Introduction
Glioma is the most common malignancy of brain tumor,
accounting for >32% of all brain tumors (1) and ~80% of malignancies in the brain
(2). The overall median survival
for patients with glioma is between 16 to 18 months (3,4).
Notable advances in the therapeutic strategies of glioma have been
made in the last decade, including neurosurgical approaches,
chemotherapy and radiotherapy; however, the prognosis of glioma
remains poor, mainly due to the fact that glioma cells are highly
aggressive and capable of infiltrating adjacent normal brain
tissue, leading to the failure of complete dissection of the tumor
by surgery in the brain (5,6).
Recurrence and resistance to chemotherapy are another two
influential factors responsible for poor prognosis (7–9).
Thus, developing novel strategies and investigating innovative
therapeutic agents for patients diagnosed with glioma is imperative
and urgently required.
Long noncoding RNAs (lncRNAs) are a class of RNAs
that are non-protein coding and longer than 200 nucleotides in
length (10–12). With the continuous advances of
research approaches, lncRNAs are widely known and have recently
undergone a rapid expansion in research and discovery. LncRNAs are
closely associated with tumor development (13), particularly in the progression of
glioma (14). For instance, lncRNA
MEG63 contributes to cisplatin-induced apoptosis via the
suppression of autophagy in human glioma (15). LncRNA CRNDE induces inflammation
via the Toll-like receptor3-nuclear factor-κB-cytokine signaling
pathway (16).
Tumor suppressive lncRNA on Chromosome 8p12 (TSLNC8)
is a novel lncRNA, which is also named LINC00589 (17). TSLNC8 has been reported to be
deleted and downregulated in human liver cancers. TSLNC8 serves its
tumor suppressive role by physically interacting with transketolase
and signal transducer and activator of transcription 3 (STAT3) and
inactivation of the interleukin-6/STAT3 signaling pathway. In
addition, TSLNC8 may inhibit the phosphorylation of STAT3,
resulting in the low transcriptional activity of STAT3 in human
liver cancer (17). Thus, TSLNC8
has been identified as a prognostic predictor for liver cancer
patients in clinic; however, the role of TSLNC8 in human glioma
requires further investigation.
In the present study, the transcript levels of
TSLNC8 were observed to be significantly decreased in human glioma
tissues compared with in noncancerous counterparts, and were
negatively associated with tumor size, distant metastasis and
tumor, node and metastasis (TNM) stage. Overexpression of TSLNC8 in
glioma cells inhibited, while knockdown of TSLNC8 promoted cell
proliferation and metastasis. TSLNC8 was also demonstrated to
regulate cell apoptosis via the intrinsic pathway. The findings
demonstrated the role of TSLNC8 in human glioma, which may
contribute to the clinical diagnosis and treatment of the
disease.
Materials and methods
Human samples
A total of 80 patients with diagnosed glioma (Male:
Female, 59:21, age) were enrolled in the present study. For all
patients, the dissected glioma tissues and their adjacent
noncancerous tissues were collected from Tianjin First Center
Hospital (Tianjin, China) during January 2014 to December 2016. All
of the tissues were frozen in liquid nitrogen following dissection
from patients during surgeries. Clinical data were also recorded
for statistical analysis. Each patient provided informed consent to
participate in the present study, which was approved by the Ethics
Committee of Tianjin First Center Hospital.
Cell culture and transfection
Human glioma cell lines BE-2C and BT325 were
purchased from the American Type Culture Collection (Manassas, VA,
USA). Human glioma cell line SWO38 was a kind gift from Jinan
University (Guangzhou, China) (18). Human glioma cell lines U251-MG,
SHG-44 and CHG-5 were purchased from the Cell Bank of Type Culture
Collection of Chinese Academy of Science (Shanghai, China). Human
normal neuronal cell line. Human normal astrocyte cells were
purchased from ScienCell Research Laboratories, Inc. (San Diego,
CA, USA; catalog no. 1830) and used as a normal control of glioma
cell. All culture media (Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) were
supplied with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher
Scientific, Inc.). Cells were maintained at 37°C in a humidified
atmosphere containing 5% CO2. Small interfering (si)RNA
against TSLNC8 was designed and synthesized by Shanghai Shenggong
Biology Engineering Technology Service, Ltd., (Shanghai, China)
with the sequence of 5′-GCACATGAACACATTGAAA-3′ and the control
siRNA sequence was 5′-GCAAAGTACACGTTACAAA-3′ with the same source
as the specific siRNAs. TSLNC8 expressing plasmid was constructed
by our own lab with XhoI and HindIII restriction
enzyme and cloned into pcDNA 3.1 vector. A total of 2 µg siRNAs or
expressing plasmid was transfected into cells with
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. After
12 h, the cell culture medium was replaced with new one. In all
conditions, the culture medium was replaced every 2 days, unless
otherwise stated.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from human tissues and all cultured cells
was extracted using TRIzol® reagent (Thermo Fisher
Scientific, Inc.). RNA was quantified by Nanodrop 2000 (Thermo
Fisher Scientific, Inc., Wilmington, DE, USA) by measuring the
optical density at a wavelength of 260 nm. RT was then immediately
performed using Prime Script TM Master Mix (Takara Bio, Inc., Otsu,
Japan) according to the manufacturer's protocol. Subsequently,
RT-qPCR was performed with SYBR® Premix EX Taq TM II
(Takara Bio, Inc.) on the real-time PCR detection system ABI7900
(Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH was
used as the internal reference, and gene mRNA expression levels
were calculated using the 2−ΔΔCq method (19). The thermocycling protocol was
conducted as follows: Initial denaturation at 95°C for 5 min,
followed by 45 cycles of a three-step cycling program consisting of
10 sec at 95°C (denaturation), 10 sec at 60°C (primer annealing)
and 10 sec at 72°C (elongation), and a final extension step for 10
min at 72°C. The primer sequences used for qPCR were as follows:
TSLNC8 forward, 5′-TGATCCTCATAGTATAATG-3′ and reverse,
5′-AGTTCTTTAGCAGTACATG-3′; GAPDH forward, 5′-GTGGACATCCGCAAAGAC-3′
and reverse, 5′-AAAGGGTGTAACGCAACTA-3′.
Cell proliferation analysis
Cell viability was determined via an MTT assay. A
total of 1,000 U251-MG and SWO38 cells were transfected with
TSLNC8-overexpressing plasmid and SHG-44 and BT325 cells were
treated with siRNAs with or without TSLNC8 knockdown for 48 h,
trypsinized and reseeded in triplicate in 96-well plates at an
initial density of 4,000 cells per well. Cell numbers were
monitored for a total of 5 consecutive days. At each indicated time
points (days 1, 2, 3, 4 and 5), cell cultures were incubated with
10 µl MTT solution (5 mg/ml, Beyotime Institute of Biotechnology,
Haimen, China) per well for 2 h at room temperature. The absorbance
was recorded at a wavelength of 570 nm. Cell viability was defined
as the cell number ratio of experimental groups to control
cells.
Transwell assay
For cell migration assays, U251-MG and SWO38 cells
were transfected with TSLNC8-overexpressing plasmid for 48 h and
then trypsinized and collected by low-speed centrifugation (1,000 ×
g, 4°C for 5 min) with serum-free DMEM. A total of 1×104
cells (~150 µl) were spread into the upper chamber. The lower
chamber was filled with 600 µl DMEM supplemented with 10% FBS.
Subsequently, the plate was incubated for 24 h at 37°C in an
incubator and the cells were allowed to grow freely. At 24 h
post-seeding, the membrane was fixed with pre-cooled methanol at
room temperature for 10 min and stained with crystal violet (1%)
for 5 min at room temperature. Cell migration was assessed by
counting the cells that had migrated through the membrane. A total
of 5 random fields were selected and images captured under a Nikon
light microscope (Nikon Corporation, Tokyo, Japan) at a
magnification of ×100. For cell invasion assays, the membrane was
pre-coated with Matrigel (Corning Incorporated, Corning, NY, USA)
for 6 h in a 37°C incubator.
Wound-healing assay
U251-MG and SWO38 cells were transfected with
TSLNC8-overexpressing plasmid for 48 h and were then cultured in
DMEM in a 6-well culture plate at a density of 5×105
cells/well overnight until 90% confluence was attained. The culture
medium was replaced with serum-free DMEM. A line was scratched in
the single cell layer using a 10 µl pipette tip and the cells were
then washed with PBS three times. Following incubation at 37°C for
24 h, images of the migrated cells were observed and images were
captured using a Nikon light microscope (magnification, ×200).
Cell apoptosis
The Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) assay was performed according to the
manufacturer's protocol (Invitrogen; Thermo Fisher Scientific,
Inc.). Briefly, a total of 10,000 U251-MG and SWO38 cells were
plated into 6-well plates and transfected with
TSLNC8-overexpressing plasmid for 48 h; SHG-44 and BT325 cells were
transfected with control or specific siRNA against TSLNC8.
Subsequently, cells were washed with pre-cooled PBS, trypsinized,
and re-suspended in 100 µl of binding buffer with 2.5 µl
FITC-conjugated Annexin V and 1 µl PI (100 µg/ml). Cells were then
incubated at room temperature for 15 min in darkness. A total of
≥10,000 cells were collected and analyzed.
Relative caspase activities
The activities of caspase-3, −8 and −9 were
determined using the caspase activity kits (Beyotime Institute of
Biotechnology, Nantong, China) according to the manufacturer's
protocols. Briefly, U251-MG and SWO38 cells were plated into 6-well
plates and transfected with TSLNC8-overexpressing plasmid for 48 h;
SHG-44 and BT325 cells were transfected with control or specific
siRNA against TSLNC8. Subsequently, cell lysates were collected by
low speed centrifugation (1,000 × g, for 5 min at 4°C). A total of
10 µl protein from each sample were added into 96-well plates and
mixed with an aliquot of 80 µl reaction buffer supplied with
caspase substrates (2 mM). Following incubation at 37°C for 4 h,
caspase activities were determined with a Tecan reader (Tecan
Group, Ltd, Mannedorf, Switzerland) at an absorbance of 450 nm.
Statistical analysis
GraphPad Prism version 5.0 (GraphPad Software, La
Jolla, CA, USA) software was used for statistical analysis. Data
are presented as the mean ± standard deviation. The two-tailed
Student's t-test was used to compare means of two groups, while
one-way analysis of variance was used for comparisons among
multiple groups (≥3 groups), followed by a Least Significant
Difference post hoc test. The Spearman correlation analysis was
used to evaluate the experimental data in Table I. P<0.05 was considered to
indicate a statistically significant difference. Each experiment
was repeated at least three times.
 | Table I.Association of TSLNC8 with clinical
variables among 80 glioma patients. |
Table I.
Association of TSLNC8 with clinical
variables among 80 glioma patients.
|
|
| Expression of
TSLNC8 |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | N | Lowd (n=43) | Highd (n=37) | P-value | Coefficient
(R2) |
|---|
| Age (years) |
|
|
| 0.245 | −0.132 |
|
<40 | 16 | 6 | 10 |
|
|
|
40-50 | 24 | 14 | 10 |
|
|
|
>50 | 40 | 23 | 17 |
|
|
| Sex |
|
|
| 0.960 | −0.006 |
|
Male | 43 | 23 | 20 |
|
|
|
Female | 37 | 20 | 17 |
|
|
| Tumor size (T) |
|
|
| 0.001c | −0.373 |
| T1 and
T2a | 38 | 13 | 25 |
|
|
| T3 and
T4b | 42 | 30 | 12 |
|
|
| Lymph node
metastasis (N) |
|
|
| 0.155 | −0.160 |
| N0 | 28 | 12 | 16 |
|
|
| N1 or
above | 52 | 31 | 21 |
|
|
| Distant metastasis
(M) |
|
|
| 0.004c | −0.317 |
| M0 | 32 | 11 | 21 |
|
|
| M1 | 48 | 32 | 16 |
|
|
| TNM stage |
|
|
| 0.003c | −0.323 |
|
I/II | 24 | 7 | 17 |
|
|
|
III/IV | 56 | 36 | 20 |
|
|
Results
LncRNA TSLNC8 is downregulated in
human glioma in vivo and in vitro
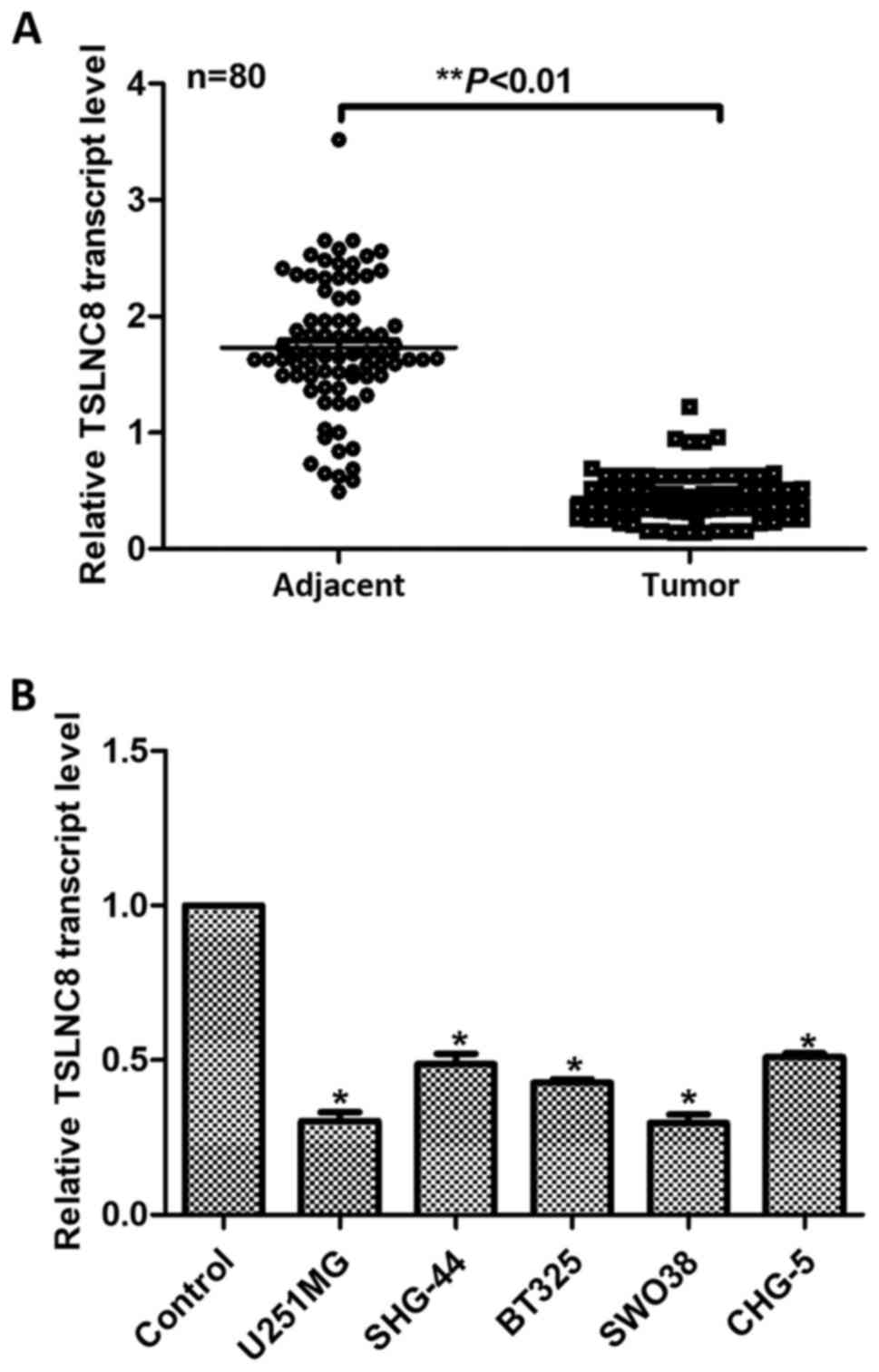
First, the relative transcript levels of TSLNC8 in
80 glioma patients were analyzed in the present study. As presented
in Fig. 1A, the expression of
TSLNC8 was significantly decreased in 95% cases of glioma patients
(76 cases) compared with in adjacent noncancerous tissues
(P<0.01). Of note, TSLNC8 transcript levels higher than the
median level were characterized as high TSLNC8 expression (n=37);
transcript levels lower than the median level were characterized as
low TSLNC8 expression (n=43). The clinical data were also analyzed
and presented in Table I, which
indicated that the relative transcript levels of TSLNC8 were
negatively associated with tumor size (P=0.001), distant metastasis
(P=0.004) and TNM stage (P=0.003), and not associated with age
(P=0.2445), sex (P=0.960) and lymph node metastasis (P=0.155).
Then, five glioma cell lines, including U251-MG, SHG-44, BT325,
SWO38, CHG-5 and normal astrocyte cell line used as a control were
analyzed. RT-qPCR analysis revealed that all of the five glioma
cell lines exhibited significantly lower transcript levels of
TSLNC8 compared with in the control astrocyte cells (Fig. 1B), of which U251-MG and SWO38
demonstrated the lowest expression levels of TSLNC8. Additionally,
U251-MG and SWO38 cells exhibited the highest potential to migrate;
SHG-44 and BT325 revealed the highest expression of TSLNC8 of the 7
glioma cell lines and the migration capacities of these cell lines
were the lowest (Fig. 1B).
Therefore, U251-MG and SWO38 were selected for the knockdown
experiments and SHG-44 and BT325 were used for overexpression
studies. All of these data showed that the transcript level of
TSLNC8 was decreased in human glioma in vivo and in
vitro.
Transcript levels of TSLNC8 are
negatively associated with cell proliferation rate in human
glioma
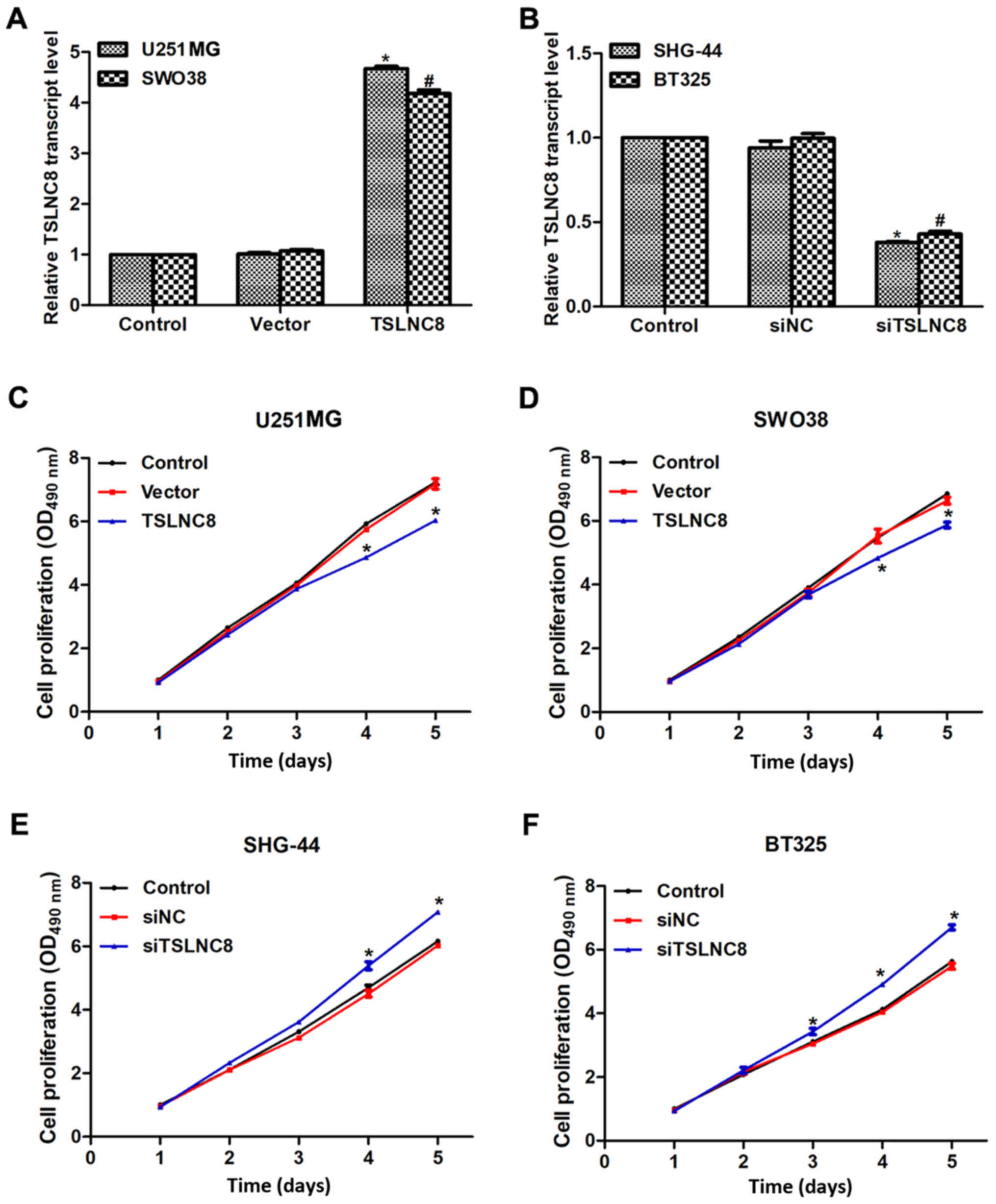
To investigate the role of TSLNC8 in human glioma,
TSLNC8 was overexpressed in U251-MG and SWO38 cells and
downregulated in SHG-44 and BT325 cells using an overexpression
plasmid or siRNAs, respectively. When U251-MG and SWO38 cells were
transfected with TSLNC8-overexpression plasmid, the transcript
levels of TSLNC8 were significantly increased by 4.5- and 4-fold,
respectively (Fig. 2A). In
addition, the expression levels of TSLNC8 in SHG-44 and BT325 cells
were significantly decreased by >50% of the original levels upon
transfection with siTSLNC8 (Fig.
2B). Subsequently, the proliferation rate of the 4 glioma cell
lines was analyzed. No significant alterations in the first 2 days
in experimental cells were observed; however, cell proliferation
decreased by 17% on day 4 and 19% on day 5 of U251 cells
transfected with TSLNC8-expressing plasmid (Fig. 2C). Furthermore, SWO38 cell
proliferation rate decreased on days 4 and 5 with TSLNC8
overexpression (Fig. 2D); the cell
proliferation rate increased on days 4 and 5 in SHG-44 and BT325
cells transfected with siRNA against TSLNC8 (Fig. 2E and F, respectively). These
results suggested that TSLNC8 may suppress cell proliferation in
human glioma cells.
Transcript levels of TSLNC8 are
negatively associated with cell metastasis in human glioma
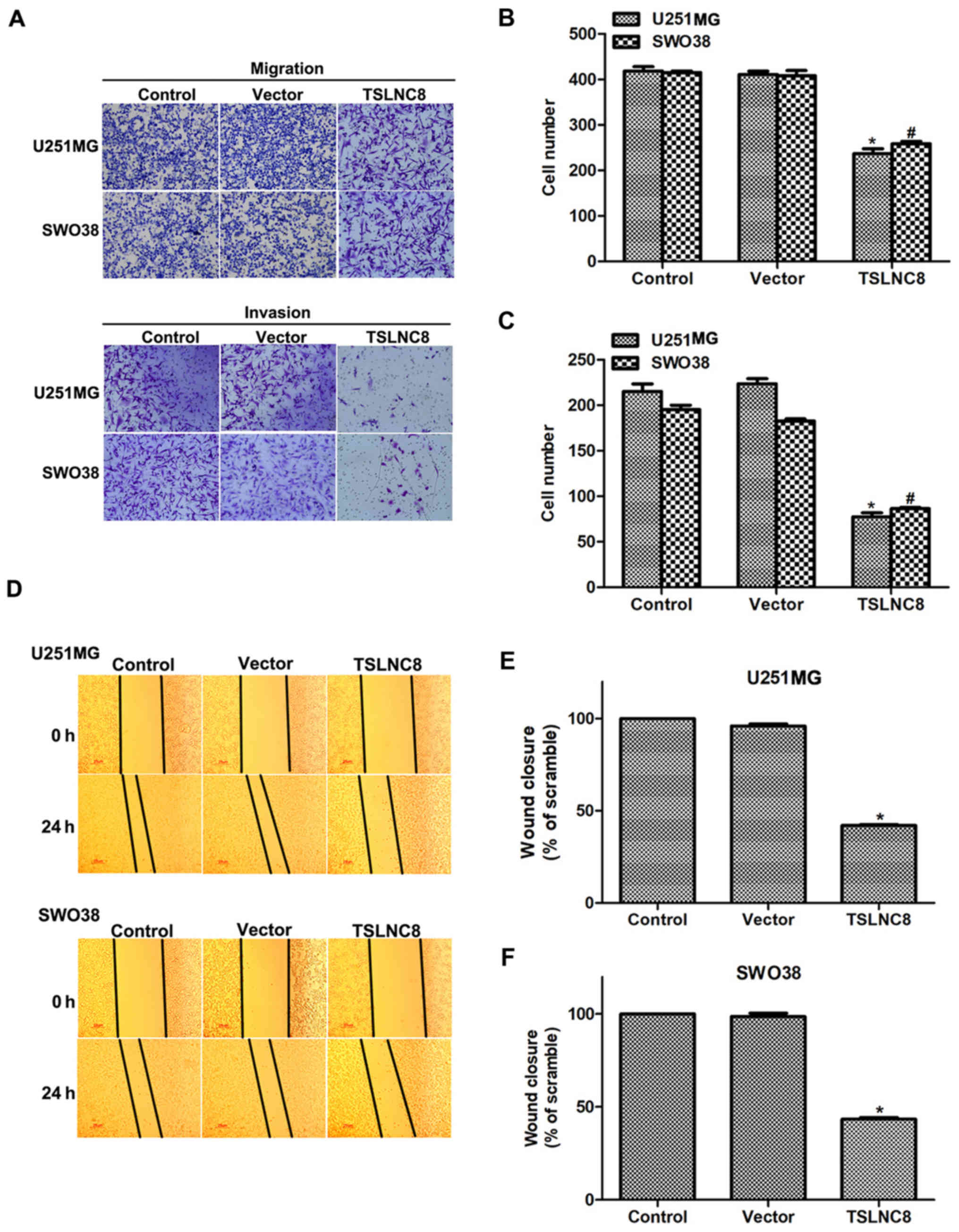
As well as cell proliferation rate, the effects of
TSLNC8 on cell metastasis were investigated. To this end,
TSLNC8-expressing plasmid was transfected into U251-MG and SWO38
cells for 48 h. Transwell assays revealed that >400 U251 and
SWO38 cells were observed to migrate via the membrane; however,
only ~200 cells were counted on the lower side of the membrane upon
transfection with TSLNC8-expressing plasmid (Fig. 3A and B). Additionally, cell
invasion was also inhibited by TSLNC8 overexpression in both
U251-MG and SWO38 cells (Fig. 3A and
C). Subsequently, wound-healing assays were also performed in
U251-MG and SWO38 cells. As presented in Fig. 3D and E, the wound closure ability
of U251-MG cells was inhibited by >50% when cells were treated
with TSLNC8-overexpression plasmid. Furthermore, the wound closure
ability of SWO38 cells was also inhibited by 50% upon TSLNC8
overexpression (Fig. 3D and F).
These data suggested that TSLNC8 suppressed cell metastasis in
human glioma.
Transcript levels of TSLNC8 are
positively associated with cell apoptosis in human glioma
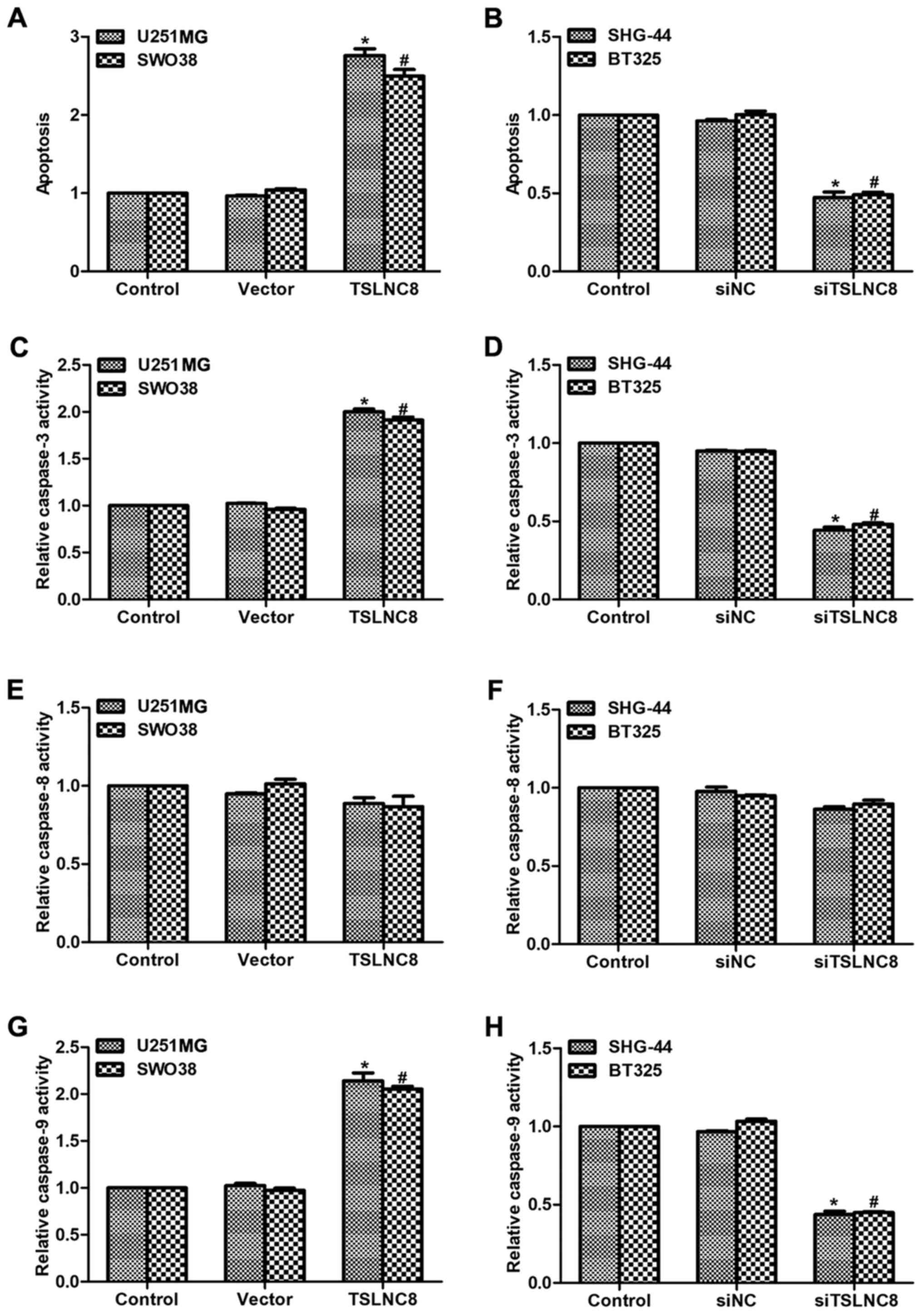
Finally, cell apoptosis was assessed in glioma
cells. As presented in Fig. 4A,
cell apoptosis was increased by 2.7-fold in U251-MG cells and
2.5-fold in SWO38 cells of the TSLNC8 treated groups compared with
in the control cells; the cell apoptotic rate of SHG-44 and BT325
cells were significantly decreased by >50% upon siTSLNC8
stimulation (Fig. 4B).
Subsequently, the relative caspase activities were analyzed.
Treatment with TSLNC8-expressing plasmid of U251-MG and SWO38 cells
increased, while stimulation of siTSLNC8 in SHG-44 and BT325 cells
decreased the activities of caspase-3, respectively (Fig. 4C and D). Interestingly, the
activity of caspase-8 remained stable upon transfection with
TSLNC8-expressing plasmid or siTSLNC8 in glioma cells (Fig. 4E and F). The relative activities of
caspase-9 were also significantly altered, following similar trends
to those exhibited by caspase-3, in the different glioma cell lines
(Fig. 4G and H). As caspase-3 and
−9 are key factors involved in the intrinsic apoptosis pathway and
caspase-8 serves significant roles in the extrinsic apoptotic
pathway (20), the findings of the
present study suggested that TSLNC8 may promote cell apoptosis via
the intrinsic pathway in human glioma.
Discussion
Deletion in the short arm of chromosome 8 is among
the most common genetic events in a variety of cancers, and
deletions include Rho guanosine 5′-triphosphate-ase activating
protein (8p22) (21), leucine
zipper tumor suppress 1 (8p21) (22) and tumor necrosis factor superfamily
member 10c (8p21) (23). The
present study demonstrated that a novel lncRNA TSLNC8, also located
at 8p12, may serve a significant role in human glioma, which was
consistent with the former findings of Zhang et al (17). The results of the present study
suggested that TSLNC8 was downregulated in human glioma, which was
associated with increased cell proliferative rate and cell
metastasis, as well as the decreased cell apoptotic capacity in
human glioma cell lines; TSLNC8 may be a potential therapeutic
target for cancer patients in clinic.
Cell proliferation and cell metastasis are two
primary manifestations in the majority of malignancies (24,25).
Initiation of metastasis requires invasion, which was examined in
the present study via Transwell and wound-healing assays. Invasion
is enabled by epithelial-mesenchymal transition (EMT) (26,27).
Cells from primary tumors lose cell-cell adhesion mediated by
E-cadherin repression and break through the basement membrane with
increased invasive capacity, thus enter the bloodstream via
intravasation (28). At novel
metastatic sites, tumor cells undergo mesenchymal-epithelial
transition by overexpressing N-cadherin (28); the protein levels of E-cadherin and
N-cadherin require further investigation in future studies, as well
as the detailed mechanism of the regulatory effects of TSLNC8 on
human glioma.
Numerous factors are involved in EMT, including
programmed death-ligand 1, twist-related protein 1 and 2, and
transforming growth factor β1 (TGFβ1). TGFβ1 may promote tumor
invasion and evasion of immune surveillance at the advanced stage
of malignancies (29). EMT is
favored and apoptosis is suppressed when TGFβ1 acts on activated
Ras-expressing mammary epithelial cells, which may be reversed by
the inducers of epithelial differentiation (30). As the inhibition of cell apoptosis
is a good basis for tumor cell growth, the role of TSLNC8 in cell
apoptosis was analyzed in the present study. TSLNC8 was observed to
promote cell apoptosis by increasing the activities of caspase-3
and caspase-9 in the intrinsic pathway in the present study. The
mitochondria-mediated intrinsic pathway is dependent on the release
of cytochrome c, which leads to the caspase-9-dependent
activation of caspase-3 (31).
Conversely, the death receptor-induced extrinsic pathway signals in
a caspase-8 dependent manner (31). The overexpression of TSLNC8
promoted, whereas knockdown of TSLNC8 inhibited the activities of
caspase-3 and caspase-9, but not caspase-8 in the present study.
However, the detailed mechanisms of the promoting effects of TSLNC8
on cell apoptosis in human glioma require further investigation in
the future.
In conclusion, the present study revealed the
importance of a novel lncRNA, TSLNC8, in human glioma. TSLNC8 was
demonstrated to inhibit cell proliferation and metastasis, and
promote cell apoptosis in glioma cells. These findings indicated
that TSLNC8 may serve as a potential therapeutic target for the
diagnosis and treatment of glioma in clinic.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
DC performed the experiments. XY designed the study,
analyzed the data and wrote the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Tianjin First Center Hospital. Each patient provided
informed consent to participate.
Patient consent for publication
Written informed consent was obtained.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Taylor LP: Diagnosis, treatment, and
prognosis of glioma: Five new things. Neurology. 75 18 Suppl
1:S28–S32. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gourlay J, Morokoff AP, Luwor RB, Zhu HJ,
Kaye AH and Stylli SS: The emergent role of exosomes in glioma. J
Clin Neurosci. 35:13–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Erdem-Eraslan L, Gravendeel LA, de Rooi J,
Eilers PH, Idbaih A, Spliet WG, den Dunnen WF, Teepen JL, Wesseling
P, Smitt Sillevis PA, et al: Intrinsic molecular subtypes of glioma
are prognostic and predict benefit from adjuvant procarbazine,
lomustine, and vincristine chemotherapy in combination with other
prognostic factors in anaplastic oligodendroglial brain tumors: A
report from EORTC study 26951. J Clin Oncol. 31:328–336. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2006. Neuro Oncol. 15
Suppl 2:ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sukumari-Ramesh S, Prasad N, Alleyne CH,
Vender JR and Dhandapani KM: Overexpression of Nrf2 attenuates
Carmustine-induced cytotoxicity in U87MG human glioma cells. BMC
Cancer. 15:1182015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu B, Emdad L, Bacolod MD, Kegelman TP,
Shen XN, Alzubi MA, Das SK, Sarkar D and Fisher PB: Astrocyte
elevated gene-1 interacts with Akt isoform 2 to control glioma
growth, survival, and pathogenesis. Cancer Res. 74:7321–7332. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Steeg PS, Camphausen KA and Smith QR:
Brain metastases as preventive and therapeutic targets. Nat Rev
Cancer. 11:352–363. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hegi ME, Diserens AC, Gorlia T, Hamou MF,
de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani
L, et al: MGMT gene silencing and benefit from temozolomide in
glioblastoma. N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reardon DA, Rich JN, Friedman HS and
Bigner DD: Recent advances in the treatment of malignant
astrocytoma. J Clin Oncol. 24:1253–1265. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chu C, Spitale RC and Chang HY:
Technologies to probe functions and mechanisms of long noncoding
RNAs. Nat Struct Mol Biol. 22:29–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Misawa A, Takayama KI and Inoue S: Long
non-coding RNAs and prostate cancer. Cancer Sci. 108:2107–2114.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li J, Meng H, Bai Y and Wang K: Regulation
of lncRNA and its role in cancer metastasis. Oncol Res. 23:205–217.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ramos AD, Attenello FJ and Lim DA:
Uncovering the roles of long noncoding RNAs in neural development
and glioma progression. Neurosci Lett. 625:70–79. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma B, Gao Z, Lou J, Zhang H, Yuan Z, Wu Q,
Li X and Zhang B: Long noncoding RNA MEG3 contributes to
cisplatininduced apoptosis via inhibition of autophagy in human
glioma cells. Mol Med Rep. 16:2946–2952. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H, Li Q, Guo T, He W, Dong C and Wang
Y: LncRNA CRNDE triggers inflammation through the
TLR3-NF-κB-Cytokine signaling pathway. Tumour Biol.
39:10104283177038212017.PubMed/NCBI
|
|
17
|
Zhang J, Li Z, Liu L, Wang Q, Li S, Chen
D, Hu Z, Yu T, Ding J, Li J, et al: Long noncoding RNA TSLNC8 is a
tumor suppressor that inactivates the interleukin-6/STAT3 signaling
pathway. Hepatology. 67:171–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang MH, Lin CL, Zhang JJ, Weng ZP, Hu T,
Xie Q and Zhong XY: Role of PTEN in cholera toxin-induced SWO38
glioma cell differentiation. Mol Med Rep. 7:1912–1918. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kerr JF, Wyllie AH and Currie AR:
Apoptosis: A basic biological phenomenon with wide-ranging
implications in tissue kinetics. Br J Cancer. 26:239–257. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zimonjic DB and Popescu NC: Role of DLC1
tumor suppressor gene and MYC oncogene in pathogenesis of human
hepatocellular carcinoma: Potential prospects for combined targeted
therapeutics (review). Int J Oncol. 41:393–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He Y and Liu X: The tumor-suppressor gene
LZTS1 suppresses hepatocellular carcinoma proliferation by
impairing PI3K/Akt pathway. Biomed Pharmacother. 76:141–146. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ganten TM, Haas TL, Sykora J, Stahl H,
Sprick MR, Fas SC, Krueger A, Weigand MA, Grosse-Wilde A, Stremmel
W, et al: Enhanced caspase-8 recruitment to and activation at the
DISC is critical for sensitisation of human hepatocellular
carcinoma cells to TRAIL-induced apoptosis by chemotherapeutic
drugs. Cell Death Differ. 11 Suppl 1:S86–S96. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Islam T and Resat H: Quantitative
investigation of MDA-MB-231 breast cancer cell motility: Dependence
on epidermal growth factor concentration and its gradient. Mol
Biosyst. 13:2069–2082. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Geng R, Tan X, Wu J, Pan Z, Yi M, Shi W,
Liu R, Yao C, Wang G, Lin J, et al: RNF183 promotes proliferation
and metastasis of colorectal cancer cells via activation of
NF-κB-IL-8 axis. Cell Death Dis. 8:e29942017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Massagué J: TGFbeta in cancer. Cell.
134:215–230. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chu IM, Lai WC, Aprelikova O, El Touny LH,
Kouros-Mehr H and Green JE: Expression of GATA3 in MDA-MB-231
triple-negative breast cancer cells induces a growth inhibitory
response to TGFß. PLoS One. 8:e611252013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weller M and Fontana A: The failure of
current immunotherapy for malignant glioma. Tumor-derived TGF-beta,
T-cell apoptosis, and the immune privilege of the brain. Brain Res
Brain Res Rev. 21:128–151. 1995. View Article : Google Scholar : PubMed/NCBI
|