Introduction
Immediate-early genes encode a type of polypeptides
that serve a significant role in cell regulation and the response
of the cell to external stimuli. The regulation of the cell cycle
by these polypeptides differs among numerous types of cells, due to
variations in expression. The response of some immediate-early gene
family members to extracellular stimuli is characterized by slow
kinetics, which delays transcription and prolongs protein half-life
(1). Immediate early response 5
(IER5) was initially reported by Williams et al (1), and is a member of the slow-kinetics
immediate-early gene family. IER5 is a gene without introns
comprising 2,350 nucleotides and is located in 1q25.3. The
predicted open reading frame encodes a 327-amino-acid protein. Its
amino terminus is rich in proline residues as previously noted for
other homology immediate-early genes, such as
pip92/IER2/ETR101. In contrast to pip92/IER2/ETR101,
the transcriptional activation of IER5 does not require
induction by phosphokinase C (2).
It has been revealed that IER5 expression was
upregulated following external stimuli, inducing cell apoptosis.
Therefore, it may be considered that IER5 is involved in the
regulation of the cell cycle. Savitz et al (3), demonstrated that the expression
levels of IER5 were increased in peripheral mononuclear
cells derived from patients with depression and mood disorders
compared with those of healthy subjects. Ishikawa et al
(4,5) and Asano et al (6) noted that IER5 was a positive
feedback regulator of heat shock factor 1 (HSF1) dephosphorylation,
following investigation of the mechanism of HSF1
transcription. In addition, Li et al (7), Kawabata et al (8) and Nakamura et al (9), also observed the cell cycle
regulation by IER5 during the progression of different
diseases. Our research group has demonstrated that radiation can
induce upregulation of IER5 in tumor cells (10,11)
and this process can modulate the transcription of cell division
cycle (CDC)25B by competitively binding to the
CDC25B promoter (12).
Additionally, we reported that decreased IER5 expression could
increase the population of cancer cells in the G2/M
phase of the cell cycle (13,14),
and that binding of a novel transcription factor and GC binding
factor (GCF) to the IER5 promoter could act as a negative
regulator of IER5 transcriptional activity (15). Furthermore, we proposed that
decreased IER5 expression significantly lowered the efficiency of
DNA double strand break repair in HeLa cells induced by ionizing
radiation (16).
Although various studies have been conducted on the
functional mechanism of IER5, only a limited number has
explored the structure of the IER5 protein. The present study aimed
to determine the structure of the IER5 protein by bioinformatics
analysis, and to explain its in vivo function and mechanism
of action, based on its structural features, so as to provide a
theoretical basis for subsequent experimental determination of its
structure.
Materials and methods
Sequence of the IER5 gene and
protein
The nucleotide sequences of 2,000 bp upstream and
1,000 bp downstream of the transcription sites of the IER5
gene (Species, Homo sapiens, accession: NC_000001.11, Gene
ID: 51278) were downloaded from the National Center for
Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/), and the amino acid
sequences of the IER5 protein (Entry: Q5VY09, Entry name:
IER5_HUMAN, Length: 327-amino-acid) were downloaded from the
UniProt database (https://www.uniprot.org/).
Prediction of the IER5 gene
sequence
The online software Promoter Scan (https://www-bimas.cit.nih.gov/molbio/proscan/; website
decommissioned March 8, 2019) was applied for the prediction of the
promoter sequence and the binding sites of the related
transcription factors. The program, which recognized ~70% of
primate promoter sequences, predicted promoter regions based on
scoring homologies with putative eukaryotic Pol II promoter
sequences. The Methprimer (http://www.urogene.org/methprimer/) was applied for
the determination of methylation sites and CpG islands at the
promoter region. The criteria for the CpG island prediction results
were the following: Island size >100, GC% (percentage of G plus
C) >50% and observed/expected values >0.5. Gene Ontology (GO)
and annotations were investigated using the GO Enrichment Analysis
using the AmiGO tool (http://amigo.geneontology.org/amigo/landing). GO
enrichment analysis identified relevant groups of genes that
functioned collectively, which reduced the thousands of molecular
changes to notably fewer biological functions in order to describe
a putative function corresponding to the mean number of molecular
changes.
Prediction of IER5 protein
features
The physical and chemical properties of the IER5
protein were predicted by the ProtParam tool (https://web.expasy.org/protparam/). The protein
parameters, including the molecular weight, theoretical isoelectric
point, amino acid composition, atomic composition, extinction
coefficient and instability index were calculated based on either
compositional data or on the N-terminal amino acid residues. The
hydrophobicity/hydrophilicity of the IER5 protein was predicted by
the ProtScale (https://web.expasy.org/protscale/) which provided 57
scales defined by a numerical value assigned to each type of amino
acid. The most frequently used scales were the hydrophobicity or
hydrophilicity scales and the secondary structure conformational
parameters scales. The O-glycosylation sites were predicted by a
genetic engineering approach. The NetOGlyc 4.0 Server (http://www.cbs.dtu.dk/services/NetOGlyc/) was used
enable a proteome-wide discovery approach of O-glycan sites by a
‘bottom-up’ ETD-based mass spectrometric analysis. The
N-glycosylation sites of the IER5 protein were predicted by the
NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) which was
based on an artificial neural network in an attempt to discriminate
between glycosylated and non-glycosylated sequences. The
phosphorylation sites of the IER5 protein were predicted by the
NetPhos 3.1 Server (http://www.cbs.dtu.dk/services/NetPhos/) which
predicted serine, threonine and tyrosine phosphorylation sites in
eukaryotic proteins using ensembles of neural networks and 17
kinases as follows: Ataxia telangiectasia-mutated, casein kinase
(CK)I, CKII, calmodulin-dependent protein kinase-II, DNA-dependent
protein kinase catalytic subunit, epidermal growth factor receptor,
glycogen synthase kinase (GSK)3, insulin receptor (INSR), protein
kinase A (PKA), protein kinase B, protein kinase C (PKC),
cGMP-dependent protein kinase, ribosomal S6 kinase, SRC,
cyclin-dependent kinase 1 (cdc2), cyclin-dependent kinase 5 (cdk5)
and p38 mitogen-activated kinase (MAPK). The subcellular
localization of the IER5 protein was predicted by the PSORT II tool
(https://psort.hgc.jp/form2.html) from
its amino acid sequences. The location of the transmembrane,
intracellular and extracellular regions was predicted by the TMHMM
Server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) by reading a
FASTA-format protein sequence. The presence and location of signal
peptide cleavage sites in the amino acid sequences were predicted
by the SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/) with a
D-cutoff score of 0.5. The nuclear localization sequence (NLS) of
the IER5 protein was predicted by the NLS mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi)
with a cut-off score of 4.0. The secondary and tertiary structures
of the IER5 protein were predicted by the PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) and I-TASSER
(https://zhanglab.ccmb.med.umich.edu/I-TASSER/)
software, respectively. These two methods used a known amino acid
sequence to match a template in a protein database. All-by-all TM
scores were calculated for the full set of putative templates and
the matrix of scores was analyzed to remove any possible outlying
templates whose structure was too dissimilar with that of the full
set of templates. The tertiary structures of the IER5 protein was
compiled and produced by PyMOL (https://pymol.org).
Results
Promoter binding and methylation
analysis of the IER5 gene
The sequences located at 2,000 bp upstream and 1,000
bp downstream of the transcription sites of IER5 were
analyzed, and the promoter was located at a region between 1,722
and 1,972 bp in the plus-strand (Table
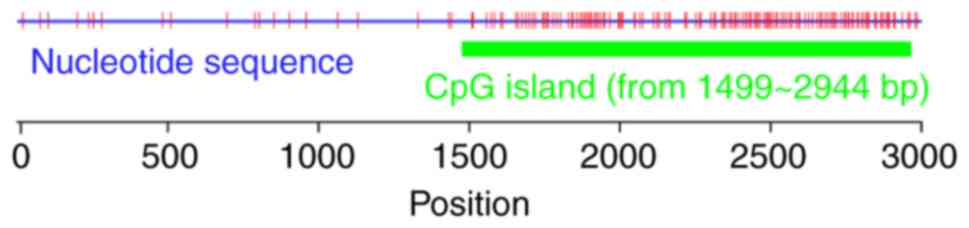
I). We identified one CpG island located at a region between
1,499 and 2,944 bp and several potential methylation sites
(Fig. 1). In addition, the
predicted promoter sequence of IER5 overlapped with the CpG
island. We further examined the transcription factors sites close
to the promoter region and identified specific transcription
factors sites associated with methylation, such as the AP-2
(Table II).
 | Table I.Promoter prediction of immediate
early response 5 gene. |
Table I.
Promoter prediction of immediate
early response 5 gene.
| Promoter | Start | End | Score | Sequence |
|---|
| Promoter 1 | 1,722 | 1,972 | 72.54 |
AATCTGTAACTCCAAACAGTAGCGCTCTCGAGCACCGTCCCGAACATTCACTCCCCACGGGCCTGGTCTGCGGCCGCAAGCCGTCGCCCCCTTTAAGAGCCTGCTCCGCGGGACTAACGTTCGAACGGGCCCTGGCGCCCCTCCTCGGGCTCCGATTGGCCGTGCGCGGCGCACGAGGGCGCGCCGGCCAGCCCCGGAACGGTTGGCGGCCCTTCGTGATTGGCGGTCGAGAAGCCTATATAAGGCGCGGG |
 | Table II.Transcription factor sites about the
promoter of immediate early response 5. |
Table II.
Transcription factor sites about the
promoter of immediate early response 5.
| Name | Strand | Location | Weight |
|---|
| AP-2 | + | 1773 | 1.355 |
| AP-2 | + | 1774 | 1.108 |
| UCE.2 | + | 1793 | 1.278 |
| UCE.2 | − | 1796 | 1.216 |
| GCF | + | 1856 | 2.361 |
| CTF | + | 1875 | 1.704 |
| UCE.2 | + | 1879 | 1.278 |
| NFI | − | 1881 | 4.221 |
| junB-US2 | − | 1882 | 1.51 |
|
TTR_inverted_repeat | − | 1892 | 3.442 |
| GCF | − | 1893 | 2.284 |
| UCE.2 | − | 1910 | 1.216 |
| AP-2 | − | 1930 | 1.672 |
| HNF1 | + | 1931 | 1.012 |
| TFIID | + | 1958 | 1.971 |
| GCF | + | 1968 | 2.361 |
| T-Ag | + | 1970 | 1.086 |
Physical and chemical properties and
hydrophobicity/hydrophilicity of the IER5 protein
A total of ~12.84% (42/327) of the amino acids (Asp
and Glu) were identified with negative charge, whereas 9.48%
(31/327) of the amino acids (Arg and Lys) were identified with
positive charge. The IER5 protein contained 11 Cys residues.
Considering that all Cys residues formed cystines, the estimated
molar extinction coefficient in the aqueous solution was 32,595
M−1 cm−1, whereas for 0.1% absorbance (1 g/1)
would be 0.967; however, providing all Cys residues could not form
cystines, the corresponding value would be 31,970 and 0.949. The
IER5 protein was predicted as an unstable protein, of which the
structural formula was
C1459H2294N428O464S14
and the total molecular weight was estimated to 33703.69. The
theoretical isoelectric point was 4.91, of which the coefficient of
instability was 61.1.
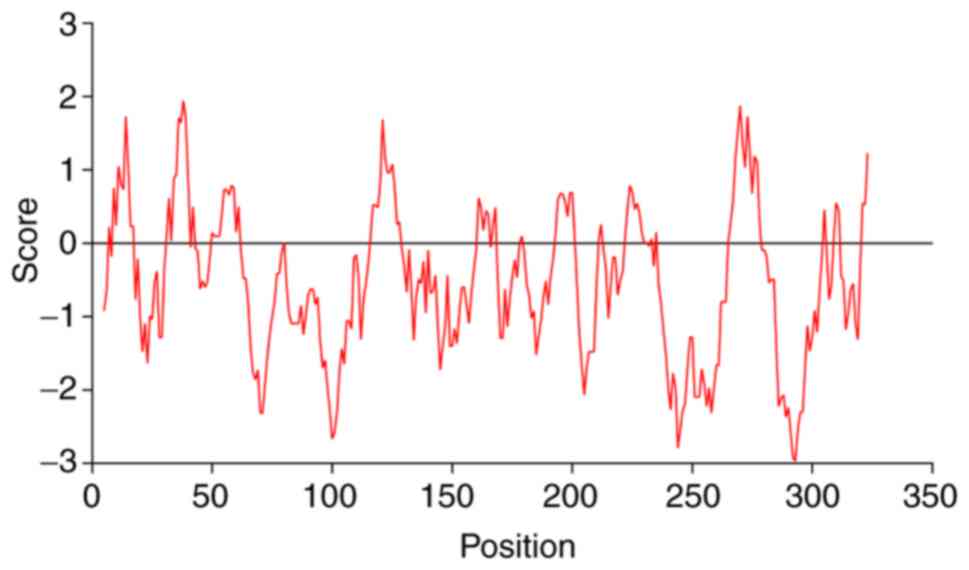
The aliphatic index of the IER5 protein was
estimated to 60.4 and the average hydrophilic value was estimated
to −0.493. Leu, the most hydrophobic amino acid, was identified at
the 38th position and with an index value of 1.944. The most
hydrophilic amino acid was reported at the 293th position and its
index was −2.976. According to the hydrophilic/hydrophobic
distribution diagram (Fig. 2), the
majority of the amino acids were hydrophilic amino acids, and
therefore IER5 was considered a hydrophilic protein.
Posttranslational modification of the
IER5 protein
Protein modifications, such as glycosylation and
phosphorylation are required to fulfill protein physiological
function. Glycosylation serves an important role in the interaction
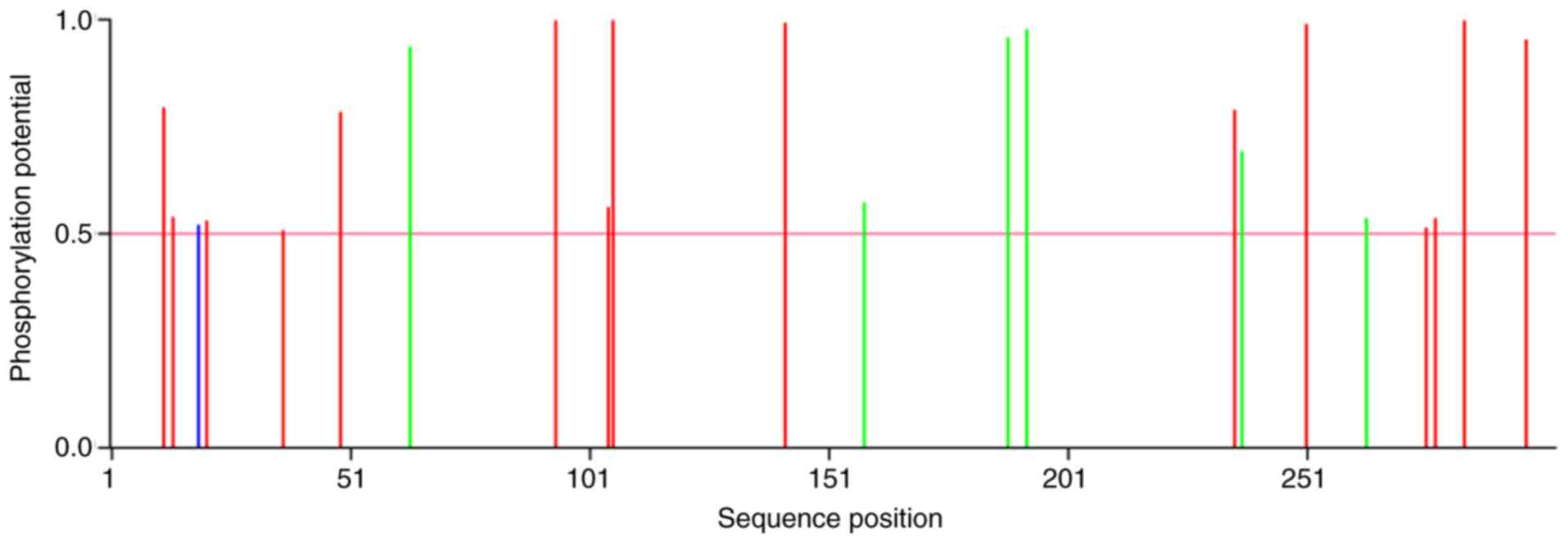
between proteins and other macromolecules (17). A total of 18 O-glycosylation sites
were identified (score >0.5), in the IER5 protein; however, no
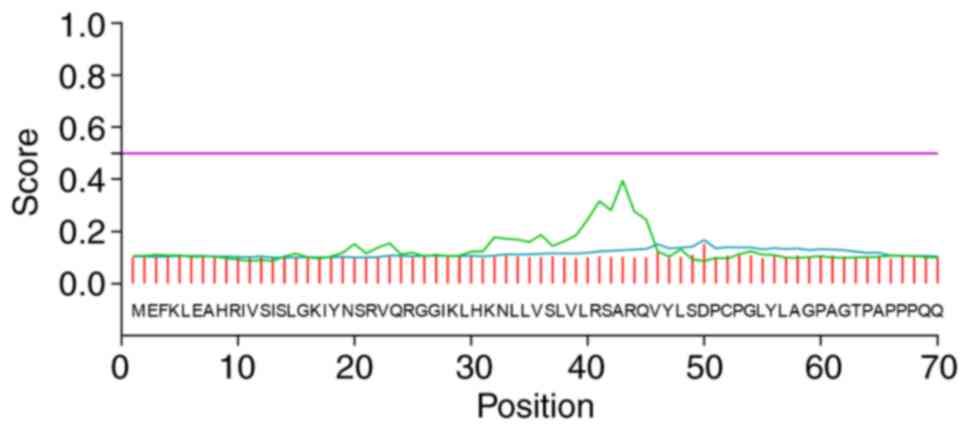
N-glycosylation sites were reported. Phosphorylation is required
for signal transduction (18). The
results indicated 15 serine (Ser), six threonine (Thr) and one
tyrosine (Tyr) protein kinase phosphorylation sites (score >0.5,
Fig. 3). A total of 9 kinases
associated with the phosphorylation reactions, including PKA, cdc2,
INSR, PKC, cdk5, GSK3, p38MAPK, CKII and CKI were reported in
addition to ‘unspecified’ types.
Subcellular localization,
transmembrane structure and signal peptide identification of the
IER5 protein
The prediction of the subcellular localization of
IER5 suggested that the protein exhibited a 56.5% probability of
localizing in the nucleus, whereas the possibility for cytoskeletal
and mitochondrial localization was notably lower (17.4 and 13.0%,
respectively). The prediction indicated a lower potential for
localization in the mitochondria, Golgi apparatus and vesicular
secretion system (4.3%). No transmembrane structures (Fig. 4) and signal peptides (Fig. 5) were present. Further analysis
indicated that the IER5 protein may possess a nuclear localization
sequence (NLS) GSTPLKKPRRNLE (the position in protein sequence from
the N terminal to C terminal is 235–247) with a score of 4.5
(threshold of 5.0).
Secondary structure of the IER5
protein
The secondary structure refers to a periodic
structure arranged along a direction, and is the regular repeated
conformation in the protein polypeptide chain. PSIPRED has been
previously used to predict protein secondary structure based on a
two-stage neural network; the average prediction accuracy was
estimated at a range of 76.5–78.3% (19). The results revealed 6 α-helixes,
but no β-sheet or β-turn motifs. The remaining structural parts of
the proteins were determined to present as disordered coils
(Fig. 6).
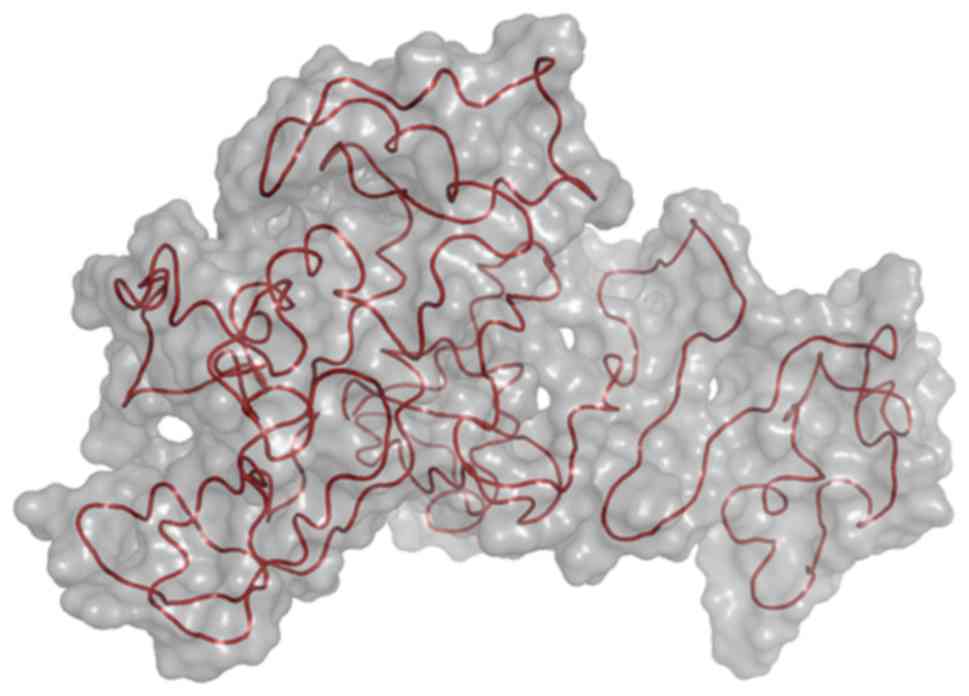
Tertiary structure of the IER5
protein
I-TASSER is an online integrated platform based on
the ‘sequence-structure-function’ model of automatic protein
structure and function prediction. Starting from the amino acid
sequence, the platform produces the three-dimensional atomic-scale
model through the comparison of multiple threading alignment
approaches and the iterative structural assembly simulation
(20,21). The platform presents five models
following the completion of predicting the tertiary structure;
default model 1 is considered as the best model based on
comprehensive analysis of the three parameters, namely the C-score,
the TM score and the RMSD. The tertiary structure was presented in
a cartoon model embedded in the surface mode (Fig. 7).
GO of the IER5 gene
GO is a database established by the GO consortium,
which is a unified induction, interpretation and analysis of the
cytological components, molecular functions and biological
approaches of genes and their products. We searched for GO terms
and annotations associated with IER5 by the AmiGO browser.
The present study reported that this gene was involved in several
primary biological and metabolic processes that require ion and
protein binding. The main distribution of IER5 has not been
predicted in terms of cellular components (Table III).
 | Table III.GO of immediate early response 5. |
Table III.
GO of immediate early response 5.
| Ontology | GO ID | Term |
|---|
| Biological
process | GO:0044238 | Primary metabolic
process |
| Cellular
component | − |
|
| Molecular
function | GO:0043167 | Ion binding |
|
| GO:0042802 | Identical protein
binding |
|
| GO:0005515 | Protein
binding |
Discussion
In the present study, we investigated the structure
and function of IER5, and its encoded protein using
bioinformatics online analysis software. Hypermethylation of CpG
islands at the promoter region has been reported to inhibit the
transcriptional activity of the gene, whereas low promoter
methylation activates gene expression (22,23).
The results of the present study indicated one CpG island and
several potential methylation sites. Liu et al (10) and Shi et al (11) demonstrated that radiation could
upregulate the expression levels of IER5. Therefore, we
proposed that the methylation levels of the wild type IER5 gene may
be low, but could notably increase following radiation exposure,
inducing its expression (24,25).
Specific transcription factors have been located at the promoter
region of IER5 and certain protein binding sites were
suggested by GO analysis. Our previous study reported two GCF
binding sites at the promoter region, which is in agreement with
the present findings (15).
Following the binding of GCF, the transcriptional activity and
radiation sensitivity of IER5 significantly decreased
(15).
Glycosylation serves an important role in cellular
immunity, signal transduction, protein translation regulation and
protein degradation. For example, the majority of transcription
factors and enzymes require glycosylation following translation
(17). In addition,
phosphorylation serves a key role in protein signal transduction,
gene expression and cell cycle regulation (18). The present study reported 18
O-glycosylation sites and 22 phosphorylation sites in the IER5
protein, which reflected the complexity of IER5 protein
function.
Based on the prediction of protein subcellular
localization, transmembrane region and signal peptide
identification, it was speculated that the IER5 protein was mainly
localized in the nucleus. The absence of the transmembrane
structure and of the signal peptide indicated that the IER5 protein
did not require entry into other membrane organelles. Following
protein expression, various hydrophilic structures and lack of the
transmembrane structure and of the signal peptide may facilitate
the free diffusion of the IER5 protein in the cell without its
modification by the endoplasmic reticulum or the Golgi apparatus
(26,27). The IER5 protein may be channeled
from the nuclear pore complex to the nucleus possibly via an
NLS.
The helix-turn-helix domain (HTH) is a relatively
conserved structure with various patterns that correspond to
different protein families (28).
HTH contains two α-helixes, which are connected by one turn, and
can recognize the specific base sequence of the DNA in order to
regulate its transcription, replication and translation (29). Previous studies suggested that
radiation increased the expression levels of the IER5 gene and
protein (10,11). Competitive binding of IER5 to the
Cdc25B promoter led to downregulated expression levels of
Cdc25B (12). We demonstrated that
the secondary structure of IER5 had only 6 α-helixes. Following
structure prediction, it was speculated that IER5 could possess an
HTH structure based on its function. Further experiments are
required to confirm this hypothesis.
Of note, the present study has certain limitations
as only bioinformatics predictions were conducted. Furthermore, we
did not conduct investigations using clinical samples, which may
verify the results reported in the present study.
We examined the features of the IER5 gene and
protein using bioinformatics analyses, which could aid future
investigation of their biological functions. Furthermore,
predicting the IER5 may provide a experimental basis for
investigation into its functions in the future.
Acknowledgements
Not applicable.
Funding
The present work was supported by grants from the
National Natural Science Foundation of China (grant nos. 31170806,
31770907 and 31640022) and the Beijing Natural Science Foundation
(grant no. 7172146).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
QX, XJ, XL, PZ and KD conceived and designed the
study. QX wrote the paper. All authors reviewed and edited the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Williams M, Lyu MS, Yang YL, Lin EP,
Dunbrack R, Birren B, Cunningham J and Hunter K: Ier5, a novel
member of the slow-kinetics immediate-early genes. Genomics.
55:327–334. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takaya T, Kasatani K, Noguchi S and Nikawa
J: Functional analyses of immediate early gene ETR101 expressed in
yeast. Biosci Biotechnol Biochem. 73:1653–1660. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Savitz J, Frank MB, Victor T, Bebak M,
Marino JH, Bellgowan PS, McKinney BA, Bodurka J, Kent Teague T and
Drevets WC: Inflammation and neurological disease-related genes are
differentially expressed in depressed patients with mood disorders
and correlate with morphometric and functional imaging
abnormalities. Brain Behav Immun. 31:161–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ishikawa Y and Sakurai H: Heat-induced
expression of the immediate-early gene IER5 and its involvement in
the proliferation of heat-shocked cells. FEBS J. 282:332–340. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ishikawa Y, Kawabata S and Sakurai H: HSF1
transcriptional activity is modulated by IER5 and PP2A/B55. FEBS
Lett. 589:1150–1155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Asano Y, Kawase T, Okabe A, Tsutsumi S,
Ichikawa H, Tatebe S, Kitabayashi I, Tashiro F, Namiki H, Kondo T,
et al: IER5 generates a novel hypo-phosphorylated active form of
HSF1 and contributes to tumorigenesis. Sci Rep. 6:191742016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li XN, Ji C, Zhou PK and Wu YM:
Establishments of IER5 silence and overexpression cervical cancer
SiHa cell lines and analysis of radiosensitivity. Int J Clin Exp
Pathol. 9:6671–6682. 2016.
|
|
8
|
Kawabata S, Ishita Y, Ishikawa Y and
Sakurai H: Immediate-early response 5 (IER5) interacts with protein
phosphatase 2A and regulates the phosphorylation of ribosomal
protein S6 kinase and heat shock factor 1. FEBS Lett.
589:3679–3685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakamura S, Nagata Y, Tan L, Takemura T,
Shibata K, Fujie M, Fujisawa S, Tanaka Y, Toda M, Makita R, et al:
Transcriptional repression of Cdc25B by IER5 inhibits the
proliferation of leukemic progenitor cells through NF-YB and p300
in acute myeloid leukemia. PLoS One. 6:e280112011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Tian M, Zhao H, He Y, Li F, Li X,
Yu X, Ding K, Zhou P and Wu Y: IER5 as a promising predictive
marker promotes irradiation-induced apoptosis in cervical cancer
tissues from patients undergoing chemoradiotherapy. Oncotarget.
8:36438–36448. 2017.PubMed/NCBI
|
|
11
|
Shi HM, Ding KK, Zhou PK, Guo DM, Chen D,
Li YS, Zhao CL, Zhao CC and Zhang X: Radiation-induced expression
of IER5 is dose-dependent and not associated with the clinical
outcomes of radiotherapy in cervical cancer. Oncol Lett.
11:1309–1314. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang C, Yang M, Feng Z, Liu X, Yin L, Zhou
P and Ding K: Radiation modulated the interaction of IER5 protein
and CDC25B promoter DNA in primary hepatocellular carcinoma. Int J
Clin Exp Pathol. 9:2888–2895. 2016.
|
|
13
|
Yang C, Wang Y, Hao C, Yuan Z, Liu X, Yang
F, Jiang H, Jiang X, Zhou P and Ding K: IER5 promotes irradiation-
and cisplatin-induced apoptosis in human hepatocellular carcinoma
cells. Am J Transl Res. 8:1789–1798. 2016.PubMed/NCBI
|
|
14
|
Ding KK, Shang ZF, Hao C, Xu QZ, Shen JJ,
Yang CJ, Xie YH, Qiao C, Wang Y, Xu LL and Zhou PK: Induced
expression of the IER5 gene by gamma-ray irradiation and its
involvement in cell cycle checkpoint control and survival. Radiat
Environ Biophys. 48:205–213. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang C, Yin L, Zhou P, Liu X, Yang M, Yang
F, Jiang H and Ding K: Transcriptional regulation of IER5 in
response to radiation in HepG2. Cancer Gene Ther. 23:61–65. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu XP, Wu YM, Liu Y, Tian M, Wang JD, Ding
KK, Ma T and Zhou PK: IER5 is involved in DNA double-strand breaks
repair in association with PAPR1 in Hela cells. Int J Med Sci.
14:1292–1300. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ohtsubo K and Marth JD: Glycosylation in
Cellular mechanisms of health and disease. Cell. 126:855–867. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Singh V, Ram M, Kumar R, Prasad R, Roy BK
and Singh KK: Phosphorylation: Implications in cancer. Protein J.
36:1–6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jones DT: Protein secondary structure
prediction based on position-specific scoring matrices. J Mol Biol.
292:195–202. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J and Zhang Y: I-TASSER server: New
development for protein structure and function predictions. Nucleic
Acids Res. 43:W174–W181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y: I-TASSER: Fully automated protein
structure prediction in CASP8. Proteins. 77 (Suppl 9):S100–S113.
2009. View Article : Google Scholar
|
|
22
|
Hashimshony T, Zhang J, Keshet I, Bustin M
and Cedar H: The role of DNA methylation in setting up chromatin
structure during development. Nat Genet. 34:187–192. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Balada E, Ordi-Ros J, Serrano-Acedo S,
Martinez-Lostao L, Rosa-Leyva M and Vilardell-Tarrés M: Transcript
levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in CD4+ T
cells from patients with systemic lupus erythematosus. Immunology.
124:339–347. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Illingworth RS and Bird AP: CpG islands-‘a
rough guide’. FEBS Lett. 583:1713–1720. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eckhardt F, Lewin J, Cortese R, Rakyan VK,
Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al:
DNA methylation profiling of human chromosomes 6, 20 and 22. Nat
Genet. 38:1378–1385. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Palmer KJ, Konkel JE and Stephens DJ:
PCTAIRE protein kinases interact directly with the COPII complex
and modulate secretory cargo transport. J Cell Sci. 118:3839–3847.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bard F, Mazelin L, Péchoux-Longin C,
Malhotra V and Jurdic P: Src regulates golgi structure and KDEL
receptor-dependent retrograde transport to the endoplasmic
reticulum. J Biol Chem. 278:46601–46606. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wintjens R and Rooman M: Structural
classification of HTH DNA-binding domains and protein-DNA
interaction modes. J Mol Biol. 262:294–313. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aravind L and Koonin EV: DNA-binding
proteins and evolution of transcription regulation in the archaea.
Nucleic Acids Res. 27:4658–4670. 1999. View Article : Google Scholar : PubMed/NCBI
|