Introduction
Diabetes mellitus is one of the most common chronic
diseases worldwide, and there were 422 million adults diagnosed
with diabetes globally in 2014, according to the World Health
Organization (1,2). As the number of novel diagnoses is
increasing, this disease is attracting increased attention.
Diabetic nephropathy (DN) is one of the principal microvascular
complications of diabetes, and it is highly prevalent in 30–40% of
hospitalized patients with diabetes (3,4). DN
is additionally one of the leading causes of end-stage kidney
disease, which only has an ~20% 5-year survival rate (5). DN has been characterized by a series
of abnormal pathological alterations, including glomerular
hypertrophy, mesangial proliferation, thickening of the glomerular
basement membrane, and accumulation of the extracellular matrix
(6,7). However, the dysregulated molecules
and the mechanisms involved in this manifestation of disease remain
poorly understood. Therefore, a better understanding of the
pathogenesis and the identification of novel factors in DN may
promote the development of novel therapeutics to address this
complex disease.
Over the past decades, the rapid improvement of
high-throughput sequencing techniques and bioinformatics methods
has led to the advent of whole human genome sequencing. Annotation
of sequencing results has revealed that <2% of the whole human
genome is protein coding genes; whereas, the majority of the rest
are non-coding genes, which yield numerous non-coding transcripts,
including microRNAs and long non-coding RNAs (lncRNAs) (8–10).
lncRNAs, novel examples of non-coding RNAs, are >200 nucleotides
in length and lack any protein coding ability (11). Previously, studies have revealed
that lncRNAs are widely expressed in almost all human tissues, and
are involved in a number of important biological process, including
X chromatin imprinting, stem cell differentiation, immune
responses, cell fate decision, proliferation, and transcriptional
and post-transcriptional regulation (12–14).
Furthermore, dysregulation of lncRNAs has been demonstrated to
contribute to the development of diverse human diseases, including
types of cancer, neurological and cardiovascular diseases, and
diabetes (15–17). For example, knockdown of lncRNA
uc.48+ improved diabetic sympathetic neuropathy in type 2 diabetic
rats through regulation of purinergic receptor P2X 7 expression and
extracellular signal-regulated kinase signaling (18).
In the case of DN, a number of lncRNAs have been
demonstrated to be dysregulated by microarray analysis. Out of
these lncRNAs, the function and underlying pathways associated with
certain ones have been characterized and reported (19). For example, lncRNA taurine
upregulated 1 alleviates extracellular matrix accumulation by
acting as an endogenous sponge for microRNA (miR)-377 and thereby
relieving the inhibition of peroxisome proliferator-activated
receptor γ in DN (20). In
addition, Wang et al (21)
reported that lncRNA CYP4B1-PS1-001 expression was significantly
downregulated in response to early DN, and CYP4B1-PS1-001
overexpression inhibited mesangial cell proliferation and fibrosis.
Furthermore, lncRNA metastasis associated lung adenocarcinoma
transcript 1 (MALAT1) expression is downregulated in kidney
cortices from streptozotocin-induced DN cases, and decreased MALAT1
is involved in high glucose-induced podocyte injury via interacting
with β-catenin (22). In the
authors' previous study, lncRNA expression patterns between a DN
model and db/m control mouse kidney tissues were analyzed using
microarray analysis (23). It was
demonstrated that hundreds of lncRNAs are dysregulated in DN, and
these lncRNAs may contribute to the pathogenesis of DN by
modulating multiple molecular pathways (23). However, the functions of these
lncRNAs in DN remain unclear. In the present study, the microarray
results were further validated, and the function of two of these
lncRNAs (Gm5524 and Gm15645) in podocytes was analyzed by loss- and
gain-of function assays.
Materials and methods
Animal model and tissue specimen
preparation
A total of 12 8-week-old male mice, including six
C57BL/KsJ db/db mice (experimental group; average weight 46.53±1.96
g) and six C57BL/KsJ m/db mice (control group; average weight
22.32±1.10 g) were purchased from the Experimental Animal Center of
Nanjing Medical University (Nanjing, China). Animal experiments
were conducted in accordance with Nanjing Medical University
guidelines and ethical standards for animal care. The mice were fed
and housed in well-ventilated plastic cages with stainless steel
grid tops, at 22±2°C and 50–60% humidity, with a 12-h light/dark
cycle and free access to water. All kidney tissues were isolated
immediately from the mice when they were sacrificed at 9 weeks old,
and were stored at −80°C. The protocol used in the present study
was approved by the Committee on the Ethics of Animal Experiments
of The Second Affiliated Hospital of Nanjing Medical University
(Nanjing, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The total RNA from tissues and cells was extracted
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. Subsequently, 1 µg total RNA was
reverse-transcribed into cDNA using HiScript Q RT SuperMix (Vazyme,
Piscataway, NJ, USA), and the RT conditions were 50°C for 15 min
and 85°C for 2 min. qPCR analysis was performed using a SYBR Green
qPCR kit (Vazyme). The thermocycling conditions were as follows:
Heating to 95°C for 5 min; 40 cycles of 95°C for 10 sec; and 60°C
for 30 sec. The 2−ΔΔCq method was used to compare the
relative expression levels of lncRNAs Gm5524 and Gm15645, and GAPDH
was used as the internal control (24). The primer sequences for Gm5524
were: Forward, 5′-GTCACAGTTTCCAGTGATAGGG-3′ and reverse,
5′-AAGTGAGGCACTCCAGATTAAC-3′. The primer sequences for Gm15645
were: Forward, 5′-GAACTCCAGACCTTTGGAAGAG-3′ and reverse,
5′-TCTGGCGACTTTCATCAATACA-3′. The GAPDH primer sequences were:
Forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse,
5′-AGGGGCCATCCACAGTCTTC-3′.
Cell culture
A separate primary mouse podocyte cell line was
obtained from the Mount Sinai School of Medicine (New York, NY,
USA). The podocytes were cultured on type I collagen in RPMI 1640
(Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; Thermo Fisher Scientific, Inc.), 10 U/ml mouse
recombinant c-interferon (PeproTech, Inc., Rocky Hill, NJ, USA) and
1% penicillin and streptomycin, under a humidified incubator with
5% CO2 at 33°C.
Cell transfection
The mouse Gm5524 and Gm15645 cDNA sequences were
synthesized and subsequently inserted into the pCDNA3.1 vector
(Invitrogen; Thermo Fisher Scientific, Inc.), and the empty
pCDNA3.1 vector was used as a control. Gm5524 and Gm15645 short
hairpin (sh)RNA oligos were synthesized, annealed and inserted into
the pLKO vector (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
All the vectors were prepared using Midiprep kits (Qiagen GmbH,
Hilden, Germany), and 2 µg plasmid were transfected into mouse
podocytes with ~70% density in six-well plates using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Gm5524, Gm15645 overexpression vectors and the shRNA lentivirus
were purchased from Genscript (Nanjing, China). A total of 48 h
following transfection, the cells were selected using puromycin for
48 h and harvested for further RT-qPCR or western blot analysis.
The sequence of scrambled (Scr) shRNA (Sigma-Aldrich; Merck KGaA;
50 ng/µl) is
5′-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3′.
Flow cytometry apoptosis assay
Podocytes transfected with sh-Gm5524, sh-Gm15645,
sh-NC vector, Gm5524 vector and Gm15645 vector were harvested 48 h
following transfection using trypsin. The cell suspension was
incubated with fluorescein isothiocyanate-Annexin V and propidium
iodide for 15 min at room temperature in the dark and analyzed
using a flow cytometer (FACScan®; BD Biosciences,
Franklin Lakes, NJ, USA) equipped with CellQuest software version
5.1 (BD Biosciences).
Evaluation of autophagy by
transmission electron microscopy
Podocytes were washed and fixed in glutaraldehyde
(2.5% in 0.1 mol/l phosphate buffer; pH 7.4) at 4°C overnight,
post-fixed in 1% osmium tetroxide for 3 h at 4°C and dehydrated.
Subsequently, the samples were embedded using a Poly/Bed 812 kit at
60°C for 24–48 h (Polysciences, Inc., Warrington, PA, USA). Samples
were sliced into 70-nm ultra-thin sections and stained with uranyl
acetate for 3 min at room temperature for transmission electron
microscopic analysis (JEOL-1010; JEOL, Ltd., Tokyo, Japan).
Western blot analysis
The total proteins from mouse podocytes were
extracted using radioimmunoprecipitation assay reagent (Beyotime
Institute of Biotechnology, Haimen, China) supplemented with a
protease inhibitor cocktail (Roche Molecular Diagnostics,
Pleasanton, CA, USA). Subsequently, 40 µg extracted protein was
separated by 8–15% SDS-PAGE, and transferred to 0.22-µm
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were blocked with 5% milk in TBS with Tween-20
at room temperature for 2 h and subsequently incubated overnight at
4°C with microtubule-associated proteins 1A/1B light chain 3B
(LC3)I, LC3II (cat. no. 4108; 1:1,000), autophagy protein 5 (Atg5;
cat. no. 12994; 1:1,000), ubiquitin-like modifier-activating enzyme
ATG7 (Atg7; cat. no. 8558; 1:1,000), caspase 3 (cat. no. 9662;
1:1,000), cellular tumor antigen p53 (p53; cat. no. 2524; 1:1,000),
apoptosis regulator BAX (Bax; cat. no. 2772; 1:1,000), apoptosis
regulator Bcl-2 (Bcl2; cat. no. 3498; 1:1,000) and GAPDH (cat. no.
5174; 1:1,000) antibodies (all from Cell Signaling Technology,
Inc., Danvers, MA, USA). Membranes were subsequently incubated with
horseradish peroxidase-conjugated secondary antibodies (cat. nos.
7077 and 7076; 1:5,000; Cell Signaling Technology, Inc.) for 2 h at
room temperature. Protein bands were visualized using enhanced
chemiluminescence (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
and a Western Blotting Detection and Imaging system (Bio-Rad
Laboratories, Inc.). Enhanced chemiluminescence chromogenic
substrate was quantified by densitometry (Quantity One 4.6
software; Bio-Rad Laboratories, Inc.).
Statistical analysis
All the statistical analyses were conducted using
SPSS Statistics 18.0 software (SPSS, Inc., Chicago, IL, USA).
One-way analysis of variance followed by the Least Significant
Difference test and Student's t-test (two-tailed) were used to
analyze the in vitro assay data. The data are presented as
the mean ± standard error of the mean of at least three independent
assays. P<0.05 was considered to indicate a statically
significant difference.
Results
Gm5524 is upregulated and Gm15645 is
downregulated in mouse DN
In the authors' previous study, lncRNA microarray
analysis of an lncRNA profile in mouse DN kidney tissues and
control tissues was performed (23). It was demonstrated that hundreds of
lncRNAs were differentially expressed in DN tissues compared with
control kidney tissues. Among these altered lncRNAs, Gm5524 was
significantly upregulated and Gm15645 was significantly
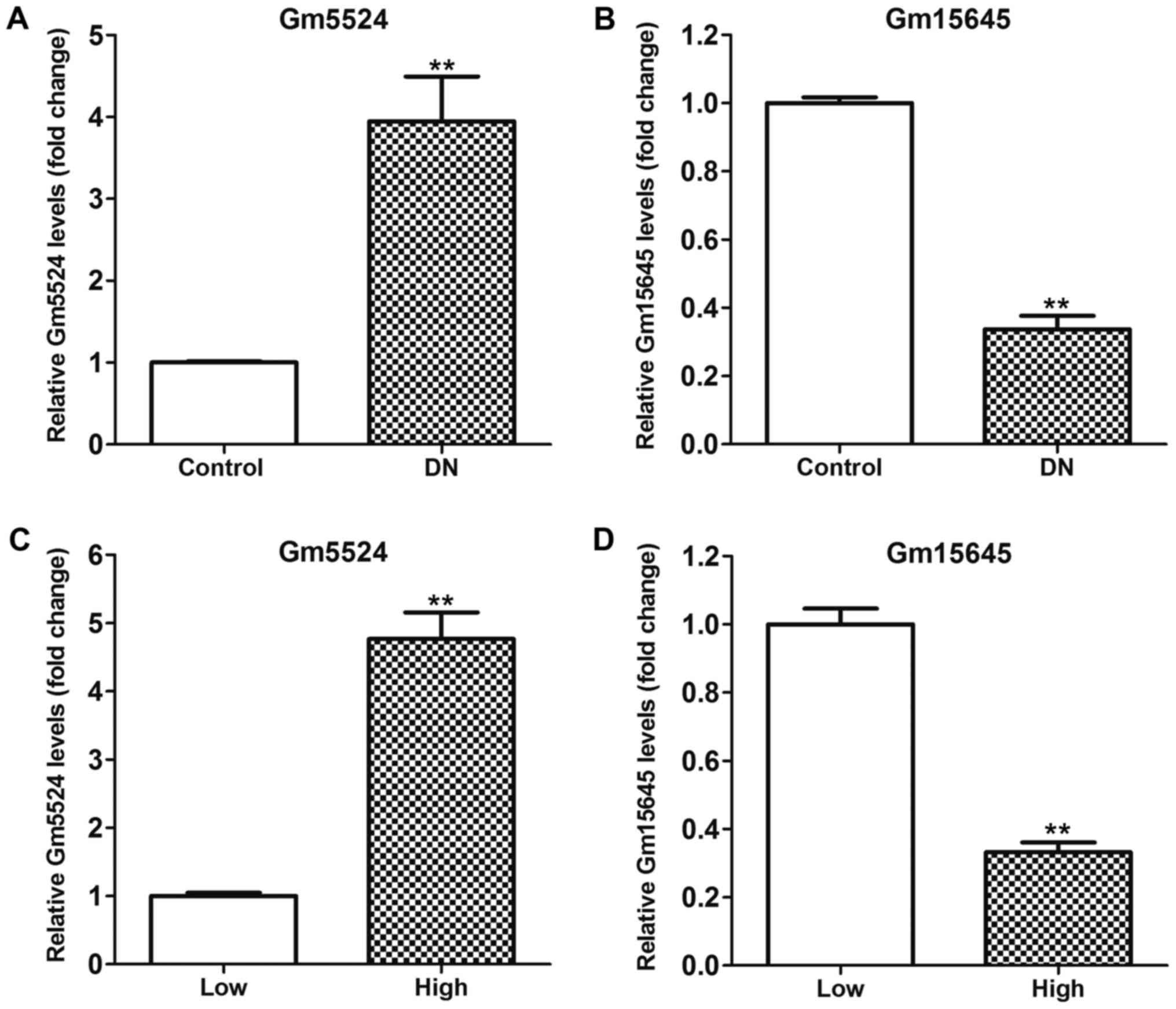
downregulated in DN tissues. RT-qPCR was used to further validate
this in DN and control tissues, and the results demonstrated that
Gm5524 was significantly upregulated and Gm15645 was significantly
downregulated in DN tissues, as was the case with the microarray
data (P<0.01; Fig. 1A and B).
Furthermore, podocytes were treated with high glucose to imitate
the in vivo DN conditions, and the expression of Gm5524 and
Gm15645 was examined. The results of the RT-qPCR demonstrated that
Gm5524 was additionally significantly upregulated and Gm15645 was
significantly downregulated in podocytes under high glucose
conditions (P<0.01; Fig. 1C and
D). Additionally, specific shRNAs were designed for the two
lncRNAs and transfected into mouse podocytes to knock down their
expression. Furthermore, Gm5524 and Gm15645 overexpression vectors
were constructed and transfected into mouse podocytes to upregulate
their expression. The results of the RT-qPCR revealed that Gm5524
and Gm15645 expression was significantly deceased or increased,
respectively, following transfection with shRNAs or overexpression
vectors compared with control cells (P<0.01; Fig. 2A and B).
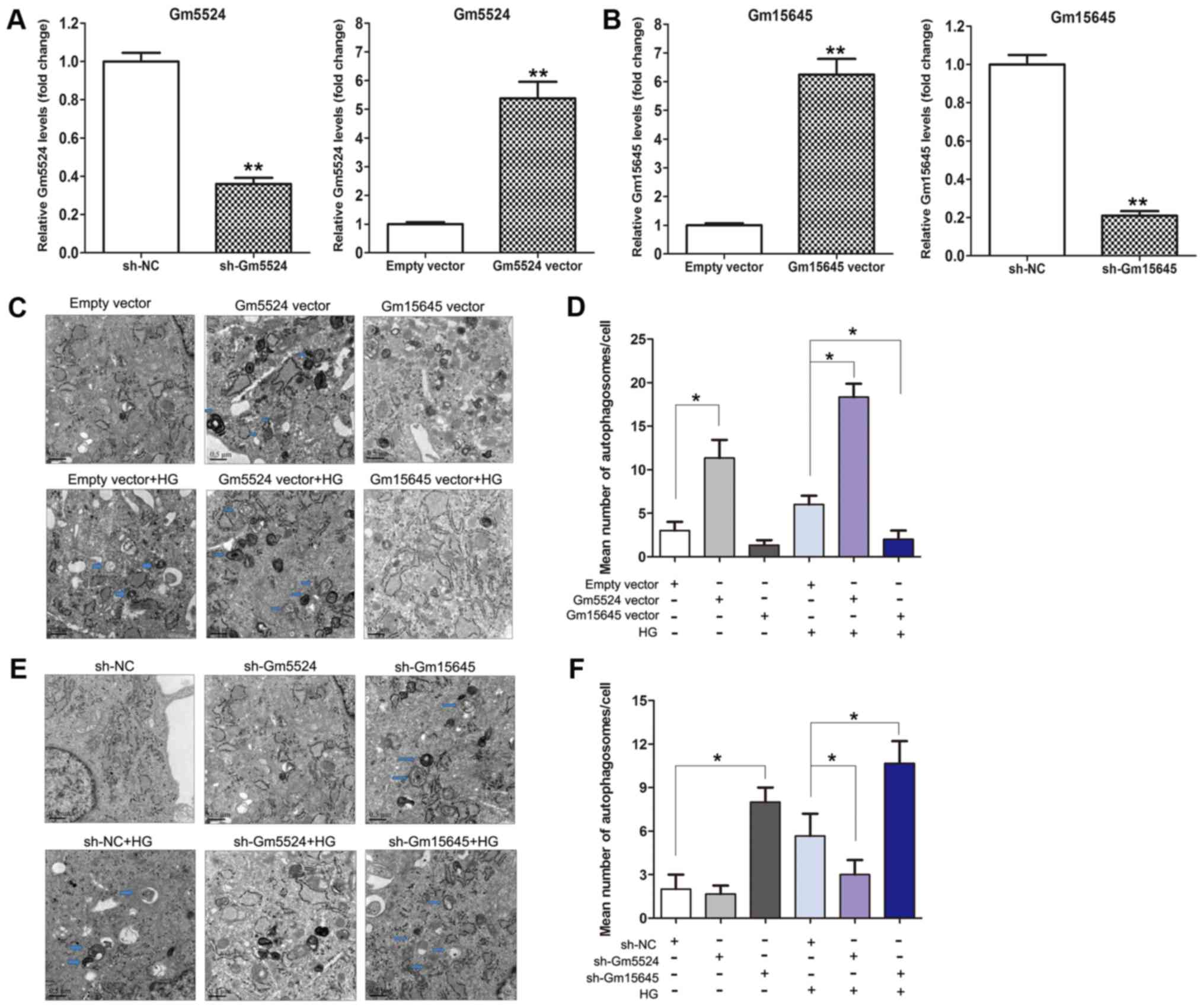 | Figure 2.Effect of Gm5524 and Gm15645 on
podocyte autophagy. (A) Relative expression levels of Gm5524 in
empty vector-, sh-Gm5524 vector- and Gm5524 overexpression
vector-treated podocytes were examined using RT-qPCR. (B) Relative
expression levels of Gm15645 in empty vector-, sh-Gm15645 vector-
and Gm15645 overexpression vector-treated podocytes were examined
using RT-qPCR. **P<0.01 vs. respective control. (C) Cell
autophagy of the empty vector-, Gm5524 and Gm15645 overexpression
vector-transfected podocytes in normal or HG medium was examined
using electron microscopy analysis (scale bar, 0.5 µM) and (D)
quantification of these results. Blue arrows indicate
autophagosomes. (E) Cell autophagy of empty vector, sh-Gm5524
vector and sh-Gm15645 vector-transfected podocytes in normal or HG
medium was examined using electron microscopy analysis (scale bar,
0.5 µM) and (F) quantification of these results. Blue arrows
indicate autophagosomes. *P<0.05. sh, short hairpin; NC,
negative control; HG, high glucose; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
Gm5524 and Gm15645 affect the podocyte
autophagy process
Increasing evidence has demonstrated that cell
autophagy serves an important role in human diseases. To
investigate whether cell autophagy may be affected by Gm5524 and
Gm15645 in DN, electron microscopy analysis was performed. Firstly,
the podocytes were cultured under normal conditions or high glucose
following transfection with Gm5524 and Gm15645 shRNAs or
overexpression vector (Fig. 2A and
B). The visualization of autophagosomes and quantification of
mean autophagosome number/cell demonstrated that podocytes exhibit
more electrodense inclusions and lipid granules following
overexpression of Gm5524 or knockdown of Gm15645, which was more
marked in high glucose-treated podocytes. In the control cells,
autophagosomes were decreased or no autophagosomes were observed
(Fig. 2C-F). These results
suggested that the presence of autophagy may contribute to DN when
Gm5524 expression is increased and Gm15645 expression is
decreased.
Effect of Gm5524 and Gm15645 on mouse
podocyte apoptosis
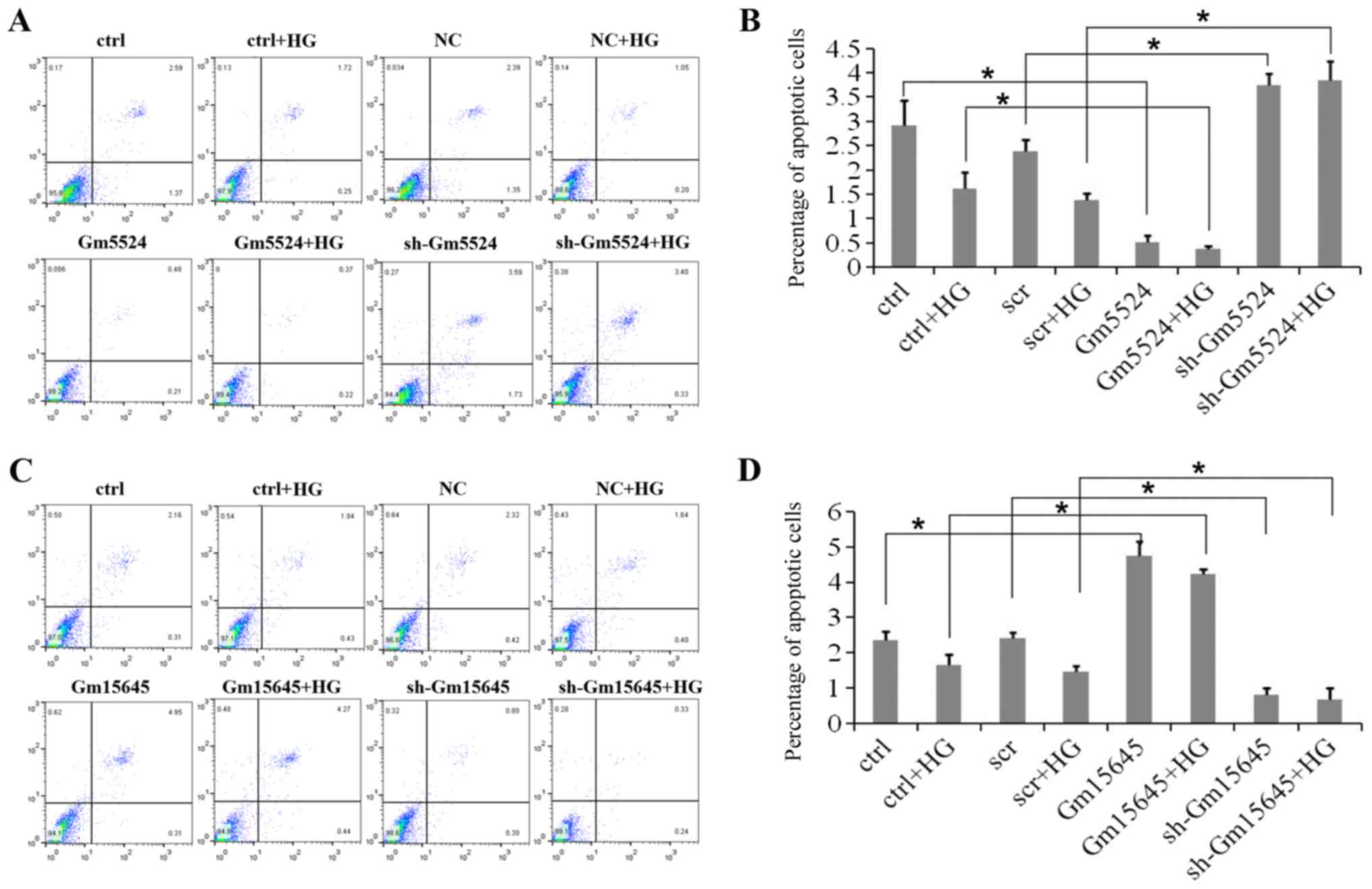
To determine whether Gm5524 and Gm15645 affected
podocyte apoptosis, flow cytometry apoptosis assays were performed.
Flow cytometry analyses of these cells demonstrated that podocytes
exhibited a significantly higher apoptotic percentage following
knockdown of Gm5524 expression under normal conditions, while the
apoptotic percentage was significantly higher in high
glucose-treated podocytes (P<0.05). Conversely, Gm5524
overexpression significantly decreased the podocyte apoptotic
percentage under the two conditions (P<0.05; Fig. 3A and B). Furthermore, podocytes
exhibited a higher apoptotic percentage following the upregulation
of Gm15645 expression under normal conditions, and the apoptotic
percentage was significantly increased under high glucose
conditions compared with the control; however, Gm15645
downregulation significantly decreased the podocyte apoptotic
percentage under the two conditions (P<0.05; Fig. 3C and D).
Gm5524 and Gm15645 affect the
expression of apoptosis and autophagy-associated factors
To determine whether Gm5524 and Gm15645 may affect
the expression levels of cellular apoptosis and
autophagy-associated regulators, western blotting was performed in
podocytes transfected with Gm5524 and Gm15645 shRNAs or an
overexpression vector. The results demonstrated that knockdown of
Gm5524 and overexpression of Gm15645 increased cleaved caspase 3,
Bax and LC3I expression, while it decreased LC3II, Atg5, Atg7 and
Bcl2 expression. Conversely, Gm5524 overexpression and Gm15645
downregulation decreased cleaved caspase 3, Bax and LC3I
expression, and increased LC3II, Atg5, Atg7 and Bcl2 protein
expression (Fig. 4). LC3II/LC3I
protein expression is a well-established biochemical assay to
determine the activation of autophagy, and the present results
demonstrated that the altered expression of Gm5524 and Gm15645
additionally affected the LC3II/LC3I ratio.
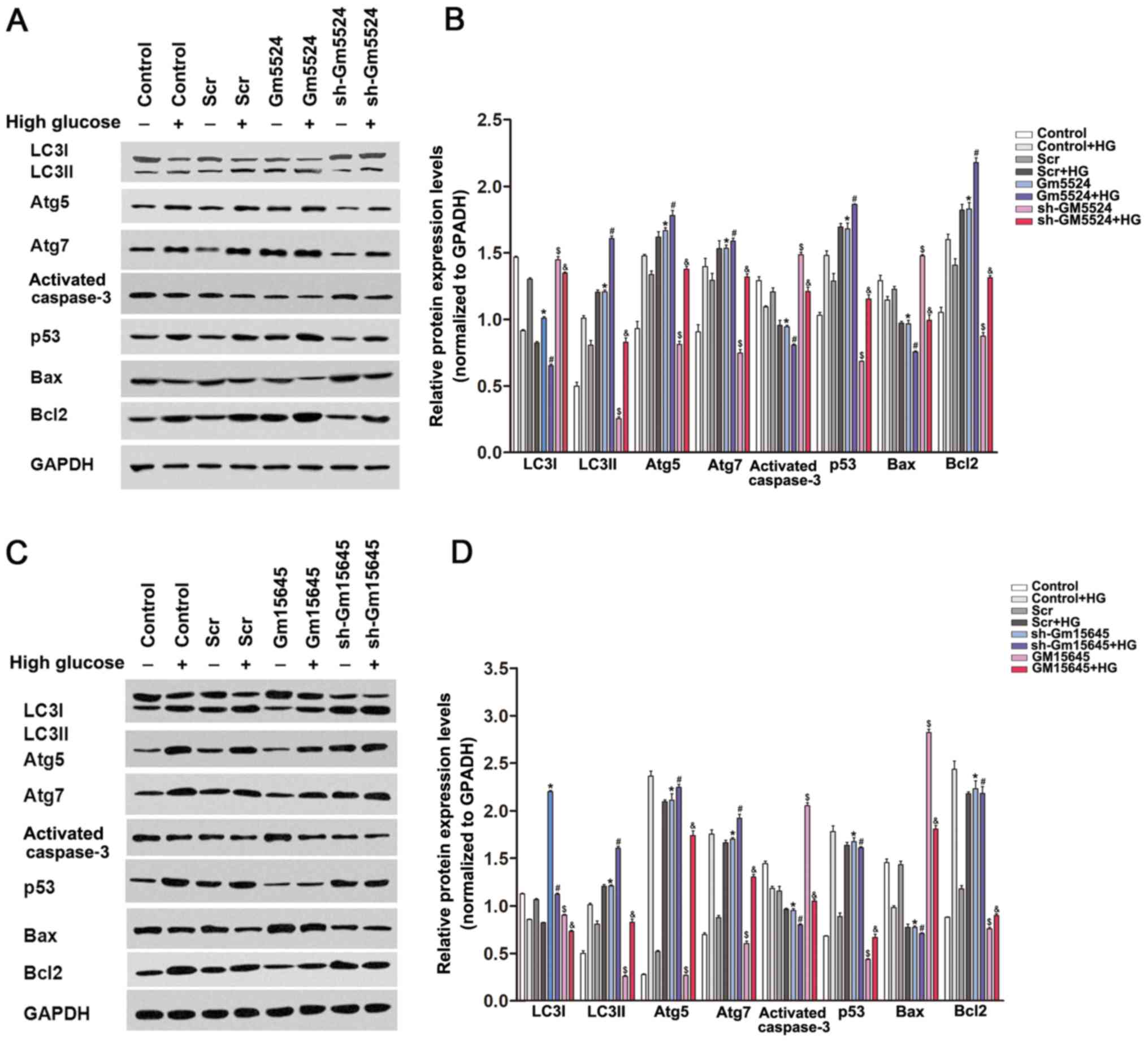 | Figure 4.Effect of Gm5524 and Gm15645 on
apoptosis and autophagy-associated protein expression levels in
podocytes. (A) Protein expression levels of LC3I, LC3II, Atg5,
Atg7, cleaved caspase 3, p53, Bax, Bcl2 and p53 were detected by
western blotting in podocytes following transfection with the empty
vector, sh-Gm5524 vector and Gm5524 overexpression vector under
normal or HG medium conditions. (B) Densitometry data from the
blots. *P<0.05 vs. control; #P<0.05 vs.
control+HG; $P<0.05 vs. Scr;
&P<0.05 vs. Scr+HG. (C) Protein expression levels
of LC3I, LC3II Atg5, Atg7, cleaved caspase 3, p53, Bax, Bcl2 and
p53 was detected by western blotting in podocytes following
transfection with the empty vector, sh-Gm15645 vector and Gm15645
overexpression vector under normal or HG medium conditions. (D)
Densitometry data from the blots. *P<0.05 vs. Scr;
#P<0.05 vs. Scr+HG; $P<0.05 vs.
control; &P<0.05 vs. control+HG. LC3,
microtubule-associated proteins 1A/1B light chain 3B; Atg5,
autophagy protein 5; Atg7, ubiquitin-like modifier-activating
enzyme ATG7; p53, cellular tumor antigen p53; Bax, apoptosis
regulator BAX; Bcl-2, apoptosis regulator Bcl-2; HG, high glucose;
sh, short hairpin; Scr, scrambled. |
Discussion
The sequencing of diverse tissue samples and
different cell lines has identified lncRNAs as a novel class of
non-coding RNAs. Subsequently, emerging evidence has revealed that
lncRNAs serve critical roles in various biological processes via
regulation of target gene expression by recruiting
chromatin-modifying complexes to their promoters in the nucleus, or
by affecting mRNA stability and translational efficiency (25,26).
Recent studies have demonstrated that lncRNAs are also critical
regulators of disease processes, in addition to their functional
roles in normal physiological processes (27–29).
In cancer, for example, lncRNAs may function as either oncogenes or
tumor suppressors, and thereby regulate cancer cell growth,
metastasis and drug resistance. Although lncRNAs have been
associated with tumor pathogenesis, the study of lncRNAs in other
human diseases requires further investigation, including in DN.
Notably, certain previous studies have identified that lncRNAs are
additionally involved in the development of diabetes and its
complications (30–32). For example, lncRNA myocardial
infarction associated transcript has been demonstrated to be
involved in diabetes-induced microvascular dysfunction by acting as
a competing endogenous RNA for miR-150-5p to antagonize its
repression of vascular endothelial growth factor in retinal
endothelial cells (33). In
addition, plasmacytoma variant translocation 1 gene is able to
increase extracellular matrix (ECM) synthesis by upregulating
transforming growth factor β1, plasminogen activator inhibitor 1
and fibronectin 1, which are the principal regulators of ECM
accumulation in DN (34).
Given the importance of lncRNAs in DN, lncRNA
microarray analyses were initially performed to identify the
differential lncRNA expression profiles between the kidney cortices
of db/db DN mice and controls, in a previous study (23). As a result, 311 lncRNAs were
demonstrated to be differentially expressed in the db/db DN
tissues, including 105 upregulated lncRNAs and 206 downregulated
lncRNAs. In addition to the results of the authors' previous study
(23), Wang et al (21) reported that 1,018 lncRNAs (221
upregulated and 797 downregulated) were differentially expressed in
kidney tissue from db/db mice with DN. Although the expression of
hundreds of lncRNAs may be dysregulated in DN, their underlying
functional roles remain unclear. In the present study, the
expression of lncRNAs was further validated in mice kidney tissues
with or without DN. Out of these lncRNAs, Gm5524 was upregulated
and Gm15645 was downregulated, in accordance with the lncRNA
microarray data and RT-qPCR validation results. Furthermore, flow
cytometry apoptosis assays suggested that downregulation of Gm5524
and overexpression of Gm15645 increased the apoptotic rate of
podocytes, a feature of the early stages of DN. In addition, the
electron microscopy analysis revealed that alterations in Gm5524
and Gm15645 expression were able to induce podocyte autophagy.
Cellular autophagy and apoptosis are basic cellular
processes, and are essential for the maintenance of normal tissue
homeostasis under physiological conditions. Disorder of these
processes has been reported in diabetes and its complications
(35). The results of the present
study suggested that Gm5524 and Gm15645 may be involved in DN by
affecting these two processes. To further investigate the molecular
mechanisms underlying the involvement of Gm5524 and Gm15645 during
the pathogenesis of DN, the effects of Gm5524 and Gm15645 on
apoptosis and autophagy-associated factors were examined. It was
demonstrated that the Bcl2 protein expression was decreased and Bax
expression was increased in Gm5524-knockdown and
Gm15645-overexpressing podocytes. Bcl-2 is an important
anti-apoptotic protein, while Bax is an important pro-apoptotic
protein. The alteration in their expression levels was consistent
with the functional results. Additionally, the ratio of LC3II/LC3I
protein expression is a well-established biochemical assay to
determine the activation of autophagy, and the results of the
present study demonstrated that the altered expression of Gm5524
and Gm15645 also affected the LC3II/LC3I ratio. These data
suggested that Gm5524 and Gm15645 may affect cellular apoptotic and
autophagy processes in the diabetic mouse kidney, and thus
contribute to the development and progression of DN. However,
further investigations are required to further elucidate the
molecular mechanisms underlying the roles of Gm5524 and Gm15645 in
the pathogenesis of DN.
In conclusion, the present study revealed that
lncRNA Gm5524 and Gm15645 may affect podocytes apoptosis and
autophagy via regulation of the Bcl2/Bax, and LC3/ATG signaling
pathway in DN. Therefore, it was hypothesized that Gm5524 and
Gm15645-mediated regulation of autophagy and apoptotic signaling
pathways may be potential, useful therapeutic targets for the
development of alternative treatment strategies for patients with
DN.
Acknowledgements
The authors would like to thank the entire staff at
the Biotech Treatment Center, The Second Affiliated Hospital of
Nanjing Medical University for their excellent technical assistance
with this research.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81270896),
Six Talent Peaks Project in Jiangsu Province (grant no.
2013-WSN-049; Jiangsu, China) and Foundation of Nanjing Medical
University (grant no. 2015NJMUZD031).
Availability of data and materials
The datasets used or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YF and YL conceived and designed the study. YF, SC,
JX, QZ and XY performed the experiments. YF, DD and WY analyzed the
data. YF and DD wrote the paper. WY and YL reviewed and edited the
manuscript. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The protocol used in the present study was approved
by the Committee on the Ethics of Animal Experiments of The Second
Affiliated Hospital of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Katsarou A, Gudbjörnsdottir S, Rawshani A,
Dabelea D, Bonifacio E, Anderson BJ, Jacobsen LM, Schatz DA and
Lernmark Å: Type 1 diabetes mellitus. Nat Rev Dis Primers.
3:170162017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bansal D, Gudala K, Muthyala H, Esam HP,
Nayakallu R and Bhansali A: Prevalence and risk factors of
development of peripheral diabetic neuropathy in type 2 diabetes
mellitus in a tertiary care setting. J Diabetes Investig.
5:714–721. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Adeshara KA, Diwan AG and Tupe RS:
Diabetes and complications: Cellular signaling pathways, current
understanding and targeted therapies. Curr Drug Targets.
17:1309–1328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gregg EW, Sattar N and Ali MK: The
changing face of diabetes complications. Lancet Diabetes
Endocrinol. 4:537–547. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Magee C, Grieve DJ, Watson CJ and Brazil
DP: Diabetic Nephropathy: A tangled web to unweave. Cardiovasc
Drugs Ther. 31:579–592. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han Q, Zhu H, Chen X and Liu Z:
Non-genetic mechanisms of diabetic nephropathy. Front Med.
11:319–332. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sharma D, Bhattacharya P, Kalia K and
Tiwari V: Diabetic nephropathy: New insights into established
therapeutic paradigms and novel molecular targets. Diabetes Res
Clin Pract. 128:91–108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE Project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
LincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
He X, Ou C, Xiao Y, Han Q, Li H and Zhou
S: LncRNAs: Key players and novel insights into diabetes mellitus.
Oncotarget. 8:71325–71341. 2017.PubMed/NCBI
|
|
16
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bhan A and Mandal SS: Long noncoding RNAs:
Emerging stars in gene regulation, epigenetics and human disease.
ChemMedChem. 9:1932–1956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu B, Zhang C, Zou L, Ma Y, Huang K, Lv Q,
Zhang X, Wang S, Xue Y, Yi Z, et al: LncRNA uc.48+ siRNA improved
diabetic sympathetic neuropathy in type 2 diabetic rats mediated by
P2×7 receptor in SCG. Auton Neurosci. 197:14–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Allison SJ: Diabetic nephropathy: A lncRNA
and miRNA megacluster in diabetic nephropathy. Nat Rev Nephrol.
12:7132016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Duan LJ, Ding M, Hou LJ, Cui YT, Li CJ and
Yu DM: Long noncoding RNA TUG1 alleviates extracellular matrix
accumulation via mediating microRNA-377 targeting of PPARγ in
diabetic nephropathy. Biochem Biophys Res Commun. 484:598–604.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang M, Wang S, Yao D, Yan Q and Lu W: A
novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation
and fibrosis in diabetic nephropathy. Mol Cell Endocrinol.
426:136–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu M, Wang R, Li X, Fan M, Lin J, Zhen J,
Chen L and Lv Z: LncRNA MALAT1 is dysregulated in diabetic
nephropathy and involved in high glucose-induced podocyte injury
via its interplay with β-catenin. J Cell Mol Med. 21:2732–2747.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen S, Dong C, Qian X, Huang S, Feng Y,
Ye X, Miao H, You Q, Lu Y and Ding D: Microarray analysis of long
noncoding RNA expression patterns in diabetic nephropathy. J
Diabetes Complications. 31:569–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang J, Zhu Y and Wang R: Long noncoding
RNAs in respiratory diseases. Histol Histopathol. 119662018.
|
|
28
|
Sallam T, Sandhu J and Tontonoz P: Long
noncoding RNA discovery in cardiovascular disease: Decoding form to
function. Circ Res. 122:155–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Saha P, Vermas S, Pathak RU and Mishra RK:
Long noncoding RNAs in mammalian development and diseases. Adv Exp
Med Biol. 1008:155–198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leung A and Natarajan R: Long noncoding
RNAs in diabetes and diabetic complications. Antioxid Redox Signal.
Oct 30–2017.(Epub ahead of print).
|
|
31
|
Sun Y and Liu YX: LncRNA HOTTIP improves
diabetic retinopathy by regulating the p38-MAPK pathway. Eur Rev
Med Phamacol Sci. 22:2941–2948. 2018.
|
|
32
|
Zhang N, Geng T, Wang Z, Zhang R, Cao T,
Camporez JP, Cai SY, Liu Y, Dandolo L, Shulman GI, et al: Elevated
hepatic expression of H19 long noncoding RNA contributes to
diabetic hyperglycemia. JCI Insight. 3(pii): 1203042018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: lncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Alvarez ML and DiStefano JK: Functional
characterization of the plasmacytoma variant translocation 1 gene
(PVT1) in diabetic nephropathy. PLoS One. 6:e186712011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hamzawy M, Gouda SAA, Rashid L, Attia
Morcos M, Shoukry H and Sharawy N: The cellular selection between
apoptosis and autophagy: Roles of vitamin D, glucose and immune
response in diabetic nephropathy. Endocrine. 58:66–80. 2017.
View Article : Google Scholar : PubMed/NCBI
|