualized to provide information
about interactions involving genes that were up- or downregulated
by BAG3 deletion.
Statistical analysis
Data are presented as the mean ± standard deviation
(SD). The statistical significance of differences between data sets
was analyzed using one-way analysis of variance (ANOVA) with
post-hoc Tukey HSD tests (R v3.2.1). P<0.05 was considered to
indicate a statistically significant difference. In microarray
analysis, raw P-values were adjusted by calculating false discovery
rate using the Benjamini-Hochberg method (GeneSpring®
software; Agilent Technologies, Inc.).
Results
Establishment and characterization of
BAG3 KO HeLa cell clones
To confirm the role of BAG3 in cell proliferation
and survival in HeLa cells, we constructed BAG3 KO HeLa cell clones
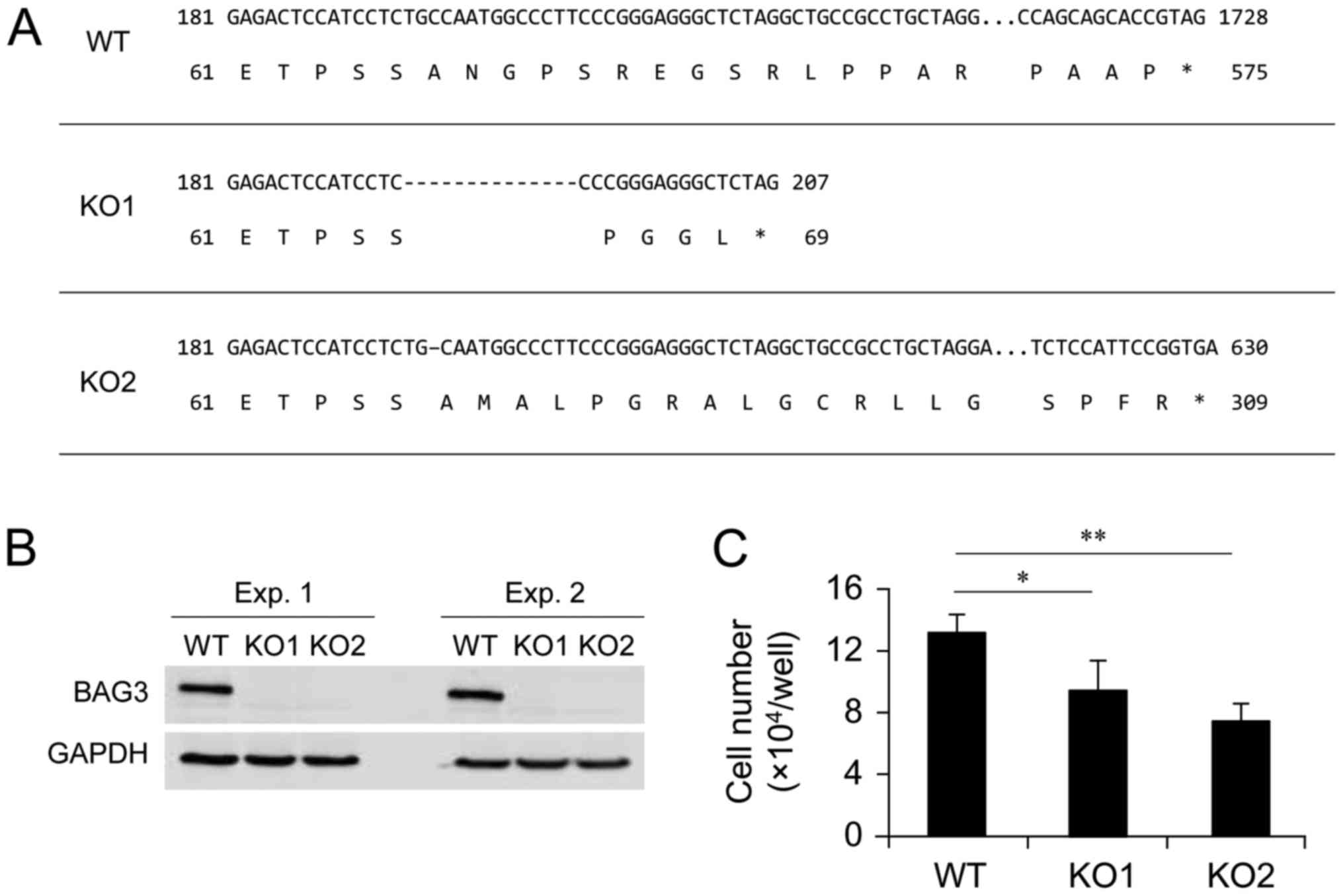
using a CRISPR-Cas9 genome editing system. Among isolated clones,
two clones, designated as KO1 and KO2, were selected as BAG3 KO
candidates. This selection was based on the result of initial
screening by western blotting using anti-BAG3 antibody, since it is
possible that translation can be initiated from an in-frame ATG in
nonsense mutation near the 5′ region of an open reading frame
(26). After the initial
screening, we confirmed the sequence of exon 1 of the BAG3 gene and
found that two clones contained a 14 or 1 bp deletion in exon 1 of
both alleles, respectively, resulting in a frame-shift and
premature termination of BAG3 translation (Fig. 1A). We confirmed that BAG3 protein
expression was diminished as a result of the deletions in the BAG3
gene by repeated western blot analysis (Fig. 1B). The two selected BAG3 KO clones
and the wild-type (WT) control cells were cultured for 7 days to
assess whether BAG3 deletion affected the number of viable cells in
culture conditions. As a result of BAG3 deletion, the number of
viable cells was significantly decreased in both clones (Fig. 1C). This result is consistent with
involvement of BAG3 with cell proliferation and/or cell survival
even under culture conditions without exogenous cytotoxic
stimulation (e.g., heat or oxidative stress).
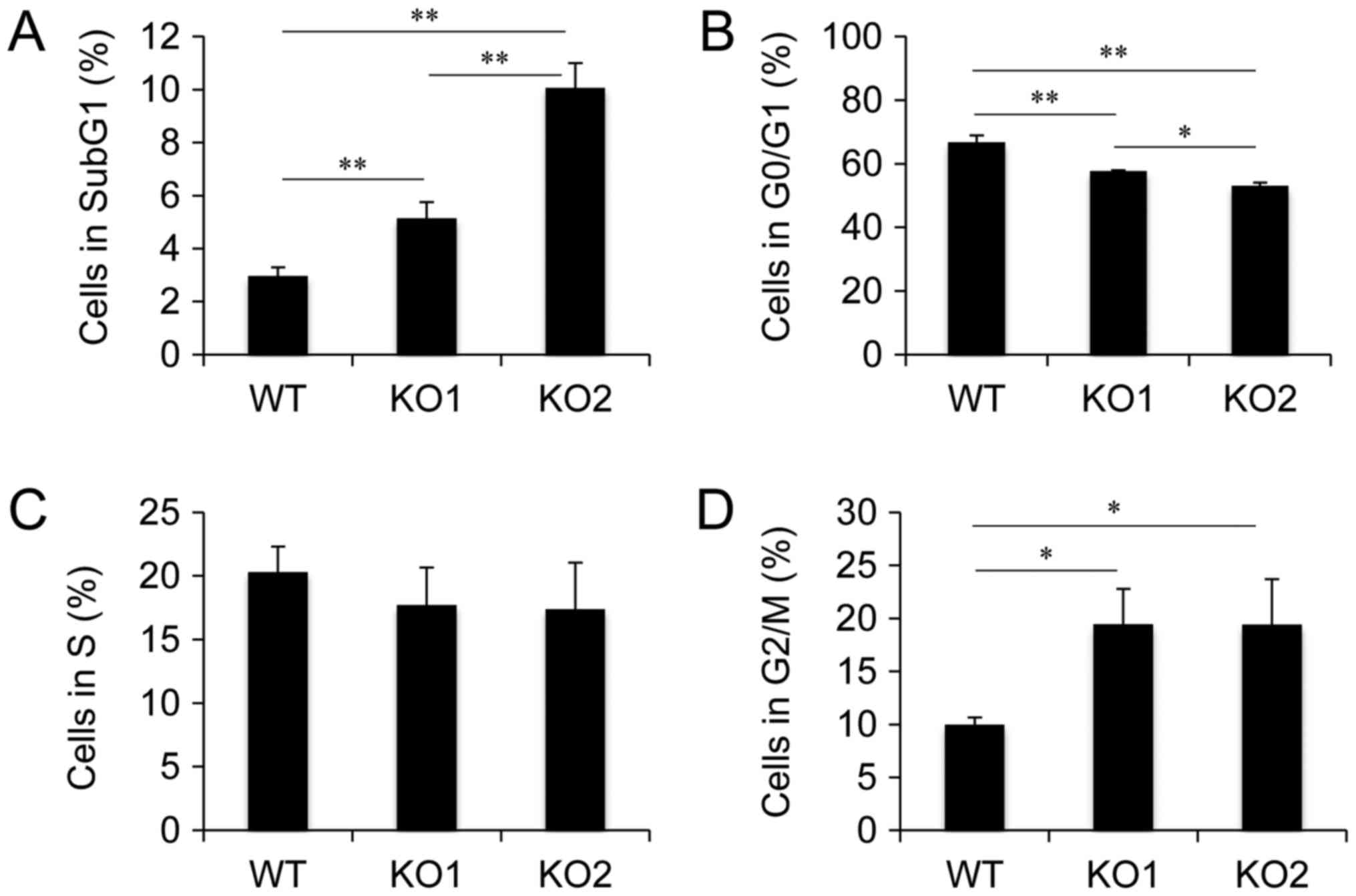
Cell cycle distribution in BAG3 KO HeLa cell clones.
Established BAG3 KO HeLa cell clones showed decreased numbers of
viable cells, indicating that BAG3 deletion led to cell cycle delay
and/or cell death. To examine the effect of BAG3 on cell cycle
progression and cell survival during cell culture, we performed
cell cycle analysis using flow cytometry. Cell cycle analysis
revealed that the populations of cells in Sub G1 and G2/M phase
were increased in both BAG3 KO clones (Fig. 2), indicating that BAG3 is involved
in both cell cycle progression and anti-apoptosis under normal
culture conditions in HeLa cells.
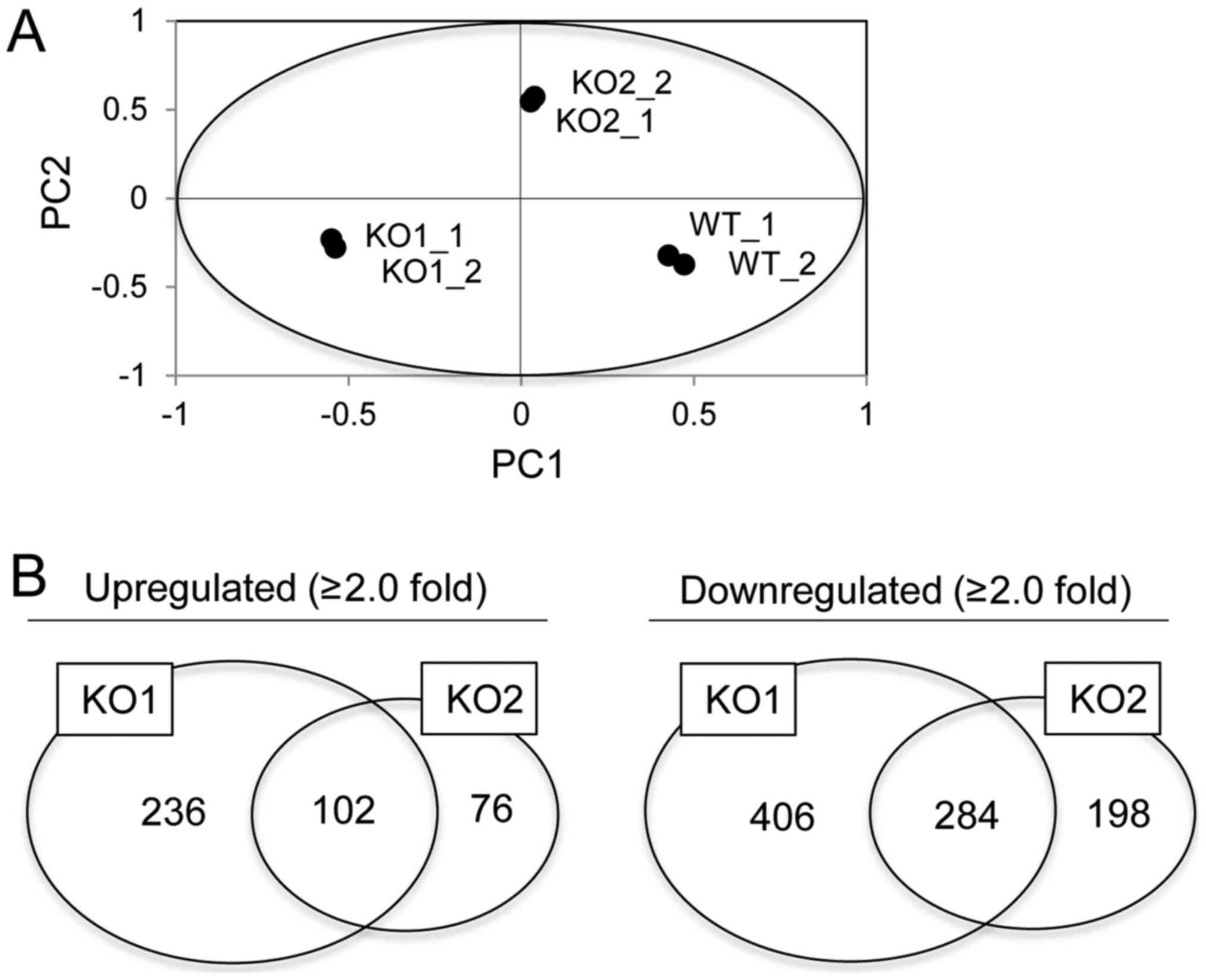
Global gene expression analysis in
BAG3 KO HeLa cell clones
In order to analyze gene expression associated with
BAG3, we performed microarray analysis in WT and two BAG3 KO HeLa
cell clones. After normalization of obtained intensities using the
MAS5.0 algorithm, we performed PCA on gene expression data. This
revealed that the gene expression patterns in WT cells were
markedly distinct from those in BAG3 KO cells (Fig. 3A). However, the gene expression
pattern in clone KO1 was also distinct from that in KO2, which we
assumed was due to normal/stochastic heterogeneity between isolated
clones. Therefore, we attempted to identify common up- or
downregulated genes between two established clones to narrow down
the potential BAG3-target genes. Of the 54,675 probe sets analyzed,
28,719 reliable probe sets were extracted using RefDIC database,
since the probes on this type of array include unreliable probes
that were designed based on a classical database. Among them, 5,436
probe sets were defined as statistically significant based on
one-way ANOVA with post-hoc Tukey HSD and Benjamini-Hochberg
procedure. Furthermore, we identified 1,274 probe sets that were
differentially expressed by a factor of 2.0 or greater in either WT
cells or BAG3 KO clones. Among them, 102 and 284 probe sets were
commonly up- or downregulated in BAG3 KO clones, respectively
(Fig. 3B).
Computational analysis of genes
responsive to BAG3 deletion and gene network analysis
Venn diagrams of differentially expressed probes
showed that 102 and 284 genes were up- and downregulated by BAG3
deletion, respectively. In the present study, we further performed
a bioinformatics analysis to identify the molecular/cellular
functions, and the genetic networks of differentially expressed
genes, in order to elucidate the mechanisms underlying cell death
and cell cycle delay in response to BAG3 deletion. From the
analysis of molecular and cellular function based on IPA knowledge
base, we found that ‘cell death and cell survival’ and ‘cellular
growth and proliferation’ were among the top five functions for up-
and downregulated genes, respectively (Table I).
 | Table I.Top five molecular and cellular
functions in up- and downregulated genes. |
Table I.
Top five molecular and cellular
functions in up- and downregulated genes.
| A, Upregulated |
|---|
|
|---|
| Molecular and
cellular function | P-value | Numbers of
molecules |
|---|
| Cell
morphology |
1.45E-02–3.31E-08 | 71 |
| Cellular
development |
1.45E-02-4.67E-06 | 70 |
| Cellular growth and
proliferation |
1.29E-02-4.67E-06 | 55 |
| Cellular assembly
and organization |
1.23E-02-4.81E-06 | 44 |
| Cellular function
and maintenance |
1.29E-02-4.81E-06 | 47 |
|
| B,
Downregulated |
|
| Molecular and
cellular function | P-value | Numbers of
molecules |
|
| Cell-to-cell
signaling and interaction |
1.25E-02-2.12E-07 | 22 |
| Cellular
movement |
1.16E-02-9.03E-07 | 27 |
| Cell death and
survival |
1.25E-02-4.49E-06 | 30 |
| Cell
morphology |
1.25E-02-2.97E-05 | 21 |
| Cellular
development |
1.21E-02-4.05E-05 | 31 |
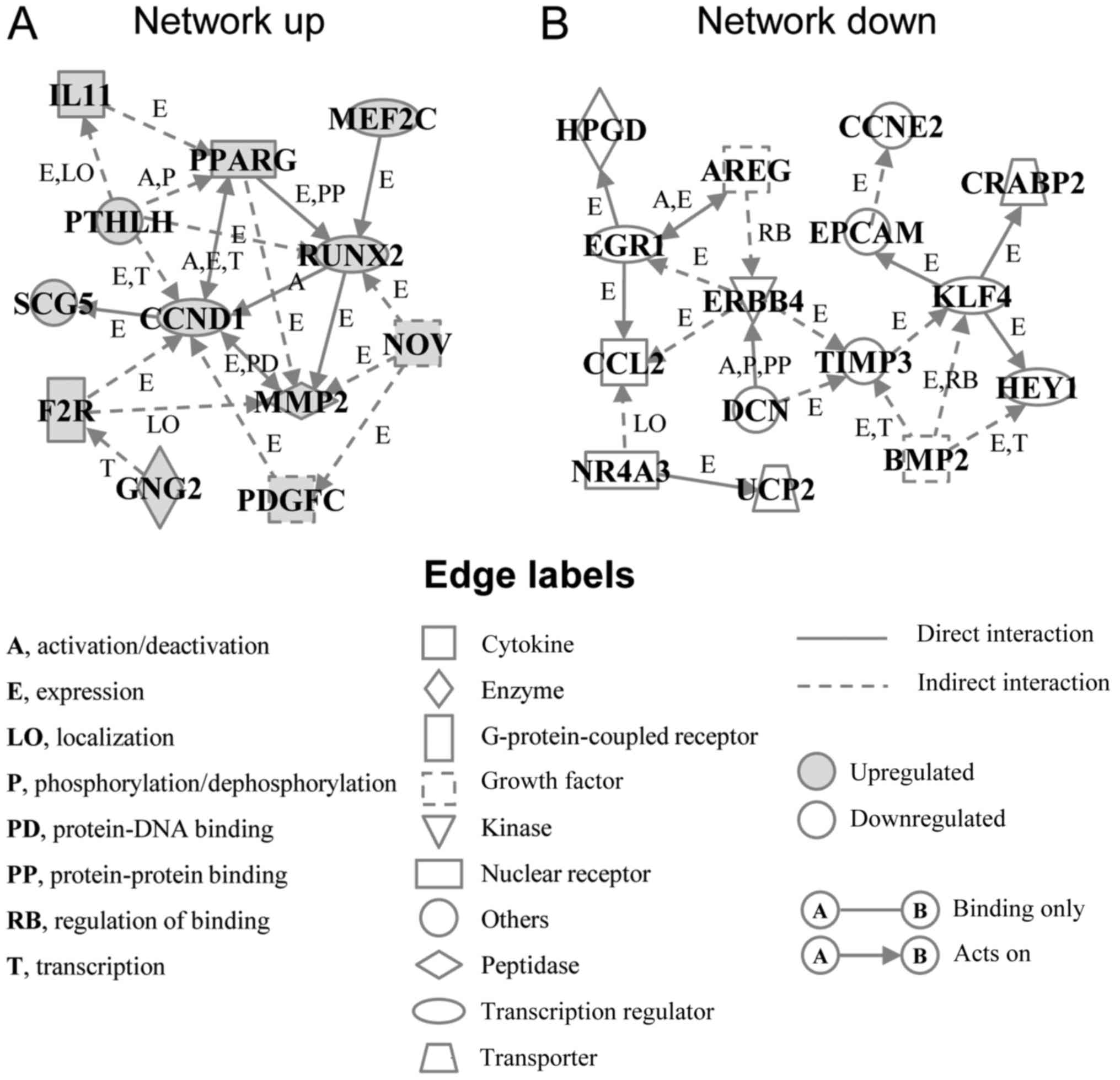
In order to elucidate the interactions between the
up- or downregulated genes, we performed a gene network analysis.
The analysis identified an upregulated gene network (Network up)
containing 12 genes, including cyclin D1 (CCND1), matrix
metalloproteinase 2 (MMP2), platelet derived growth factor C
(PDGFC), runt-related transcription factor 2 (RUNX2), peroxisome
proliferator-activated receptor γ (PPARG) and coagulation factor II
thrombin receptor (F2R) (Fig. 4A
and Table II). The downregulated
gene network (Network down) contained 15 genes, including TIMP
metallopeptidase inhibitor 3 (TIMP3), Krupper like factor 4 (KLF4),
epithelial cell adhesion molecule (EPCAM), erb-b2 receptor tyrosine
kinase 4 (ERBB4), and bone morphogenetic protein 2 (BMP2) (Fig. 4B and Table II). Among them, BAG3 has already
been reported to interact with MMP2 and PDGFC (6,21),
but not with other genes. In addition, the BioGRID database, a
depository of interaction datasets including the results of recent
interactome analyses, also did not report interaction between BAG3
and the genes in the networks we identified, other than MMP2 and
PDGFC, suggesting that the expression changes of most transcripts
identified here may have been independent of BAG3 interaction with
proteins coded by these genes.
 | Table II.Genes in two identified genetic
networks. |
Table II.
Genes in two identified genetic
networks.
| A, Upregulated
(network up) |
|---|
|
|---|
|
| Fold change (vs.
WT) |
|
|---|
|
|
|
|
|---|
| Gene symbol | KO1 | KO2 | Gene title |
|---|
| MMP2 | 3.41 | 5.28 | Matrix
metallopeptidase 2 |
| SCG5 | 7.03 | 5.12 | Secretogranin
V |
| F2R | 2.93 | 3.29 | Coagulation factor
II thrombin receptor |
| NOV | 2.47 | 6.06 | Nephroblastoma
overexpressed |
| PTHLH | 3.85 | 2.68 | Parathyroid hormone
like hormone |
| IL11 | 3.00 | 3.76 | Interleukin 11 |
| PPARG | 2.97 | 3.02 | Peroxisome
proliferator activated receptor γ |
| CCND1 | 5.87 | 5.00 | Cyclin D1 |
| MEF2C | 2.20 | 2.55 | Myocyte enhancer
factor 2C |
| PDGFC | 4.09 | 2.58 | Platelet derived
growth factor C |
| GNG2 | 3.48 | 3.03 | G protein subunit γ
2 |
| RUNX2 | 14.4 | 6.96 | Runt related
transcription factor 2 |
|
| B, Downregulated
(network down) |
|
|
| Fold change (vs.
WT) |
|
|
|
|
|
| Gene
symbol | KO1 | KO2 | Gene
title |
|
| TIMP3 | 0.49 | 0.28 | TIMP
metallopeptidase inhibitor 3 |
| EGR1 | 0.11 | 0.32 | Early growth
response 1 |
| EPCAM | 0.07 | 0.16 | Epithelial cell
adhesion molecule |
| CRABP2 | 0.09 | 0.27 | Cellular retinoic
acid binding protein 2 |
| HPGD | 0.10 | 0.26 |
15-hydroxyprostaglandin dehydrogenase |
| AREG | 0.22 | 0.43 | Amphiregulin |
| BMP2 | 0.18 | 0.45 | Bone morphogenetic
protein 2 |
| NR4A3 | 0.15 | 0.35 | Nuclear receptor
subfamily 4 group A member 3 |
| UCP2 | 0.27 | 0.32 | Uncoupling protein
2 |
| DCN | 0.30 | 0.20 | Decorin |
| CCNE2 | 0.49 | 0.46 | Cyclin E2 |
| ERBB4 | 0.18 | 0.26 | Erb-b2 receptor
tyrosine kinase 4 |
| CCL2 | 0.24 | 0.16 | C-C motif chemokine
ligand 2 |
| KLF4 | 0.44 | 0.49 | Kruppel like factor
4 |
| HEY1 | 0.20 | 0.45 | Hes related family
bHLH transcription factor with YRPW motif 1 |
Discussion
The DNA microarray has been a standard technology
for elucidating genome-wide gene expression signatures in life
science research fields. In this study, we addressed the role of
BAG3 in gene transcription by combining transcriptome and
computational analysis in two stable BAG3 KO HeLa cell clones.
Currently, more than 400 BAG3 partner proteins are listed in public
and commercial databases. Among them, we identified MMP2 and PDGFC
as transcriptionally upregulated by BAG3 deletion. MMP2 is known to
contribute to tumor cell apoptosis, probably through the
degradation of poly (ADP-ribose) polymerase, which repairs DNA
single-strand breaks (27,28). In contrast, PDGFC shows
anti-apoptotic effect and promotes cell proliferation (29,30).
These genes seem to compete with each other at the level of
transcription and probably also at the post-translational level by
interacting with BAG3. Interestingly, only these two genes were
identified as BAG3-related genes in our gene networks. It is
possible that the upregulation of MMP2 and PDGFC transcripts
resulted from destabilization of MMP2 and PDGFC (negative feedback
through protein degradation) in the absence of BAG3. Also of note,
we found that the transcription factors RUNX2 and PPARG, not
previously reported to interact with BAG3, were markedly
upregulated by BAG3 deletion in two BAG3 KO clones established in
this study. RUNX2 was reported to be involved in cell proliferation
under normal conditions as well as cell survival under conditions
of stress (31,32). PPARG was overexpressed in cancer
but its excessive activation led to growth inhibition and apoptosis
(33). Neither transcription
factor has yet been reported to interact with BAG3, but both are
known to enhance the expression of MMP2, indicating that BAG3 may
be indirectly involved in RUNX2- and PPARG-dependent transcription
of MMP2. BAG3 deletion also upregulated expression of CCND1, known
to be a downstream target of RUNX2 and PPARG (34,35).
CCND1 is well known as a G1 cyclin, and degradation of this protein
results in G1 arrest. However, overexpression of CCND1 perturbs
normal replication and induces DNA damage (36), resulting in apoptotic cell death.
Parathyroid hormone like hormone (PTHLH) is positive regulator of
CCND1 transcription through RhoA/ROCK signaling (37). F2R is also known to promote CCND1
expression through the transcription factor c-Fos (38). Nephroblastoma overexpressed (NOV)
inhibits cell proliferation when overexpressed in Ewing's sarcoma
cells (39). It is conceivable
that BAG3 indirectly suppresses transcription of the genes in the
Network up to promote cell cycle progression and cell survival in
HeLa cells.
In contrast to genes identified in the Network up,
some genes were downregulated by BAG3 deletion. Among downregulated
genes, ERBB4, TIMP3, KLF4, and BMP2 were located in the central
region of the Network down. ERBB4 encodes HER4, a tyrosine kinase
receptor belonging to the epidermal growth factor receptor family.
ERBB4 is overexpressed in Ewing's sarcoma cells and activates the
PI3K-Akt cascade, resulting in the promotion of cell growth and
survival (40). In addition, ERBB4
is known to enhance the expression of downstream molecules such as
early growth response 1 (EGR1) and TIMP3 (41). EGR1 is a transcription factor that
plays a critical role in cell growth and survival (42,43).
TIMP3 acts as a tissue inhibitor of MMP2 (44). Thus, downregulation of ERBB4 and
TIMP3 may lead to the activation of overexpressed MMP2 in BAG3 KO
HeLa cells. TIMP3 also regulates the expression of KLF4. KLF4 is a
transcription factor linked to tumor cell growth by a study showing
that its downregulation inhibits the proliferation of cancer cells
(45). Mutation of KLF4 leads to a
decrease in the level of EPCAM (46), high expression of which in gastric
cancer is linked to proliferation (47). BMP2 inhibits apoptosis through the
activation of BMP receptor 2 (48). Furthermore, BMP2 was reported to
upregulate expression of TIMP3 and KLF4 (49). The BMP2-TIMP3 and BMP2-KLF4
signaling axes may also contribute to proliferation and cell
survival through BAG3-dependent transcription.
In this study, we identified two BAG3-dependent
genetic networks associated with cellular growth and proliferation
as well as cell death and survival. These findings will provide a
molecular basis for understanding BAG3-dependent transcriptional
regulation of genes in cancer cells. Further investigation is
needed to identify the BAG3-binding, up-stream transcriptional
regulators of the genes listed in the genetic networks we
identified.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the JSPS
KAKENHI (grant nos. 16K20309 and 17K01353).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YF and YT conceived the study, designed the
experiments, wrote the manuscript and performed the experiments. TH
and SM also performed the experiments. HI and HM provided the
materials and performed genome editing. TY and AH provided
materials for microarray analysis and were involved in data
analysis.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BAG
|
BCL2-associated athanogene
|
|
BMP2
|
bone morphogenetic protein 2
|
|
CCND1
|
cyclin D1
|
|
Cas9
|
CRISPR associated protein 9
|
|
CRISPR
|
clustered regularly interspaced short
palindromic repeats
|
|
EPCAM
|
epithelial cell adhesion molecule
|
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4
|
|
EGR1
|
early growth response 1
|
|
F2R
|
coagulation factor II thrombin
receptor
|
|
IPA
|
Ingenuity Pathways Analysis
|
|
KLF4
|
Kruppel like factor 4
|
|
NOV
|
nephroblastoma overexpressed
|
|
PCA
|
principal component analyses
|
|
PDGFC
|
platelet derived growth factor C
|
|
PPARG
|
peroxisome proliferator activated
receptor γ
|
|
PTHLH
|
parathyroid hormone like hormone
|
|
QUICK
|
quantitative immunoprecipitation
combined with knockdown
|
|
RUNX2
|
runt-related transcription factor
2
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
References
|
1
|
Behl C: Breaking BAG: The co-chaperone
BAG3 in health and disease. Trends Pharmacol Sci. 37:672–688. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rosati A, Ammirante M, Gentilella A,
Basile A, Festa M, Pascale M, Marzullo L, Belisario MA, Tosco A,
Franceschelli S, et al: Apoptosis inhibition in cancer cells: A
novel molecular pathway that involves BAG3 protein. Int J Biochem
Cell Biol. 39:1337–1342. 2017. View Article : Google Scholar
|
|
3
|
Kassis JN, Guancial EA, Doong H, Virador V
and Kohn EC: CAIR-1/BAG-3 modulates cell adhesion and migration by
downregulating activity of focal adhesion proteins. Exp Cell Res.
312:2962–2971. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Falco A, Festa M, Basile A, Rosati A,
Pascale M, Florenzano F, Nori SL, Nicolin V, Di Benedetto M,
Vecchione ML, et al: BAG3 controls angiogenesis through regulation
of ERK phosphorylation. Oncogene. 31:5153–5161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi H, Xu H, Li Z, Zhen Y, Wang B, Huo S,
Xiao R and Xu Z: BAG3 regulates cell proliferation, migration, and
invasion in human colorectal cancer. Tumour Biol. 37:5591–5597.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Suzuki M, Iwasaki M, Sugio A, Hishiya A,
Tanaka R, Endo T, Takayama S and Saito T: BAG3 (BCL2-associated
athanogene 3) interacts with MMP-2 to positively regulate invasion
by ovarian carcinoma cells. Cancer Lett. 303:65–71. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kathage B, Gehlert S, Ulbricht A, Lüdecke
L, Tapia VE, Orfanos Z, Wenzel D, Bloch W, Volkmer R, Fleischmann
BK, et al: The cochaperone BAG3 coordinates protein synthesis and
autophagy under mechanical strain through spatial regulation of
mTORC1. Biochim Biophys Acta. 1864:62–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bonelli P, Petrella A, Rosati A, Romano
MF, Lerose R, Pagliuca MG, Amelio T, Festa M, Martire G, Venuta S,
et al: BAG3 protein regulates stress-induced apoptosis in normal
and neoplastic leukocytes. Leukemia. 18:358–360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tabuchi Y, Ando H, Takasaki I, Feril LB
Jr, Zhao QL, Ogawa R, Kudo N, Tachibana K and Kondo T:
Identification of genes responsive to low intensity pulsed
ultrasound in a human leukemia cell line Molt-4. Cancer Lett.
246:149–156. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jung SE, Kim YK, Youn DY, Lim MH, Ko JH,
Ahn YS and Lee JH: Down-modulation of Bis sensitizes cell death in
C6 glioma cells induced by oxygen-glucose deprivation. Brain Res.
1349:1–10. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pagliuca MG, Lerose R, Cigliano S and
Leone A: Regulation by heavy metals and temperature of the human
BAG-3 gene, a modulator of Hsp70 activity. FEBS Lett. 541:11–15.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yunoki T, Kariya A, Kondo T, Hayashi A and
Tabuchi Y: The combination of silencing BAG3 and inhibition of the
JNK pathway enhances hyperthermia sensitivity in human oral
squamous cell carcinoma cells. Cancer Lett. 335:52–57. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yunoki T, Tabuchi Y, Hayashi A and Kondo
T: Network analysis of genes involved in the enhancement of
hyperthermia sensitivity by the knockdown of BAG3 in human oral
squamous cell carcinoma cells. Int J Mol Med. 38:236–242. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Franceschelli S, Rosati A, Lerose R, De
Nicola S, Turco MC and Pascale M: Bag3 gene expression is regulated
by heat shock factor 1. J Cell Physiol. 215:575–577. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Colvin TA, Gabai VL, Gong J, Calderwood
SK, Li H, Gummuluru S, Matchuk ON, Smirnova SG, Orlova NV,
Zamulaeva IA, et al: Hsp70-Bag3 interactions regulate
cancer-related signaling networks. Cancer Res. 74:4731–4740. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Staibano S, Mascolo M, Di Benedetto M,
Vecchione ML, Ilardi G, Di Lorenzo G, Autorino R, Salerno V, Morena
A, Rocco A, et al: BAG3 protein delocalisation in prostate
carcinoma. Tumour Biol. 31:461–469. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang JT, Wang JL, Du W, Hong J, Zhao SL,
Wang YC, Xiong H, Chen HM and Fang JY: MicroRNA 345, a
methylation-sensitive microRNA is involved in cell proliferation
and invasion in human colorectal cancer. Carcinogenesis.
32:1207–1215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ammirante M, Rosati A, Arra C, Basile A,
Falco A, Festa M, Pascale M, D'Avenia M, Marzullo L, Belisario MA,
et al: IKK{gamma} protein is a target of BAG3 regulatory activity
in human tumor growth. Proc Natl Acad Sci USA. 107:pp. 7497–7502.
2010; View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yunoki T, Tabuchi Y, Kondo T, Ishii Y and
Hayashi A: Overexpression of the anti-apoptotic protein BAG3 in
human choroidal melanoma: A case report. Oncol Lett. 13:4169–4172.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Boiani M, Daniel C, Liu X, Hogarty MD and
Marnett LJ: The stress protein BAG3 stabilizes Mcl-1 protein and
promotes survival of cancer cells and resistance to antagonist
ABT-737. J Biol Chem. 288:6980–6990. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Yang LN, Cheng L, Tu S, Guo SJ, Le
HY, Xiong Q, Mo R, Li CY, Jeong JS, et al: Bcl2-associated
athanogene 3 interactome analysis reveals a new role in modulating
proteasome activity. Mol Cell Proteomics. 12:2804–2819. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ito T, Hayashida M, Kobayashi S, Muto N,
Hayashi A, Yoshimura T and Mori H: Serine racemase is involved in
d-aspartate biosynthesis. J Biochem. 160:345–353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Furusawa Y, Yamanouchi Y, Iizumi T, Zhao
WL, Mitsuhashi Y, Morita A, Enomoto A, Tabuchi Y and Kondo T:
Checkpoint kinase 2 is dispensable for regulation of the p53
response but is required for G2/M arrest and cell survival in cells
with p53 defects under heat stress. Apoptosis. 22:1225–1234. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Watson JV, Chambers SH and Smith PJ: A
pragmatic approach to the analysis of DNA histograms with a
definable G1 peak. Cytometry. 8:1–8. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hijikata A, Kitamura H, Kimura Y, Yokoyama
R, Aiba Y, Bao Y, Fujita S, Hase K, Hori S, Ishii Y, et al:
Construction of an open-access database that integrates
cross-reference information from the transcriptome and proteome of
immune cells. Bioinformatics. 23:2934–2941. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Makino S, Fukumura R and Gondo Y:
Illegitimate translation causes unexpected gene expression from
on-target out-of-frame alleles created by CRISPR-Cas9. Sci Rep.
6:396082016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kwan JA, Schulze CJ, Wang W, Leon H,
Sariahmetoglu M, Sung M, Sawicka J, Sims DE, Sawicki G and Schulz
R: Matrix metalloproteinase-2 (MMP-2) is present in the nucleus of
cardiac myocytes and is capable of cleaving poly (ADP-ribose)
polymerase (PARP) in vitro. FASEB J. 18:690–692. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aldonyte R, Brantly M, Block E, Patel J
and Zhang J: Nuclear localization of active matrix
metalloproteinase-2 in cigarette smoke-exposed apoptotic
endothelial cells. Exp Lung Res. 35:59–75. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McDermott U, Ames RY, Iafrate AJ,
Maheswaran S, Stubbs H, Greninger P, McCutcheon K, Milano R, Tam A,
Lee DY, et al: Ligand-dependent platelet-derived growth factor
receptor (PDGFR)-alpha activation sensitizes rare lung cancer and
sarcoma cells to PDGFR kinase inhibitors. Cancer Res. 69:3937–3946.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang Z, Arjunan P, Lee C, Li Y, Kumar A,
Hou X, Wang B, Wardega P, Zhang F, Dong L, et al: Survival effect
of PDGF-CC rescues neurons from apoptosis in both brain and retina
by regulating GSK3beta phosphorylation. J Exp Med. 207:867–880.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lucero CM, Vega OA, Osorio MM, Tapia JC,
Antonelli M, Stein GS, van Wijnen AJ and Galindo MA: The
cancer-related transcription factor Runx2 modulates cell
proliferation in human osteosarcoma cell lines. J Cell Physiol.
228:714–723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sugimoto H, Nakamura M, Yoda H, Hiraoka K,
Shinohara K, Sang M, Fujiwara K, Shimozato O, Nagase H and Ozaki T:
Silencing of RUNX2 enhances gemcitabine sensitivity of
p53-deficient human pancreatic cancer AsPC-1 cells through the
stimulation of TAp63-mediated cell death. Cell Death Dis.
6:e19142015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Krishnan A, Nair SA and Pillai MR: Biology
of PPAR gamma in cancer: A critical review on existing lacunae.
Curr Mol Med. 7:532–540. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Owens TW, Rogers RL, Best S, Ledger A,
Mooney AM, Ferguson A, Shore P, Swarbrick A, Ormandy CJ, Simpson
PT, et al: Runx2 is a novel regulator of mammary epithelial cell
fate in development and breast cancer. Cancer Res. 74:5277–5286.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sharma C, Pradeep A, Pestell RG and Rana
B: Peroxisome proliferator-activated receptor gamma activation
modulates cyclin D1 transcription via beta-catenin-independent and
cAMP-response element-binding protein-dependent pathways in mouse
hepatocytes. J Biol Chem. 279:16927–16938. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shimura T, Ochiai Y, Noma N, Oikawa T,
Sano Y and Fukumoto M: Cyclin D1 overexpression perturbs DNA
replication and induces replication-associated DNA double-strand
breaks in acquired radioresistant cells. Cell Cycle. 12:773–782.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang G, Woods A, Sabari S, Pagnotta L,
Stanton LA and Beier F: RhoA/ROCK signaling suppresses hypertrophic
chondrocyte differentiation. J Biol Chem. 279:13205–13214. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Parrales A, Palma-Nicolás JP, López E and
López-Colomé AM: Thrombin stimulates RPE cell proliferation by
promoting c-Fos-mediated cyclin D1 expression. J Cell Physiol.
222:302–312. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Benini S, Perbal B, Zambelli D, Colombo
MP, Manara MC, Serra M, Parenza M, Martinez V, Picci P and
Scotlandi K: In Ewing's sarcoma CCN3(NOV) inhibits proliferation
while promoting migration and invasion of the same cell type.
Oncogene. 24:4349–4361. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mendoza-Naranjo A, El-Naggar A, Wai DH,
Mistry P, Lazic N, Ayala FR, da Cunha IW, Rodriguez-Viciana P,
Cheng H, Tavares Guerreiro, Fregnani JH, et al: ERBB4 confers
metastatic capacity in Ewing sarcoma. EMBO Mol Med. 5:1087–1102.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Capone C, Dabertrand F, Baron-Menguy C,
Chalaris A, Ghezali L, Domenga-Denier V, Schmidt S, Huneau C,
Rose-John S, Nelson MT and Joutel A: Mechanistic insights into a
TIMP3-sensitive pathway constitutively engaged in the regulation of
cerebral hemodynamics. Elife. 5(pii): e175362016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zins K, Pomyje J, Hofer E, Abraham D,
Lucas T and Aharinejad S: Egr-1 upregulates Siva-1 expression and
induces cardiac fibroblast apoptosis. Int J Mol Sci. 15:1538–1553.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Baron V, De Gregorio G, Krones-Herzig A,
Virolle T, Calogero A, Urcis R and Mercola D: Inhibition of Egr-1
expression reverses transformation of prostate cancer cells in
vitro and in vivo. Oncogene. 22:4194–4204. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jackson HW, Defamie V, Waterhouse P and
Khokha R: TIMPs: Versatile extracellular regulators in cancer. Nat
Rev Cancer. 17:38–53. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tien YT, Chang MH, Chu PY, Lin CS, Liu CH
and Liao AT: Downregulation of the KLF4 transcription factor
inhibits the proliferation and migration of canine mammary tumor
cells. Vet J. 205:244–253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu YN, Abou-Kheir W, Yin JJ, Fang L,
Hynes P, Casey O, Hu D, Wan Y, Seng V, Sheppard-Tillman H, et al:
Critical and reciprocal regulation of KLF4 and SLUG in transforming
growth factor β-initiated prostate cancer epithelial-mesenchymal
transition. Mol Cell Biol. 32:941–953. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kroepil F, Dulian A, Vallböhmer D, Geddert
H, Krieg A, Vay C, Topp SA, Am Esch JS, Baldus SE, Gires O, et al:
High EpCAM expression is linked to proliferation and lauren
classification in gastric cancer. BMC Res Notes. 6:2532013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu Z, Shen J, Pu K, Katus HA, Plöger F,
Tiefenbacher CP, Chen X and Braun T: GDF5 and BMP2 inhibit
apoptosis via activation of BMPR2 and subsequent stabilization of
XIAP. Biochim Biophys Acta. 1793:1819–1827. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li Q, Kannan A, Das A, Demayo FJ, Hornsby
PJ, Young SL, Taylor RN, Bagchi MK and Bagchi IC: WNT4 acts
downstream of BMP2 and functions via β-catenin signaling pathway to
regulate human endometrial stromal cell differentiation.
Endocrinology. 154:446–457. 2013. View Article : Google Scholar : PubMed/NCBI
|