Introduction
Cervical cancer is the second most common malignant
tumor in women worldwide, and a leading cause of cancer deaths of
females in developing countries (1). Human papillomavirus (HPV) infection
is the etiologic agent for virtually all cases of cervical squamous
cell carcinoma (SCC) and a large portion of endocervical
adenocarcinoma. Among >200 human papillomavirus phenotypes, 14
high-risk HPV (hrHPV) phenotypes have been reported to be closely
associated with the initiation and progression of cervical cancer
(2). Two genotypes of hrHPV,
including HPV 16 and 18, are responsible for ~75% of all cases of
cervical cancer (2). Continuous
production of the E7 protein from oncogenic genotypes of HPV is
required for progression of malignancy (2). Thus, sensitive and specific detection
of E7 HPV protein expression in the clinical samples of exfoliated
epithelial cells or biopsies from cervix may provide a clinical
benefit for early detection of precancerous conditions.
The HPV genome consists of six early open reading
frames (E1, E2, E4, E5, E6, and E7), two late open reading frames
(L1 and L2). Among six proteins encoded by early open reading
frames, E6 and E7 are critical for the development of cervical
cancer by regulating cervical epithelial cell immortalization
(3). Transient viral infection
usually resolves spontaneously within 6 to 12 months without
increasing the risk of cervical cancer (4). However, in certain cases, viral DNA
can integrate into the host genome to cause persistent HPV
infection, resulting in an abnormal accumulation of HPV E6 and E7
proteins within host cells (5).
Overexpression of E7 protein can competitively bind to the tumor
suppressor protein, retinoblastoma, to cause the disassociation of
E2F proteins, leading to the abnormal proliferation of cervical
cells (6,7) and indispensably contributing to the
development of cervical intraepithelial neoplasia (CIN),
adenocarcinoma in situ and invasive cervical carcinoma
(4,8).
Viral genotyping analysis to identify the oncogenic
HPV has not produced good predictive values for the development of
intraepithelial CIN lesions (8).
It has been reported that 12–14% of low-grade squamous
intraepithelial lesions (LSIL) progress to high-grade squamous
intraepithelial lesions (HSIL) (9); without treatment, a few of those
lesions progress to invasive cancer (10). Further investigation is required to
identify other parameters with clinically meaningful predictive
values.
Protein biomarkers involved in the development of
cervical cancer, including SCC antigen, serum fragments of
cytokeratin, carcinoma embryonic antigen, soluble CD44 and matrix
metalloproteinases, have been considered as potential diagnostic
markers for cervical cancer screening (11). However, these potential protein
biomarkers are encoded by the human genome; therefore, diagnostic
use is inhibited by the endogenous expression of these biomarkers
within noncancerous cervical cells. The oncoprotein E7 encoded by
the hrHPV genome with specific expression in HPV-transformed human
cervical cells may serve as an ideal biomarker for the early
detection of cervical cancer (12).
In the present study, investigation of the E7
oncoprotein marker via immunostaining of exfoliated epithelial
cells and formalin-fixed paraffin-embedded biopsies for the
identification of pre-malignant and malignant lesions in squamous
cervical carcinoma was performed.
Materials and methods
Cervical cancer cell lines
CaSki cell line (HPV16 positive; CRL-1550™), HeLa
cell line (HPV18 positive; CCL-2™) and C-33A cell line (HPV
negative; HTB-31™) were purchased from the American Type Culture
Collection (ATCC; Manassas, VA, USA). CaSki cells were cultured in
RPMI-1640 medium (SH30027; Hyclone; GE Healthcare Life Sciences,
Logan, UT, USA) containing 10% heat-inactivated fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100
U/m penicillin and 100 g/ml streptomycin. HeLa and C-33A cells were
cultured in Minimum Essential Medium (SH30024; Hyclone; GE
Healthcare Life Sciences) containing 10% heat-inactivated FBS, 100
U/ml penicillin and 100 g/ml streptomycin. For immunocytochemical
(ICC) staining, cells (3×104/well) were transferred to a
24-well plate and were cultured to reach approximately 60–70%
confluency. Then cells were rinsed twice with 10 mM PBS (pH 7.4)
and fixed in 4% paraformaldehyde for 15 min at room temperature.
For other assays, adherent cells were harvested following digestion
by trypsin (25200; Gibco; Thermo Fisher Scientific, Inc.).
Following two washes with PBS, the harvested cells were fixed in 4%
paraformaldehyde for 10 min at room temperature for liquid based
cytology (LBC).
Preparation of E7 recombinant
proteins
DNA fragments encoding E7 proteins in hrHPV16, 18,
31, 33, 35, 39, 45, 52, 58 and 59 strains, and low-risk HPV (lrHPV)
6 and 11 strains were cloned and incorporated into pET-21a(+)
(Novagen; Merck KGaA, Darmstadt, Germany) or pGEX-4T-1 (GE
Healthcare Life Sciences) plasmids, and then were expressed in
Escherichia coli (E. coli) BL21/DE3 (American
Type Culture Collection, Manassas, VA, USA) to purify His-tagged or
glutathione S-transferase-tagged fusion protein. Protein
purification was performed as described previously (13,14).
The aforementioned proteins tagged with green fluorescent protein
were prepared by transient expression in 293 mammalian cell line
(ATCC; CRL-3216). Coding sequences of E7 proteins were optimized
from original HPV genomic sequences. The accession numbers in
Genbank were as follows: AHK23257.1 (HPV16), AGM34461.1 (HPV18),
AGM34454.1 (HPV31), AGM34459.1 (HPV33), ACV53985.1 (HPV35),
AGU90520.1 (HPV39), AGM34464.1 (HPV45), AET07150.1 (HPV52),
AFO63477.1 (HPV58) and ACL12335.1 (HPV59).
Preparation of E7 monoclonal
antibodies (mAbs)
The present study was approved by the ethics
committee of The First Affiliated Hospital of Soochow University
(Soochow, China). mAbs against hrHPV E7 proteins were generated by
immunizing mice or rabbits with purified HPV16 or 18 recombinant
proteins, followed by multistage procedures of established
technologies for screening antibody clones from hybridoma (15) and from phage displayed antibody
libraries (16–18). Individual clones of mAbs were
analyzed by various assays including, ELISA with E.
coli-derived recombinant hrHPVs and lrHPVs E7 proteins, western
blot analysis using E. coli or mammalian-expressed E7
proteins or endogenous E7 proteins from cervical cancer cell lines,
ICC using cervical cancer cell lines, and immunohistochemistry
(IHC) using human cancer tissue sections. Immunoreactivity of
antibodies generated from HPV16 E7 or HPV18 E7 immunized mice or
rabbit were tested against E7 proteins from other HPV strains by
ELISA using the aforementioned recombinant proteins. Subsequently,
mAb clones with desirable properties from either murine or rabbit
origins were selected for molecular cloning of immunoglobulin G
(IgG) genes. Cloning of IgG genes was accomplished by isolating the
coding sequence of immunoglobulin heavy chain and light chain from
hybridoma cells, or scFv sequences in the M13 phage, as described
previously (19). The intact IgG
molecules for mAb from murine (clone no. E7MuB6 and E7MuH1) and
from rabbit (clone no. E7Rb04 and E7Rb19) were produced by
large-scale 293 cell transfection using Lipofectamine®
LTX reagent (Thermo Fisher Scientific, Inc.) according to
manufacturers instructions. Secreted IgG protein in culture medium
was subjected to affinity purification by chromatography using a
Protein-A Sepherose 4B column (cat. no. 101041; Thermo Fisher
Scientific, Inc.). Protein concentration of purified IgG was
determined by measuring the absorbance at 280 nm and the purity was
analyzed via SDS-PAGE. Immunological activities of antibodies were
analyzed by ELISA, and ICC and IHC staining.
The cyclin dependent kinase inhibitor 2A
(p16INK4A; clone no. E6H4; cat. no. 705-4713) antibody
was purchased from Ventana Medical Systems, Inc. (Oro Valley, AZ,
USA) and secondary antibody reagent EnVision+ Peroxidase system
(cat. no. K5007) was from Dako (Agilent Technologies, Inc., Santa
Clara, CA, USA).
Specimens
The use of human tissue in the present study was
approved by the Institutional Review Board at the First Affiliated
Hospital of Soochow University (Soochow, China). Tissue slides
(three slides from each sample) used for the evaluation of the IHC
staining were prepared using formalin-fixed and paraffin-embedded
(FFPE) tissue blocks. Specimens with cervical LSIL (45 samples),
HSIL (64 samples) and invasive SCC (7 samples) were selected from
the tissue archives in the Pathology Department at the First
Affiliated Hospital of Soochow University. The age of the patients
ranged from 28–66 years with an mean age of 41.8 years. All
specimens were cut into 4–5 mm sections. Three slides from each
tissue sample were selected for hematoxylin and eosin staining, E7
antibody immunostaining and p16INK4A antibody
immunostaining.
Exfoliated cervical epithelial cells were obtained
from 13 patients with HSIL at the Cancer Hospital at the Chinese
Academy of Medical Sciences (Beijing, China). Samples from 10
healthy individuals were obtained from Peking University Hospital
(Beijing, China). Cells were preserved in an LBC fixative solution
and used within 6 months post-fixation. Those specimens were
previously stained in Pap-stain and graded according to the 2001
Bethesda System (20) by the
pathologists at Peking University First Hospital and Cancer
Hospital Chinese Academy of Medical Sciences (Beijing, China).
Slides were treated with a graded series of ethanol (100, 70, 50%
and water; 1 min each), followed by staining with hematoxylin
solution, Harris modified (Sigma-Aldrich; Merck KGaA) at room
temperature for 5 min. Slides were washed with water for 10 sec and
incubated with 0.5% hydrochloric acid for 8 sec, followed by a
further wash with tap water for 5 min. Subsequently, slides were
treated with a graded series of ethanol (50, 70, 80 and 96%; 30 sec
each), followed by EA-50 staining at room temperature for 2.5 min
and washing with 95% ethanol for 1 min and 100% ethanol for 1 min.
Finally, slides were washed twice with xylene for 2 min each and
mounted with VectaMount™ Permanent Mounting Medium (Vector
Laboratories, Burlingame, CA, USA). The study protocol was proved
by Institutional Review Board at the Peking University First
Hospital and Cancer Hospital Chinese Academy of Medical
Sciences.
Western blotting
Western blot analysis of endogenous E7 proteins was
performed by immunoprecipitation of cell lysates. Protein was
extracted from CaSki, HeLa and C-33A cervical cancer cell lines
using radioimmunoprecipitation lysis and extraction buffer (Thermo
Fisher Scientific, Inc.). Immunoprecipitation was performed as
described previously (21).
Protein concentration was determined with the bicinchoninic acid
method. Protein (20 µg/lane) was subjected to 10% SDS-PAGE,
transmembrane and immunoblotting with anti-E7 antibodies. Membranes
were blocked at room temperature for 10 min with TBS-Tween (TBST;
50 mM Tris-HCl, 150 mM NaCl, 0.5% Tween 20, pH 7.6) containing 5%
non-fat milk. Following washing, membranes were incubated with the
aforementioned primary monoclonal E7 antibodies (1:1,000) overnight
at 4°C. Following washing, membranes were incubated with the
horseradish peroxidase (HRP)-labeled goat anti-mouse secondary
antibody (1:1,000; cat. no. A2554, Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) or HRP-labeled mouse anti-rabbit secondary
antibody (1:1,000; cat. no. A1949, Sigma-Aldrich; Merck KGaA) at
room temperature for 2 h. Finally, signals were detected by adding
HRP substrate solution containing 3,3′-diaminobenzidine chromogen
(cat. no. ZLT-9033; OriGene Technologies, Inc., Beijing,
China).
ICC
For ICC analysis, the prepared Pap-smear slides and
LBC (ThinPrep®; Hologic China, Inc., Beijing, China)
slides prepared according to the manufacturer's protocols were
immediately fixed in 99% ethanol at room temperature for 1 h, and
were air-dried at room temperature overnight. Prior to antibody
staining, slides were rehydrated in 50% ethanol for 10 min and
distilled water for ≥30 sec. Antigen retrieval was performed by
heat treatment in epitope retrieval buffer (Tris-EDTA, pH 9.0) at
95–99°C for 10 min and then slides in epitope retrieval buffer were
maintained at room temperature for 20 min until the temperature
decreased below 50°C. Cells were fixed in 4% paraformaldehyde at
room temperature for 2 h. and permeabilized in 0.3% Triton X-100
for 15 min at room temperature, followed by washing twice with
TBST. ICC staining was then conducted. Briefly, slides were rinsed
once in TBST and treated with 3% H2O2 at room
temperature for 10 min to block endogenous peroxidase. Slides were
blocked in 10% newborn bovine serum (Thermo Fisher Scientific,
Inc.) at room temperature for 1 h, followed by incubation with
primary E7 antibody, E7MuB6 or E7MuH1 (1:1,000) at room temperature
(20–25°C) for 4 h. Slides were subsequently treated with secondary
antibody reagent EnVision+ Detection Systems
Peroxidase/DAB, Rabbit/Mouse (K5007; Dako; Agilent Technologies,
Inc.) according to the manufacturer's protocols. Finally, slides
were stained with hematoxylin (0.5%) at room temperature for 2 h,
and dehydrated in a series of graded ethanol concentrations (75,
85, 95 and 100%; 10 min each) at room temperature. Finally, slides
were mounted with coverslips for observation under an optical
microscope (Olympus Corporation, Tokyo, Japan). Positive cervical
cancer cells revealed staining of the cytoplasm. CaSki was used as
the positive control, and C-33A was used as the negative
control.
IHC
IHC staining with anti-E7 rabbit mAbs (clone nos.
E7R04 and E7R19) or with anti-p16INK4A antibody (clone
no. E6H4; cat. no. 705-4713; Ventana Medical Systems, Inc.) were
performed on tissue sections. Briefly, slides carrying cervical
tissue sections were dewaxed in xylene for 5 min and rehydrated in
a descending ethanol gradient at room temperature (95, 85, 50 and
30%; 2 min each). Epitope retrieval was performed by heating the
slides at 121°C for 2 min in citrate buffer (10 mmol/l, pH 6.0).
Endogenous peroxidase activity was blocked with treatment with 3%
H2O2 solution at room temperature for 20 min.
Blocking was performed by incubation with TBST containing 10%
newborn bovine serum at room temperature for 1 h. Following
washing, primary antibodies (1:1,000) were incubated with the
slides at 20–25°C for 1 h, followed by washing with TBST and
incubation with HRP-labeled secondary antibody (Dako EnVision+
Peroxidase system) at room temperature for 30 min. DAB chromogen
system (Dako; Agilent Technologies, Inc.) was used for color
development according to the manufacturer's protocols. Slides were
washed with xylene three times, for 3 min each time. Subsequently,
slides were treated with a series of graded ethanol (100, 80, 50
and 30%; 2 min each) at room temperature. Then slides were stained
with hematoxylin (0.5%) at room temperature for 20 min. Slides were
washed with tap water for 5 min and then treated with acid alcohol
(1% HCl in 70% EtOH) until the slides turned pink. Following a
further wash with tap water for 5 min, slides were treated with
ammonia water (1 ml NH4OH in 1l H2O) until
the sections noticeably darkened. Slides were washed again with tap
water for 5 min and stained with Eosin Y (0.5%) at room temperature
for 1 min. Following rinsing in tap water for 1 min, slides were
dehydrated in a graded ethanol series (50, 80, 90 and 100%; 2 min
each) and mounted on coverslips. The HPV-positive cervical cancer
tissue was used as a positive control, and sham-treated slide
treated with blank blocking buffer instead of primary antibody was
used as negative control.
Interpretation of E7 and
p16INK4A immunostaining
Stained slides were analyzed by two pathologists for
variable representation of epithelial and stromal components and
immunoreactivity. Squamous epithelial cells, with brown-colored
cytoplasmic staining for E7 protein, or with nuclear and/or
cytoplasmic staining for p16INK4A protein, were
identified and graded as follows: 0, no staining; 1, 1–25%
staining; 2, 26–50% staining; 3, 51–75% staining; and 4, 76–100%
staining. Cases were classified as positive staining (2+ for E7; 3+
for p16INK4A) vs. negative or focal staining. The
staining intensity for each marker was not taken into
consideration. The examination and scoring of immunohistochemistry
slides were confirmed by a panel of more than two certified
professionals in pathology.
In situ hybridization (ISH) analysis
for HPV E7 mRNA
ISH analysis for HPV E7 mRNA was performed using
RNAscope HPV kit (Advanced Cell Diagnostics, Inc., Newark, CA, USA)
according to the manufacturer's protocols. Briefly, FFPE tissue
sections (4 mm-thick) were heated (65°C) for 5 min and pretreated
with protease K (10 µg/ml) at 37°C for 20 min prior to
hybridization. For each case, three slides from adjacent tissue
sections were separately hybridized with probes targeting ubiquitin
C (cat. no. 310041; Advanced Cell Diagnostics, Inc.), DapB (cat.
no. 310043; Advanced Cell Diagnostics, Inc.) and a cocktail of 18
genotypes of hrHPV (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56,
58, 59, 66, 68, 73 and 82; cat. no. 312591). A HRP-based signal
amplification system was subsequently used to hybridize the target
probes using a peroxidase activity assay kit (Sigma-Aldrich), and
signal generation followed by incubation with the chromogenic
substrate 3,3′-diaminobenzidine (0.05%) at room temperature for 30
min. Counter staining with hematoxylin was performed using the
method described above. A ubiquitin C probe was used as an
endogenous control for RNA integrity assessment. The bacterial gene
DapB was used as a negative control to assess background staining.
A positive stain was defined by a typical pattern of punctate brown
colored granules (precipitates) in or around the nucleus, or in the
cytoplasm of the cells. For the paired IHC analysis, adjacent
tissue sections were processed by staining with the aforementioned
anti-E7 antibody cocktail.
Results
E7 mAb recognizes endogenous E7
protein in cervical cancer cells
mAbs with high specificity towards E7 recombinant
proteins from the hrHPV strains (HPV16, 18, 31, 33, 35, 39, 45, 52,
58 and 59), and without cross-reactivity to E7 proteins from the
lrHPV strains (HPV6 and 11) were selected via ELISA. Positive
antibody clones were further characterized by their abilities to
bind the endogenous E7 protein in cervical cancer cells. As
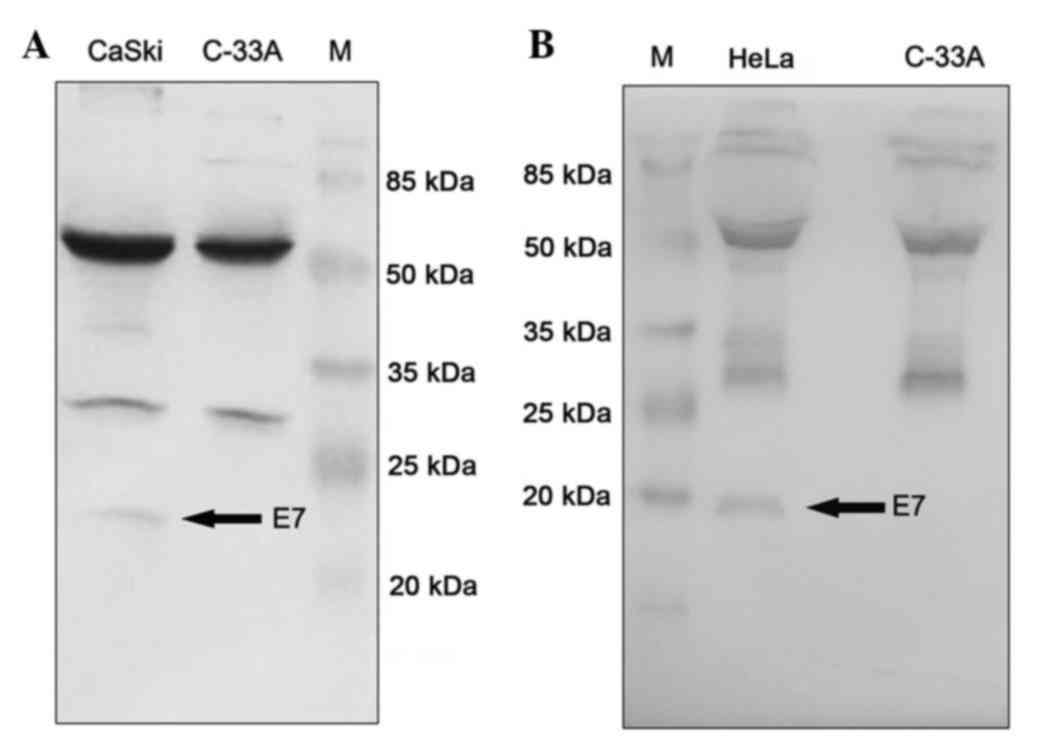
presented in Fig. 1, antibodies
immunoprecipitated the endogenous E7 protein from cell lysates
prepared from cervical cancer cell lines CaSki (HPV16 positive) and
HeLa (HPV18 positive). The C-33A cell lysate was included in the
parallel experiment as a negative control. Protein samples from
immunoprecipitation were analyzed by western blotting. A single
specific band with expected molecular weight was detected in CaSki
cell lysate (Fig. 1A) and in HeLa
cell lysate (Fig. 1B), indicating
that mAbs may specifically recognize endogenous E7 proteins
expressed in cancer cell lines. Two upper bands in each lane with
apparent molecular weight of 55 and 28 kDa respectively represented
the IgG heavy chain and light chain of mAb used in the
immunoprecipitation.
E7 mAb specifically stained cervical
cells by ICC
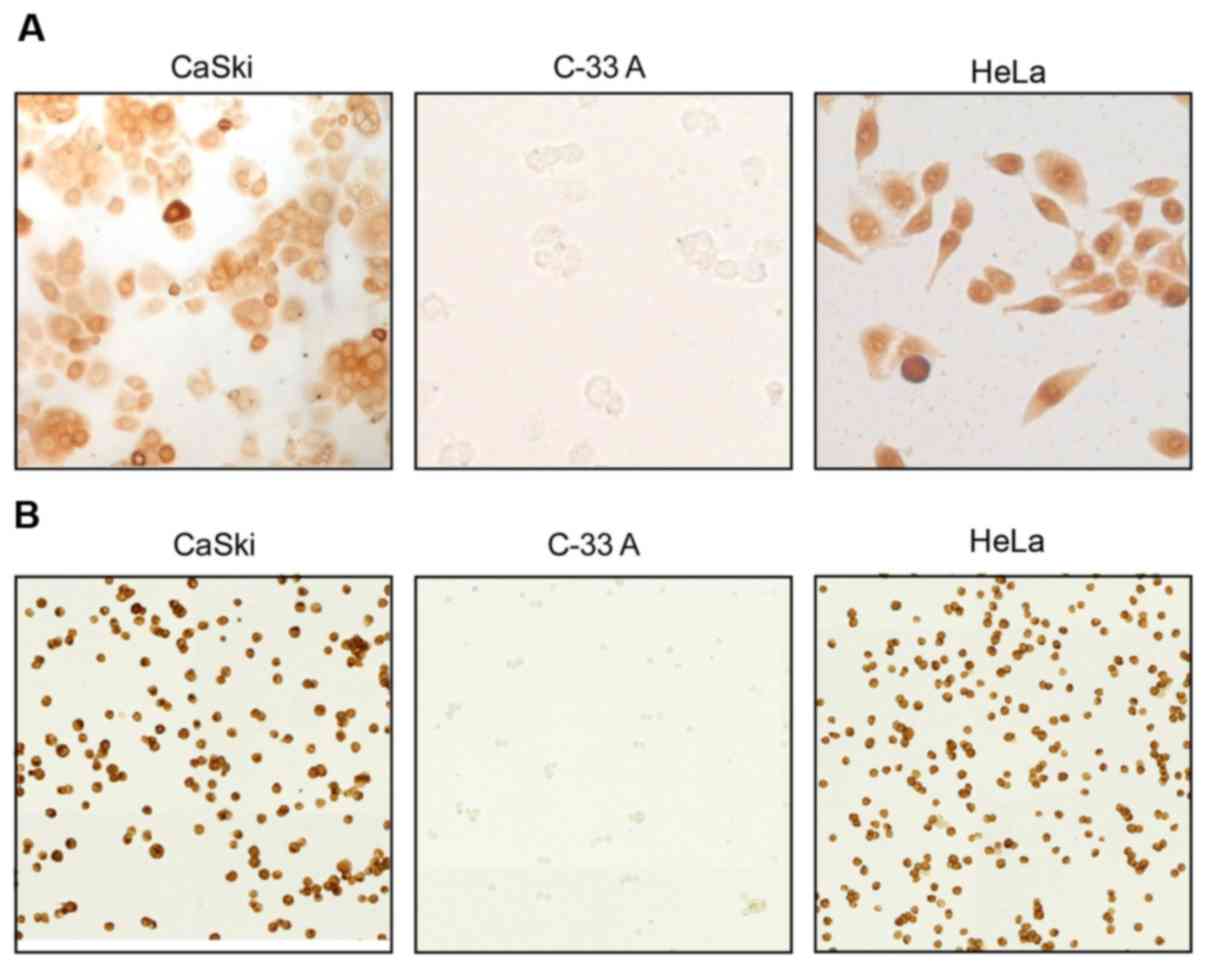
For further confirmation of the specificity of mAbs
against E7 proteins expressed in cervical cells, three cervical
cancer cell lines C-33A, HeLa and CaSki were cultured and analyzed
via ICC (Fig. 2). HeLa cells
exhibited high expression of HPV18 E7 protein, while CaSki cells
demonstrated overexpression of HPV16 E7 proteins. C-33A cells were
used as a negative control as HPV16 E7 and HPV18 E7 proteins are
not expressed within these cells. HeLa cells were exclusively
stained by mAb E7MuH1 against HPV18 E7 protein, while HPV16 E7
protein was specifically detected in CaSki cells using mAb E7MuB6
against HPV16 E7 proteins; neither antibody recognized C-33A cells.
Collectively, the results of the present study indicated that the
mAbs generated have high specificity for E7 proteins expressed in
cervical cancer cells.
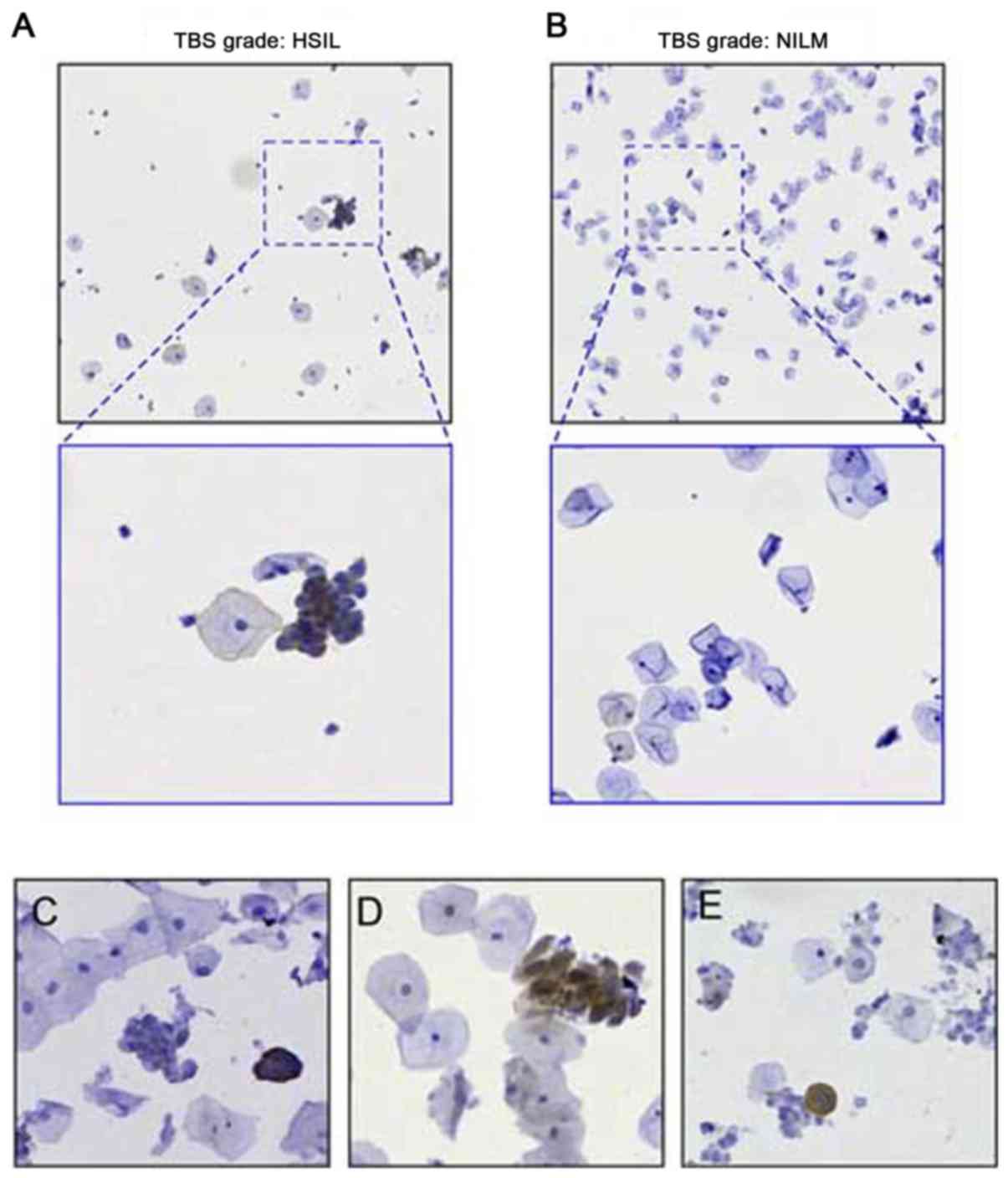
Application of mAb in ICC staining of
exfoliated cervical epithelial cells
LBC or thin-layer cytology has been widely applied
in clinical testing for the examination of exfoliated cervical
epithelial cells. To investigate the compatibility of E7 mAb for
immunostaining on LBC slides, fixed cervical epithelial cells were
treated with mAb E7MuB6. Antibody activity and specificity were
demonstrated using positive samples of HSIL and positive HPV16
genotyping results. Normal cervical cell slides negative for
intraepithelial lesion and malignancy (NILM) were used as negative
controls. As presented in Fig. 3,
the slides treated with murine mAb revealed dark brown colored
cells, indicating E7 positive expression within the transformed
cells, consistent with their HSIL cytological morphology features
(Fig. 3A). None of cells on the
slides NILM were stained by the E7 mAb (Fig. 3B). In addition, LBC slides with
varying lesion grades [HSIL (Fig.
3C), LSIL (Fig. 3D), and
atypical squamous cells of undetermined significance (ASC-US;
Fig. 3E)] could be identified via
a direct evaluation based on the expression of the oncoprotein E7.
This demonstrated the feasibility of using a specific biomarker for
the interpretation of the cytopathological results. As presented in
Fig. 3E, a case classed as ASC-US
case by the conventional cytology test was identified as positive
for precancerous cells based on the identification of E7
oncoprotein expression.
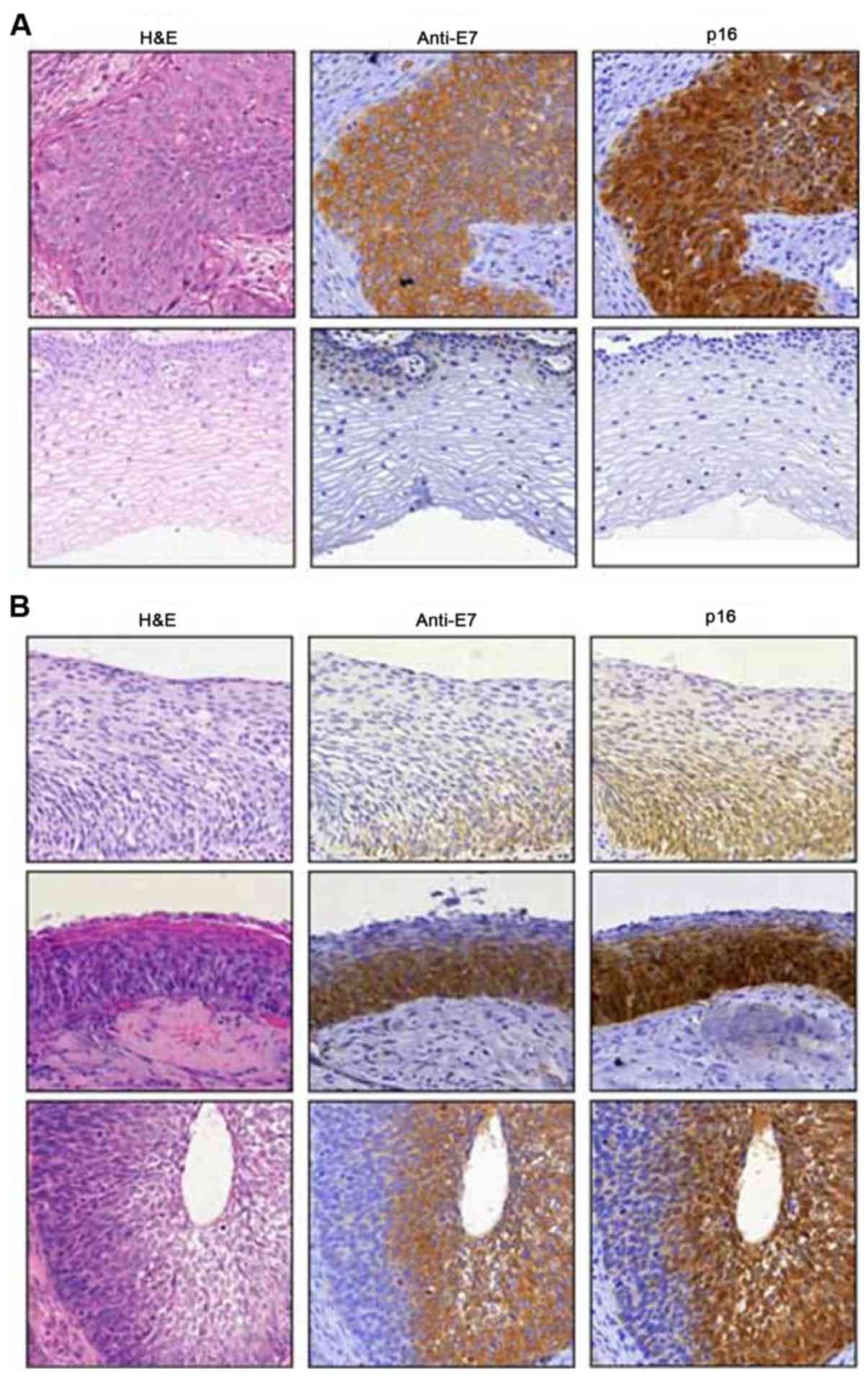
IHC reactivity with E7 protein and
comparison with surrogate p16INK4A marker
Pathology tests are considered to be the most
important test for the clinical diagnosis of cervical cancer or
precancerous lesions. To evaluate the utility of E7 antibodies for
histopathological examination of cervical cancer, the IHC staining
capacity of the rabbit monoclonal anti-E7 cocktail was investigated
and compared with the anti-E7 immunostaining patterns of the
established surrogate protein marker p16INK4A. As
presented in Fig. 4A, rabbit
monoclonal anti-E7 mAbs exhibited intense granular staining in
cytoplasm of human cervical carcinoma lesions (top), while no brown
staining in normal cervical tissues was observed (bottom). In
addition, E7 mAbs demonstrated a similar staining pattern to that
of anti-p16INK4A antibody in different premalignant and
malignant lesions (Fig. 4A and
B).
It has been speculated that the tumorigenic
functions of E7 protein may be associated with its expression
levels during the development and progression of cancer. To further
investigate whether the E7 antibodies used in the present study may
reveal the expression levels of HPV E7 proteins in cervical lesions
of increasing pathological grades, tissue sections from cervical
intraepithelial neoplasia (CIN)1, CIN2 and CIN3 samples were
subjected to IHC staining using the novel rabbit anti-E7 mAbs
(Fig. 4B). The staining results
indicated an increased amount of E7 positive cells on the slides as
the pathological stage was increased. This observation of E7
staining is consistent with the increased severity of pathological
stages, from CIN1 to CIN2, and then to CIN3 lesions. Comparison of
the E7 and p16INK4A antibody staining results in normal
tissue (negative) and cervical carcinoma (positive) demonstrated
similar staining patterns as presented in Fig. 4A. A notable difference is that the
p16 INK4A antibody stained numerous foci in
morphologically normal cervical cells, whereas the E7 antibody
demonstrated an overall similar staining pattern, but with a
distinct specificity to the cervical cancer lesions (Fig. 4B).
To assess the robustness of the E7 immunostaining in
a larger number of pathology specimens, an antibody reagent
cocktail of anti-E7 mAbs was optimized and used to test tissue
slides from 116 cases with pathology reports in a double-blind
experiment. For comparison, p16INK4A immunostaining was
performed in parallel experiments. The results were analyzed
against the gold standard of pathology. As presented in Table I, the staining of E7 among the
three pathological groups, LSIL, HSIL and SCC exhibited a trend of
increasing E7-positive rates: 31.1, 90.6 and 100%, respectively.
The p16INK4A group (Table
I) revealed similar results to those of the anti-E7 test group.
Highly similar results were reported between the two protein
biomarkers.
 | Table I.E7 and p16INK4A
immunostaining in tissue slides via immunohistochemistry. |
Table I.
E7 and p16INK4A
immunostaining in tissue slides via immunohistochemistry.
|
| LSIL | HSIL | SCC |
|---|
|
|
|
|
|
|---|
| Protein
expression | Cases | % | Cases | % | Cases | % |
|---|
| E7 |
|
|
|
|
|
|
|
Negative | 31 | 68.9 | 6 | 9.4 | 0 | 0 |
|
Positive | 14 | 31.1 | 58 | 90.6 | 7 | 100 |
|
Total | 45 | | 64 |
| 7 |
|
|
p16INK4A |
|
|
|
|
|
|
|
Negative | 35 | 77.8 | 4 | 6.3 | 0 | 0 |
|
Positive | 10 | 22.2 | 60 | 93.7 | 7 | 100 |
|
Total | 45 |
| 64 | | 7 |
|
ISH analysis of HPV E7 mRNA
In the present study, antibody-binding specificity
to E7 antigen was investigated via IHC staining. In addition, HPV
E7 mRNA expression levels were investigated via ISH using adjacent
sections from the same tissue blocks used for E7 antibody staining
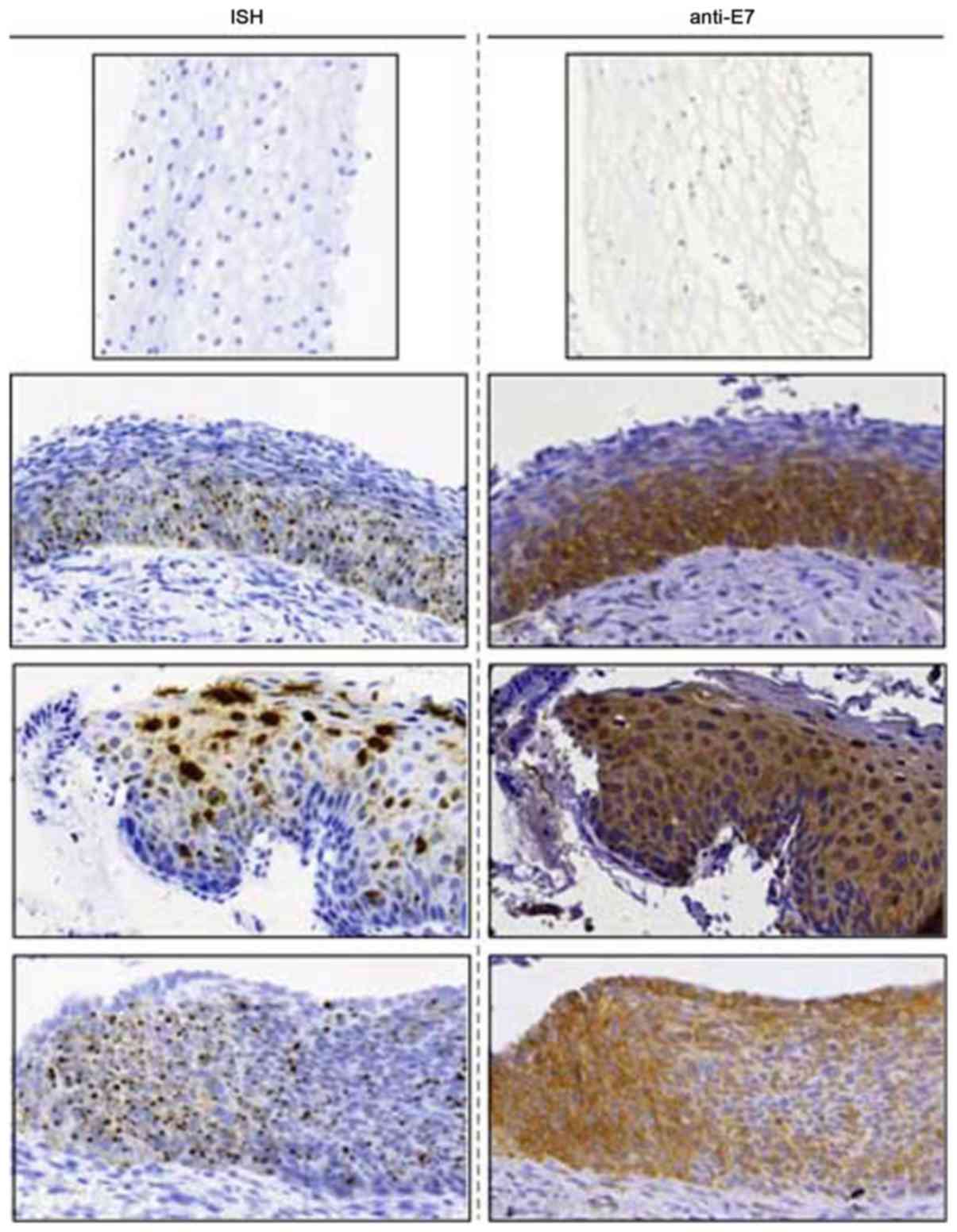
by IHC. As presented in Fig. 5, E7
mAbs and E7 mRNA probes revealed similar localization of positively
stained loci or regions on the slides, indicating the accurate
recognition of the antibodies to the endogenous E7 antigen in the
immunohistochemistry assay. Morphologically aberrant cells
exhibited the typical punctate dots following the application of
the E7 mRNA probe via ISH; staining intensity also appeared to be
increased with the increasing degree of cancer. In this set of
experiments, the mRNA and protein staining patterns appeared to be
co-localized in the same areas in the lesions. Antibody staining
indicated that the E7 mRNA expression levels were associated with
the intensity and location of E7 protein expression. The findings
of the present study further confirmed the specific immunoreaction
of E7 antibodies and the endogenous tumor antigen E7 within
cervical lesion tissue.
Discussion
Cervical cancer is a leading cause of
cancer-associated mortality in females worldwide and the second
most common cancer in females in developing countries (1). According to the data released by the
National Central Cancer Registry of China, the incidence of
cervical cancer in China was 15/100,000 in 2015, which is still
listed as one of the major malignant tumors threatening female
health in China (22).
Fortunately, cervical cancer is preventable and curable at the
early stages due to the relatively slow progression of cervical
carcinoma (1). The morbidity and
mortality of cervical cancer has been remarkably reduced by the
early screening tests, including Pap-smear and liquid-based
cytology (9). Low sensitivity and
false positive results from the abnormal morphology of ASC-US are
major limitations of cytology tests used in primary screening.
According a meta-analysis of Pap-smear tests and LBC, the
sensitivity and accuracy of cervical cytology exhibited wide
variation in different regions and institutions (23). In addition, a study of a large
population revealed that 30–60% of patients with invasive cervical
cancer were reported to be normal in Pap-smear testing 3–5 years
prior to diagnosis, indicating that the cervical cytology test has
a poor negative predictive value (24). By contrast, the development of
molecular HPV testing has largely improved the detection
sensitivity, however, it is limited by its low overall specificity
and low positive predictive value (10,25).
A desirable improvement in the early screening is to identify the
real-time phenotype of precancerous cervical lesions, and to avoid
over diagnosis and/or treatment of patients with transient HPV
infection only, or ASC-US and a low-grade LSIL cervical lesion.
Biomarkers can potentially improve the diagnostic
interpretation of cytology tests for early stage cervical cancer
(11). Despite the benefit of
decreasing inter-observer variations for the diagnosis of LSILs,
the clinical significance of p16 INK4A is limited by its
intrinsic properties, including the inconsistent expression in
tumors, lack of tumor specificity and the lack of uniformity in
scoring systems.
E7 is a pivotal oncoprotein, with expression
required to maintain the transformed phenotype in hrHPV-induced
human cancers. The conformational, structural and biochemical
behavior of E7 protein has been extensively characterized (18). Several conformational E7 species
have been identified, including a nuclear dimer and high molecular
weight spherical oligomers in the cytoplasm of transformed cells
(26). It has been hypothesized
that the predominant species of E7 oligomers may exist in
transformed cells, and contain epitopes that are not exposed in the
dimer. The complexity of conformational changes in E7 protein made
the ‘rational design’ and construction of an E7 antibody extremely
difficult.
Considering its highly specific expression in
transformed cervical cells, HPV E7 protein has been well reported
as an appropriate biomarker candidate for the detection of cervical
cancer (12). A number of studies
reported the detection of E7 proteins using either polyclonal
antibodies or mAbs (12,26–32).
Lidqvist et al (18)
described the production of mouse mAbs to HPV E7 protein and use
for testing LBC slides. However, data from a clinical evaluation
was not reported. Faoro et al (26) reported a new E7 antibody with
limited antibody specificity data and indiscriminative
immunoreactivity in LSIL and HSIL. Collectively, available reports
have demonstrated a lack of thorough characterization of antibody
specificity, or the evaluation of E7 protein as a biomarker for
diagnostic testing.
The present study reported an empirical approach to
develop several unique mAbs that recognize E7 proteins from HPV16,
HPV18 as well as structurally-associated E7 proteins from other
strains of hrHPVs. This may increase the range of diagnostic
application for the analysis of clinical specimens. mAb were
constructed using protein antigens of E7 from hrHPV16 and 18 and
subsequently used to immunize mice and rabbits. mAb clones were
screened and selected based on binding specificity to E7 proteins
from numerous hrHPV strains, including HPV16 and 18, and
structurally-associated hrHPV31, 33, 35, 39, 45, 52, 58, and 59.
Antibody candidate clones demonstrating cross-reactivity with low
risk strain HPV E7 proteins (HPV6 and 11) were excluded from
subsequent analysis in the present study. Using a variety of
characterization assays, it was confirmed that the selected mAbs
specifically bind to the endogenous hrHPV E7 proteins in cervical
cancer cells (CaSki and HeLa). Finally, the applications of these
anti-E7 mAbs for the specific recognition of E7-expressing cervical
cells in clinical specimens were demonstrated, via LBC slides and
FFPE tissue slides by ICC or IHC, respectively. Of particular
interest is investigation of the use of anti-E7 staining in ASC-US
slides, which may contribute to the interpretation of
cytology-based early screening tests. In addition, IHC analysis
using the E7 antibody cocktail reagent produce similar results to
p16INK4A immunostaining, indicating the feasibility of
using E7 as a specific biomarker for identification of premalignant
cervical epithelial lesions.
In summary, the HPV E7 protein encoded by the viral
genome, but only expressed in transformed human cells, may be
considered as an ideal biomarker for the early detection of
precancerous lesions. The detection of E7 protein expression may
effectively distinguish malignant transformations from transient
HPV infections. This E7 protein identification technique may be
used in either ICC or IHC analyses of cervical samples, and provide
more reliable results for improved efficiency in cervical cancer
screening and early diagnosis.
Acknowledgements
The authors acknowledge the support and critical
reading of this manuscript from Dr Jiliang Hu of the College of
Pharmacology, Guangzhou University (Guangzhou, China).
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LS and XC designed the experiments. LS, FH and CS
performed the experiments. YH and YL analyzed the data. All authors
read the manuscript.
Ethics approval and consent to
participate
The use of human tissue in the present study was
approved by the Institutional Review Board at the First Affiliated
Hospital of Soochow University, prior to data collection. Written
informed consent was obtained from all participants involved in
this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McGuire S: World Cancer Report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for Research on Cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Crosbie EJ, Einstein MH, Franceschi S and
Kitchener HC: Human papillomavirus and cervical cancer. Lancet.
382:889–899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Choi YJ and Park JS: Clinical significance
of human papillomavirus genotyping. J Gynecol Oncol. 27:e212016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Groves IJ and Coleman N: Pathogenesis of
human papillomavirus-associated mucosal disease. J Pathol.
235:527–538. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gravitt PE: The known unknowns of HPV
natural history. J Clin Invest. 121:4593–4599. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yugawa T and Kiyono T: Molecular
mechanisms of cervical carcinogenesis by high-risk human
papillomaviruses: Novel functions of E6 and E7 oncoproteins. Rev
Med Virol. 19:97–113. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rashid NN, Rothan HA and Yusoff MS: The
association of mammalian DREAM complex and HPV16 E7 proteins. Am J
Cancer Res. 5:3525–3533. 2015.PubMed/NCBI
|
|
8
|
Egawa N, Egawa K, Griffin H and Doorbar J:
Human papillomaviruses; epithelial tropisms, and the development of
neoplasia. Viruses. 7:3863–3890. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saslow D, Solomon D, Lawson HW, Killackey
M, Kulasingam SL, Cain J, Garcia FA, Moriarty AT, Waxman AG, Wilbur
DC, et al: American Cancer Society, American Society for Colposcopy
and Cervical Pathology, and American Society for Clinical Pathology
screening guidelines for the prevention and early detection of
cervical cancer. Am J Clin Pathol. 137:516–542. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goodman A: HPV testing as a screen for
cervical cancer. BMJ. 350:h23722015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dasari S, Wudayagiri R and Valluru L:
Cervical cancer: Biomarkers for diagnosis and treatment. Clin Chim
Acta. 445:7–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stiasny A, Kuhn C, Mayr D, Alexiou C,
Janko C, Wiest I, Jeschke U and Kost B: Immunohistochemical
evaluation of E6/E7 HPV oncoproteins staining in cervical cancer.
Anticancer Res. 36:3195–3198. 2016.PubMed/NCBI
|
|
13
|
Hoffmann A and Roeder RG: Purification of
his-tagged proteins in non-denaturing conditions suggests a
convenient method for protein interaction studies. Nucleic Acids
Res. 19:6337–6338. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Harper S and Speicher DW: Purification of
proteins fused to glutathione S-transferase. Methods Mol Biol.
681:259–280. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Morbini P, Alberizzi P, Tinelli C, Paglino
C, Bertino G, Comoli P, Pedrazzoli P and Benazzo M: Identification
of transcriptionally active HPV infection in formalin-fixed,
paraffin-embedded biopsies of oropharyngeal carcinoma. Hum Pathol.
46:681–689. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pirog EC: Immunohistochemistry and in situ
hybridization for the diagnosis and classification of squamous
lesions of the anogenital region. Semin Diagn Pathol. 32:409–418.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Szarewski A, Mesher D, Cadman L, Austin J,
Ashdown-Barr L, Ho L, Terry G, Liddle S, Young M, Stoler M, et al:
Comparison of seven tests for high-grade cervical intraepithelial
neoplasia in women with abnomal smears: The predictors 2 study. J
Clin Microbiol. 50:1867–1873. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lidqvist M, Nilsson O, Holmgren J, Hölters
S, Röijer E, Dürst M and Fermér C: Detection of human
papillomavirus oncoprotein E7 in liquid-based cytology. J Gen
Virol. 93:356–363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marks JD and Bradbury A: PCR cloning of
human immunoglobulin genes. Methods Mol Biol. 248:117–134.
2004.PubMed/NCBI
|
|
20
|
Solomon D, Davey D, Kurman R, Moriarty A,
O'Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T Jr, et
al: The 2001 Bethesda System: Terminology for reporting results of
cervical cytology. JAMA. 287:2114–2119. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szalmás A, Tomaić V, Basukala O, Massimi
P, Mittal S, Kónya J and Banks L: The PTPN14 tumor suppressor is a
degradation target of human papillomavirus E7. J Virol.
91:e00057–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nanda K, McCrory DC, Myers ER, Bastian LA,
Hasselblad V, Hickey JD and Matchar DB: Accuracy of the
Papanicolaou test in screening for and follow-up of cervical
cytologic abnormalities: A systematic review. Ann Intern Med.
132:810–819. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Katki HA, Kinney WK, Fetterman B, Lorey T,
Poitras NE, Cheung L, Demuth F, Schiffman M, Wacholder S and Castle
PE: Cervical cancer risk for women undergoing concurrent testing
for human papillomavirus and cervical cytology: A population-based
study in routine clinical practice. Lancet Oncol. 12:663–672. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zazove P, Reed BD, Gregoire L, Ferenczy A,
Gorenflo DW and Lancaster WD: Low false-negative rate of PCR
analysis for detecting human papillomavirus-related cervical
lesions. J Clin Microbiol. 36:2708–2713. 1998.PubMed/NCBI
|
|
26
|
Faoro V, Barbazza R, Bonin S, Brunetti D,
Sulfaro S and Stanta G: Detection of HPV E7 oncoviral protein in
cervical lesions by a new antibody. Appl Immunohistochem Mol
Morphol. 21:341–350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ramirez N, Guerra F, Camporeale G,
Quintana S, Diaz LB, Cuneo N, Villacorta Hidalgo J, Tatti SA,
Alonso LG, Borkosky SS, et al: Expressions of E2 and E7-HPV16
proteins in pre-malignant and malignant lesions of the uterine
cervix. Biotech Histochem. 90:573–580. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arbyn M, Paraskevaidis E, Martin-Hirsch P,
Prendiville W and Dillner J: Clinical utility of HPV-DNA detection:
Triage of minor cervical lesions, follow-up of women treated for
high-grade CIN: An update of pooled evidence. Gynecol Oncol. 99 3
Suppl 1:S7–S11. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nasiell K, Roger V and Nasiell M: Behavior
of mild cervical dysplasia during long term follow-up. Obstet
Gynecol. 67:665–669. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Holowaty P, Miller AB, Rohan T and To T:
Natural history of dysplasia of the uterine cervix. J Natl Cancer
Inst. 91:252–258. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
van Bogaert LJ: P16INK4a
immunocytochemistry/immunohistochemistry: Need for scoring
uniformization to be clinically useful in gynecological pathology.
Ann Diagn Pathol. 16:422–426. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dantur K, Alonso L, Castaño E, Morelli L,
Centeno-Crowley JM, Vighi S and de Prat-Gay G: Cytosolic
accumulation of HPV16 E7 oligomers supports different
transformation routes for the prototypic viral oncoprotein: The
amyloid-cancer connection. Int J Cancer. 125:1902–1911. 2009.
View Article : Google Scholar : PubMed/NCBI
|