Introduction
Psoriasis is a common chronic inflammatory skin
disease and affects 0.1–3% of the world population (1). This disease is stubborn, recurrent
and difficult to cure, and seriously affects the quality of life of
patients (2). The immunological
mechanism of the onset of psoriasis is of great concern. Psoriasis
is a chronic inflammatory condition, where psoriatic lesions are
formed through the interactions between keratinocytes and immune
cells. Abnormal human keratinocyte proliferation is an important
feature of psoriasis. The hyperproliferation and abnormal
differentiation of keratinocytes comprise the typical
histopathological basis for the formation of epidermal hypertrophy
and parakeratosis (3).
Typical Notch signaling pathways have been
introduced. Mammals possess four types of Notch receptors (Notch-1,
Notch-2, Notch-3 and Notch-4) and five types of ligands
(Jagged-1/-2 and Delta-like 1, 3 and 4). The Notch signaling
pathway serve an important role in cell differentiation,
proliferation and apoptosis (4),
and it is correlated with inflammation. This pathway is active in
numerous inflammatory diseases, including psoriasis, rheumatoid
arthritis and systemic lupus erythematosus (5,6).
Jagged-1 expression is markedly upregulated in patients with
psoriatic lesions as the binding of the Jagged-1 protein to the
Notch receptor activates the Notch signaling pathway and promotes
epidermal hyperplasia (7).
Jagged-1 protein is a transmembrane protein on the cell surface and
is widely present in different cells of the peripheral immune
system. In particular, this protein serves important roles in the
activation, proliferation and differentiation of peripheral
CD4+ T cells (8).
Therefore, the Jagged-1 protein-mediated Notch signaling pathway
may be involved in the hyperproliferation of psoriatic
keratinocytes and the abnormal activation of the immune system
(9).
MicroRNAs (miRNAs) are single-stranded, non-coding,
short RNA molecules. They are gene expression regulators that
trigger target mRNA degradation and inhibit translation, thereby
regulating biological processes, including cell proliferation,
differentiation, apoptosis and tumorigenesis. Therefore, miRNAs
serve critical roles in epidermal disorders, including psoriasis.
miR-31 was observed in lesional and non-lesional skin of patients
with psoriasis and was triggered by activating NF-κB to promote
keratinocyte hyperproliferation in psoriasis (10,11).
Additionally, miR-21 inhibits the apoptosis of activated T cells in
psoriasis (12). Furthermore,
miR-125b is becoming a popular area of molecular research. It is
highly expressed in skin stem cells and in an inducible mouse
system, and is associated with stem cell differentiation and
inhibition (13). By identifying
the potential role of miR-125b in psoriasis pathogenesis,
researchers have noted that the most notably downregulated miRNA in
psoriatic skin is miR-125b. As indicated by in situ
hybridization results, the cell type mainly involved in the
downregulation of miR-125b in psoriatic lesions is the
keratinocyte, and a decrease or loss of miR-125b in psoriatic skin
may cause abnormality in the hyperproliferation and differentiation
of such cells (14,15).
When miRNA was detected in the sera of 32 patients
with psoriasis in previous studies, the expression of miR-125b was
markedly decreased. Bromodomain-containing protein 4 (BRD4), a
member of the Bromodomain and extra-terminal domain (BET) family,
activates the NF-κB signaling pathway and is associated with
inflammation and cancer (16,17).
The association between miR-125b and the BRD4/Notch signaling
pathway with regard to psoriasis has been poorly reported. The
present study aimed to investigate the mechanism of action between
miR-125b and the BRD4/Notch signaling pathway, and to investigate
novel targets and ideas for treating psoriasis.
Materials and methods
The present study was approved by the Ethics
Committee of the Northern Jiangsu Province Hospital (Jiangsu,
China).
Human serum
A total of 32 subjects (18 males and 14 females;
aged 18–39 years; mean age, 27.28±6.33 years) were enrolled in the
present study. The sera of 10 healthy volunteers (5 males and 5
females; aged 20–35 years; mean age, 28.1±5.53 years) for the
control group were provided by the Department of Dermatology of
Northern Jiangsu Province Hospital (Jiangsu, China). The sera were
sampled, centrifuged (1,000 × g for 10 min at 4°C) and stored at
−70°C.
Cell culture
The HaCaT and 293T cells were purchased from Chinese
Academy of Sciences Cell Bank (shanghai, China). The cells were
cultured with RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and 1%
streptomycin-penicillin (Gibco; Thermo Fisher Scientific,
Inc.).
Cell Counting Kit-8 (CCK-8)
Cells were inoculated at 1×105/ml in
96-well culture plates (~5,000 cells in 100 µl medium per well),
100 µl serum-free medium was then added, and cells were starved for
6 h. The medium was replaced with complete medium containing
different concentrations of DAPT (5, 10 and 20 nmol/l; cat. no.
HY-13027; MedChemExpress LLC, Monmouth Junction, NJ, USA). Controls
included three wells without cells, and three wells with
saline-treated cells. The complete medium was replaced with 100 µl
serum-free medium containing 10% CCK-8 dye after 1, 2 and 3 days,
and cells were cultured for an additional 1 h. The optical
absorbance at 450 nm for each sample was measured using an
absorbance microplate reader (ELx800 Absorbance Microplate Reader;
BioTek Instruments, Inc., Winooski, VT, USA). Survival of neurons
in the saline control group was defined as 100% and the results are
expressed as percentage relative to the control values.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using an RNA extraction kit
(RNA Quickly Extraction Kit, BioTeke Corporation, Beijing, China).
Following quantification, RNA was reverse transcribed into the
first strand of cDNA using an an iScript™ cDNA kit (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) at 37°C for 15 min and 85°C
for 5 sec, prior to storage at 4°C. Real-Time PCR Detection was
performed using iTaq™ SYBR-Green (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The samples were treated with recombinant DNase
I (DNA-free DNA removal kit; Ambion; Thermo Fisher Scientific,
Inc.) to remove possible DNA contamination. β-actin was used as an
internal control. The primer sequences used during the present
study are presented in Tables I
and II. The thermocycling
conditions were as follows: Initial pre-denaturation at 95°C for 1
min, denaturation for 15 sec, annealing at 55–65°C for 20 sec and
extension at 72°C for 30 sec. A total of 50 cycles were performed.
Based on the Cq value and relative standard curve of the PCR
product, the amount of RNA template contained in each specimen was
determined and compared with the amount of β-actin. The specific
value of the amount of RNA template to that of β-actin was adopted
as the final statistical value. The results were processed using
the 2−ΔΔCq method (18).
 | Table I.miRNA primer sequences used in
reverse transcription-quantitative polymerase chain reaction. |
Table I.
miRNA primer sequences used in
reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer
sequence |
|---|
| β-actin | Forward:
5′-CATGTACGTTGCTATCCAGGC-3′ |
|
| Reverse:
5′-CTCCTTAATGTCACGCACGAT-3′ |
| miRNA-125b | Forward:
5′-CTTGCCAGAAACGTCAATGGA-3′ |
|
| Reverse:
5′-GTGCAACTACGTCATAGCCTG-3′ |
| miRNA-200c | Forward:
5′-CTTGCCAGAAACGTCAATGGA-3′ |
|
| Reverse:
5′-GTGCAACTACGTCATAGCCTG-3′ |
| miRNA-200b | Forward:
5′-ACTGGTGGGTATGGGCATTG-3′ |
|
| Reverse:
5′-GCGCAGATGAACACGAACAG-3′ |
| miRNA-19b | Forward:
5′-GAGCCTGGGTTCGACGATG-3′ |
|
| Reverse:
5′-CCTGCTCTCGCTTATCTCCA-3′ |
| miRNA-152 | Forward:
5′-ACAGGCAGACACTAACGTCC-3′ |
|
| Reverse:
5′-CTGGGGTAGGATGCGAGGA-3′ |
| miRNA-296-5p | Forward:
5′-GAAGGGCCCCCCCTCA-3′ |
|
| Reverse:
5′-GTGCGTGTCGTGGAGTCG-3′ |
| miRNA-30d | Forward:
5′-CCTGTTGGTGCACTTCCTAC-3′ |
|
| Reverse:
5′-TGCAGTAGTTCTCCAGCTGC-3′ |
| miRNA-611 | Forward:
5′-AGGCGAGGACCCCT-3′ |
|
| Reverse:
5′-GTCCAGTTTTGTCAG-3′ |
| miRNA-419-3p | Forward:
5′-GCACTGGATACGACGTAGAA-3′ |
|
| Reverse:
5′-GCCCCTTATGCAAGATTCCC-3′ |
 | Table II.mRNA primer sequences used in reverse
transcription-quantitative polymerase chain reaction. |
Table II.
mRNA primer sequences used in reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer
sequence |
|---|
| β-actin | Forward:
5′-CATGTACGTTGCTATCCAGGC-3′ |
|
| Reverse:
5′-CTCCTTAATGTCACGCACGAT-3′ |
| Hes1 | Forward:
5′-TCAACACGACACCGGATAAAC-3′ |
|
| Reverse:
5′-CTCCTTAATGTCACGCACGAT-3′ |
| Hey1 | Forward:
5′-GTTCGGCTCTAGGTTCCATGT-3′ |
|
| Reverse:
5′-CGTCGGCGCTTCTCAATTATTC-3′ |
| Hey2 | Forward:
5′-CCTAACAGAAGTTGCGCGGTA-3′ |
|
| Reverse:
5′-GAGGCGACAAGGGGTTGAC-3′ |
| Jagged-1 | Forward:
5′-GTCCATGCAGAACGTGAACG-3′ |
|
| Reverse:
5′-GCGGGACTGATACTCCTTGA-3′ |
| Jagged-2 | Forward:
5′-TGGGCGGCAACTCCTTCTA-3′ |
|
| Reverse:
5′-GCCTCCACGATGAGGGTAAA-3′ |
| DLL1 | Forward:
5′-GATTCTCCTGATGACCTCGCA-3′ |
|
| Reverse:
5′-TCCGTAGTAGTGTTCGTCACA-3′ |
| BRD4 | Forward:
5′-GAGCTACCCACAGAAGAAACC-3′ |
|
| Reverse:
5′-GAGTCGATGCTTGAGTTGTGTT-3′ |
Western blot analysis
Cells were placed on ice immediately following
treatment and washed with ice-cold Hank's balanced salt solution
(HBSS). All the wash buffers and final resuspension buffer included
1X protease inhibitor cocktail (Pierce; Thermo Fisher Scientific,
Inc.), NaF (5 mM) and Na3VO4 (200 mM). The
protein concentration of the lysate was measured using a
Bicinchoninic Acid Protein Assay kit (Thermo Fisher Scientific,
Inc.). Equal amounts (30 µg) of total protein were subjected to
8–12% SDS-PAGE separation and transferred via electroblotting to
nitrocellulose membranes (Bio-Rad Laboratories, Inc.). The
membranes were blocked in non-fat dry milk solution (5%, room
temperature, 2 h) and incubated overnight at 4°C with primary
antibodies (all from Abcam, Cambridge UK) against Hes1 (1:1,000;
cat. no. ab71559), Hey1 (1:500; cat. no. ab22614), Hey2 (1:500;
cat. no. ab25404), Jagged-1 (1:1,000; cat. no. ab109536), BRD4
(1:1,000; cat. no. ab75898), β-tubulin (1:1,000; cat. no.
ab179513), GAPDH (1:1,000; cat. no. ab181602) and Notch1
intracellular domain 1 (NICD1; 1:1,000; cat. no. ab83232).
Membranes were then incubated with horseradish
peroxidase-conjugated anti-mouse immunoglobulin G (IgG; 1:5,000;
cat. no. G-21040, Thermo Fisher Scientific, Inc.) and anti-rabbit
IgG antibodies (1:5,000; cat. no. 31460, Thermo Fisher Scientific,
Inc.) for 2 h at room temperature. Afterwards, the membranes were
developed using the enhanced chemiluminescence substrate LumiGLO
(EMD Millipore, Billerica, MA, USA) and exposed to X-ray film. The
bands were analyzed using Gel-Pro Analyzer 4.0 (Media Cybernetics,
Inc., Rockville, MD, USA).
Bioinformatics analysis
The target genes of miR-125b were analyzed using the
target prediction tool TargetScan version 7.1 (http://www.targetscan.org/vert_71/) and miRanda
version 3.3a (http://www.microrna.org/microrna/home.do).
Transient transfection
The cells (50% confluence) were transfected with
miR-125b mimic (50, 100 and 150 nmol/l), inhibitor (25, 50 and 100
nmol/l) or BRD4 small interfering RNA (siRNA; 50 nmol/l; all
Guangzhou RiboBio Co., Ltd., Guangzhou, China), according to the
manufacturer's protocols. For experiments where only one
concentration was used, cells were transfected with 50 nmol/l
mimic/inhibitor/siRNA. miR-negative control (NC; 50 nmol/l),
inhibitor NC (50 nmol/l) and si-NC (50 nmol/l) were used as
respective transfection controls. The transfection reagent used was
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
The sequence of BRD4 siRNA was
5′-GACUAGAAACUUCCCAAAUGUCUUUCAAGAGAAGACAUUUGGGAAGUUUCUAGUC-3′. The
sequence of miR-125b mimic was sense, 5′-UCACAAGUUAGGGUCUCAGGGA-3′
and antisense, 3′-AGUGUUCAAUCCCAGAGUCCCU-5′. The sequence of
miR-125b inhibitor was 5′-UCACAAGUUAGGGUCUCAGGGA-3′. The sequence
of miR-NC was 5′-GGCUGCCUACUUAGCUUGAGAGUG-3′. The sequence of
inhibitor NC was 5′-ACGGGUGUGACCACUCCAGGCUGC-3′. The sequence of
si-NC was 5′-CCAUCUCCCGGUACAAAAUCUGCU-3′. At 48 h following
transfection, the cells were used for protein extraction.
Luciferase reporter assays
The wild-type 3′-UTR (3′-UTR-WT) of BRD4 containing
the miR-125b binding sites was purchased from Guangzhou RiboBio
Co., Ltd. Mutant BRD4 3-'UTR (3′-UTR-MUT), in which mutations occur
in the conserved binding sites for miR-125b, was generated using
overlapping extension PCR. The fragment of BRD4 3′-UTR-WT and the
mutant 3′-UTR fragments were inserted downstream of the
Renilla luciferase gene in psiCHECK™-2 vectors (Guangzhou
RiboBio Co., Ltd.). The sequence of 3′-UTR-WT of BRD4 included the
putative binding sites of miR-125b, whereas the sequence of
3′-UTR-MUT of BRD4 did not. Subsequently, the psiCHECK-2 vectors
with 3′-UTR-WT or 3′-UTR-MUT regions of BRD4 were co-transfected
with miR-125b mimic or mimic control using Lipofectamine 2000 into
293T cells. At 2 days following transfection, luciferase assays
were performed according to the manufacturer's protocols (Guangzhou
RiboBio Co., Ltd.) in triplicate. Renilla luciferase
activity was normalized firefly luciferase activity for each well,
yielding a gene expression ratio. Duplicate readings were averaged
to generate a single ratio value per well. Values were then
normalized to the average value of the control within the same cell
line and experiment to yield the relative luciferase activity.
Statistical analysis
SPSS 19.0 statistical software (IBM Corp., Armonk,
NY, USA) was used for all statistical analysis. Data are presented
as the mean ± standard deviation, and data were analyzed using
one-way analysis of variance, followed by a Bonferroni's post hoc
test for multiple comparisons. All experiments were independently
repeated 3 times and P<0.05 was considered to indicate a
statistically significant difference.
Results
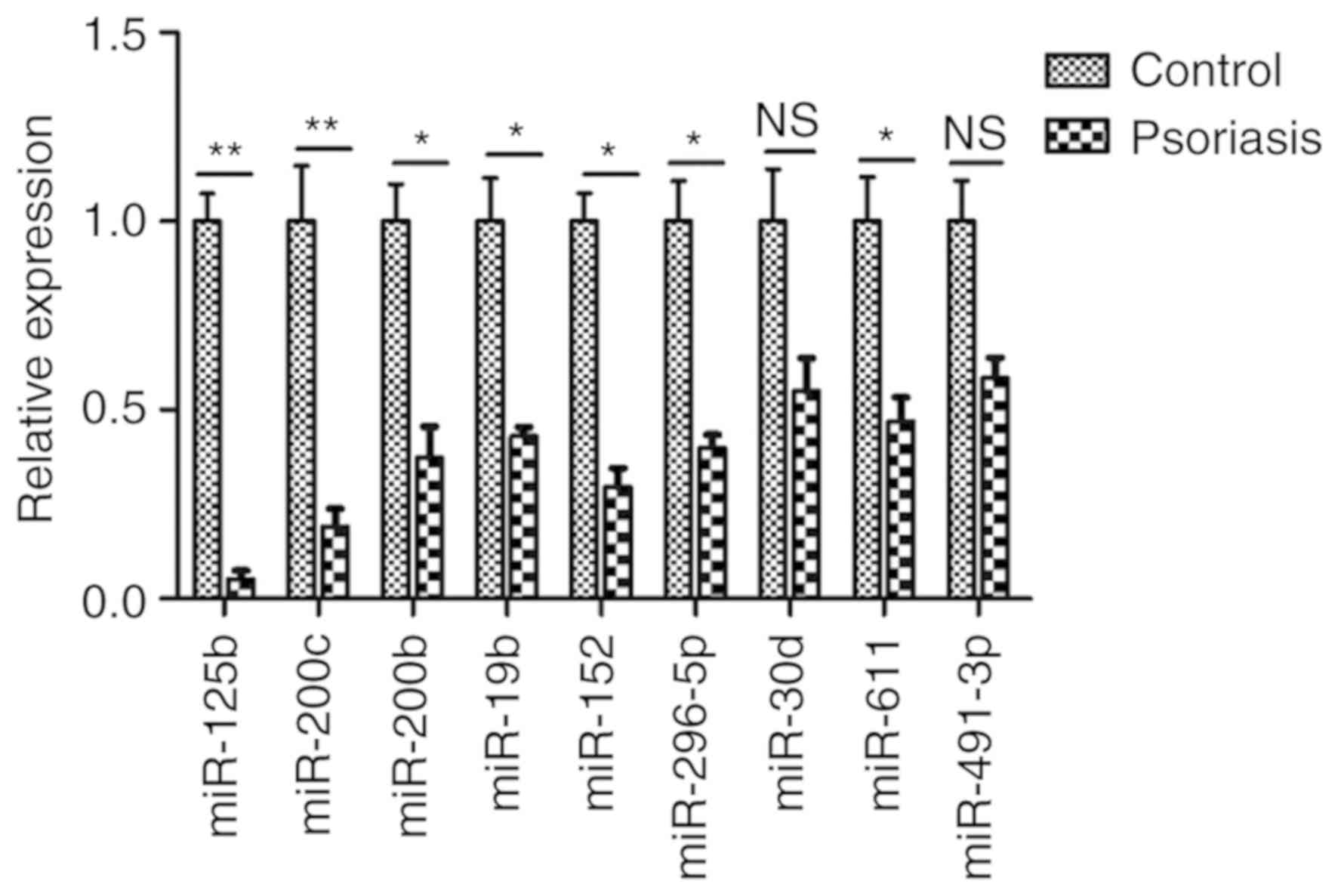
Analysis of miRNA in the serum
Certain miRNAs, including miR-125b, miR-200c,
miR-200b, miR-19b, miR-152, miR-296-5p, miR-30d, miR-611,
miR-491-3p, miR-23b, miR-95, miR-210, miR-224, miR-26a, miR-200a,
miR-27b, miR-328 and miR-376a, are associated with the in
vitro and in vivo differentiation of human keratinocytes
and are hypothesized to serve a role in the proliferation of
psoriatic keratinocytes. As detected by RT-qPCR in the sera of 32
patients with psoriasis, the miR-125b expression in the group of
patients with psoriasis was more markedly downregulated than that
in the control group of healthy volunteers (Fig. 1). The expression levels of miR-23b,
miR-95, miR-210, miR-224, miR-26a, miR-200a, miR-27b, miR-328 and
miR-376a were also upregulated to varying degrees (data not
shown).
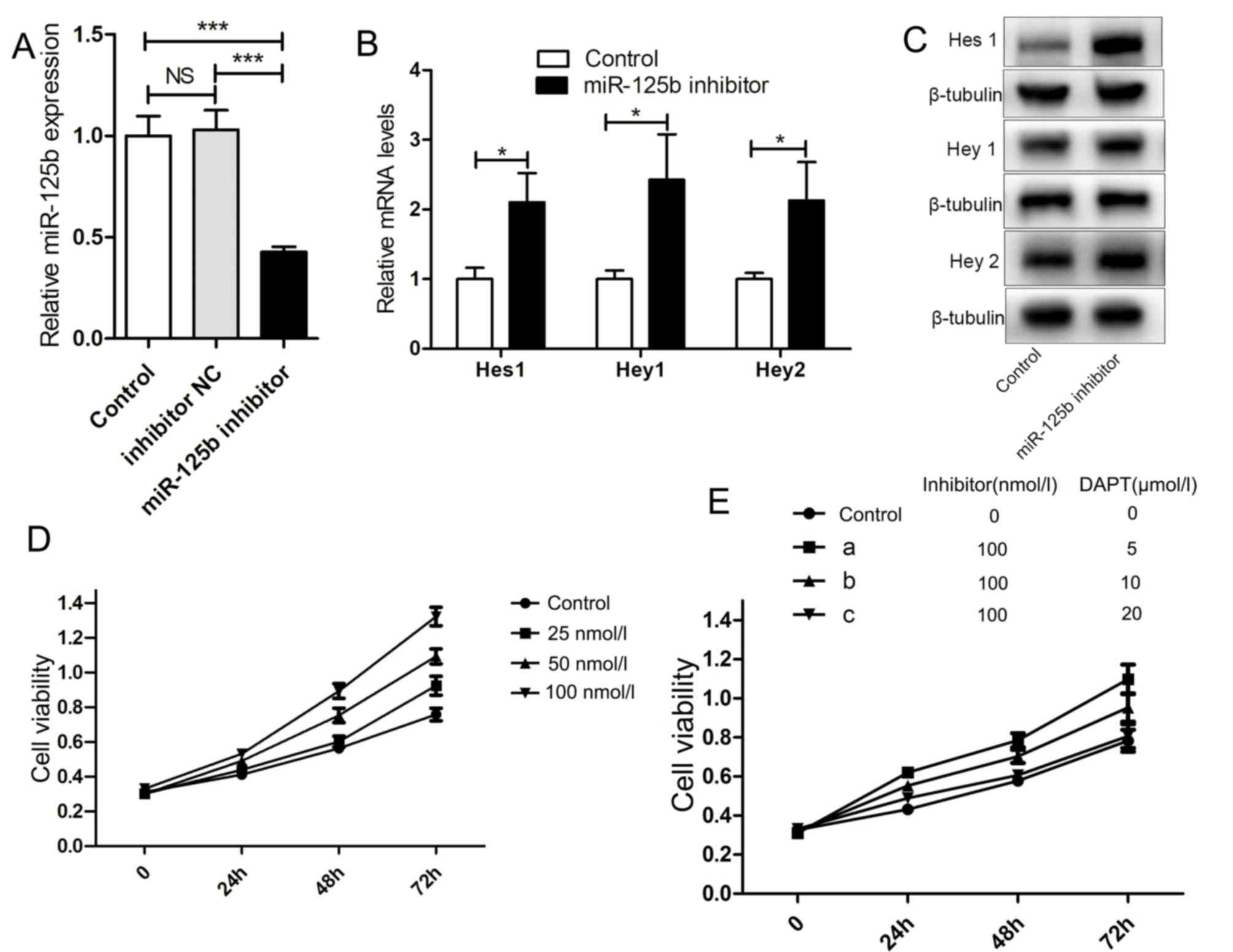
Inhibiting miR-125b upregulates the
mRNA and protein expression of downstream target genes of the Notch
signaling pathway and increases the proliferation of HaCaT
cells
Analysis of miRNA contents from the sera of patients
with psoriasis revealed that miR-125b was downregulated. The
abnormal proliferation of psoriatic keratinocytes is associated
with the activation of the Notch signaling pathway. Therefore, the
association between miR-125b and the Notch signaling pathway was
investigated. miR-125b inhibitor was added to HaCaT cell culture
fluid. Transfection with miR-125b inhibitor significantly decreased
the expression of miR-125b in HaCaT cells (Fig. 2A). Additionally, after 48 h, the
mRNA and protein expression of downstream genes (Hes1, Hey1
and Hey2) of the Notch signaling pathway was upregulated
with markedly increased cell proliferation (Fig. 2B-D). The inhibitor DAPT of the
Notch signaling pathway was added with the miR-125b inhibitor to
the HaCaT cell culture fluid; consequently, HaCaT cell
proliferation was inhibited (Fig.
2E).
Decreasing the abundance of miR-125b
upregulates Jagged-1 expression and thereby activates the Notch
signaling pathway
As indicated, the miR-125b inhibitor may upregulate
the downstream genes of the Notch signaling pathway, and the cell
proliferation mediated by the miR-125b inhibitor may be suppressed
by an inhibitor of the Notch signaling pathway. Therefore, miR-125b
may control HaCaT cell proliferation by regulating the Notch
signaling pathway. Inhibiting miR-125b upregulated the expression
of ligands of the Notch signaling pathway, including Jagged-1,
Jagged-2 and δ-like canonical Notch ligand 1 (DLL1; Fig. 3A-C). Jagged-1 is the most markedly
upregulated ligand among the five ligand types (Fig. 3A). Simultaneously, the Jagged-1
protein expression was increased by three different concentrations
of miR-125b inhibitor (Fig. 3D).
Furthermore, NICD1 is induced through miR-125b inhibitor (Fig. 3E). To further investigate the
association between miR-125b and Notch signaling, cells were
transfected with miR-125b mimic, which significantly upregulated
miR-125b expression in HaCaT cells (Fig. 3F). When the miR-125b mimic was
added, Jagged-1 was inhibited, and the release of active
intracellular domain NICD1 from Notch1 was concurrently inhibited
(Fig. 3G and H). Notch2
intracellular domain (NICD2) was upregulated, but to a lesser
extent than NICD1 (data not shown). The miR-125b inhibitor and DAPT
were then gradually added to the HaCaT cell culture fluid at
increasing concentrations. As a result, Jagged-1 was upregulated,
while NICD1 was downregulated. When DAPT was added to the inhibitor
at the highest concentration, NICD1 expression was lowest (Fig. 3I), indicating that the Notch
signaling pathway activation by miR-125b downregulation may be
completely inhibited by DAPT, and miR-125b may suppress the
activation of the Notch signaling pathway. When miR-125b mimic and
soluble Jagged-1 fragment were added concomitantly, NICD1 was
upregulated (Fig. 3J), implying
that the Notch signaling pathway, as inhibited by miR-125b, may be
activated by an ectogenic Jagged-1 fragment.
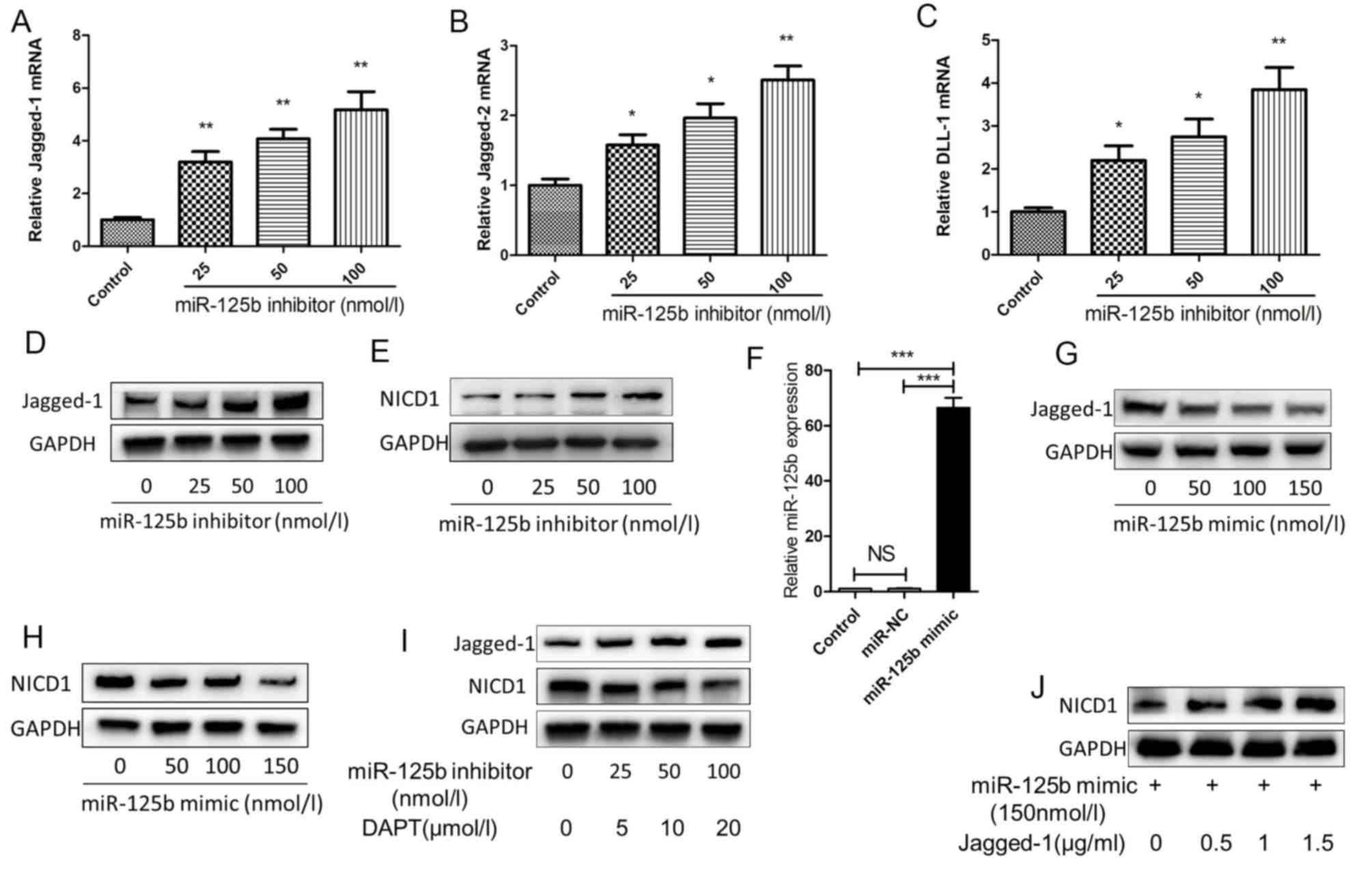 | Figure 3.miR-125b inhibits the activation of
the Jagged1/Notch signaling pathway. At 48 h after the HaCaT cells
being transfected with the three different concentrations of
miR-125b inhibitor, (A) Jagged-1, (B) Jagged-2 and (C) DLL1 mRNA
expression was detected by reverse transcription-quantitative
polymerase chain reaction analysis. (D and E) HaCaT cells were
transfected with three different concentrations of miR-125b
inhibitor, and the expression of (D) Jagged-1 and (E) NICD1 was
detected by western blotting. (F) Following transfection of HaCaT
cells for 48 h with the miR-125b mimic (50 nmol/l) or miR-NC (50
nmol/l), the levels of the miR-125b were detected via reverse
transcription-quantitative polymerase chain reaction analysis to
verify the transfection. Following HaCaT cells being transfected
with three different concentrations of miR-125b mimic, the
expression of (G) Jagged-1 and (H) NICD1 was detected by western
blotting. (I) Following three different concentrations of miR-125b
inhibitor and DAPT being added to HaCaT cells, the expression of
Jagged-1 and NICD1 was detected by western blotting. (J) Following
miR-125b mimic and different concentrations of Jagged-1 being added
to HaCaT cells, the expression of NICD1 was detected by western
blotting. *P<0.05, **P<0.01 vs. control; ***P<0.001. DLL1,
δ-like canonical Notch ligand 1; miR, microRNA; NICD1, Notch1
intracellular domain 1. |
miR-125b directly regulates the
upstream protein BRD4 of Jagged-1
miR-125b directly or indirectly regulates Jagged-1
expression, thereby activating the Notch signaling pathway. By
using two computational bioinformatics methods, namely TargetsScan
and miRanda, it was verified that Jagged-1 was not a direct target
of miR-125b, and that the BRD4 3′-UTR may include a direct binding
site of miR-125b (Fig. 4A). To
confirm the effect of miR-125b on the expression of BRD4, HaCaT
cells were transfected with the miR-125b inhibitor for 48 h. The
mRNA and protein expression levels of BRD4 were markedly
upregulated (Fig. 4B and C). By
contrast, with the upregulated expression of miR-125b in HaCaT
cells, the mRNA and protein expression levels of BRD4 were markedly
decreased (Fig. 4D and E).
Therefore, it was concluded that miR-125b inhibits BRD4.
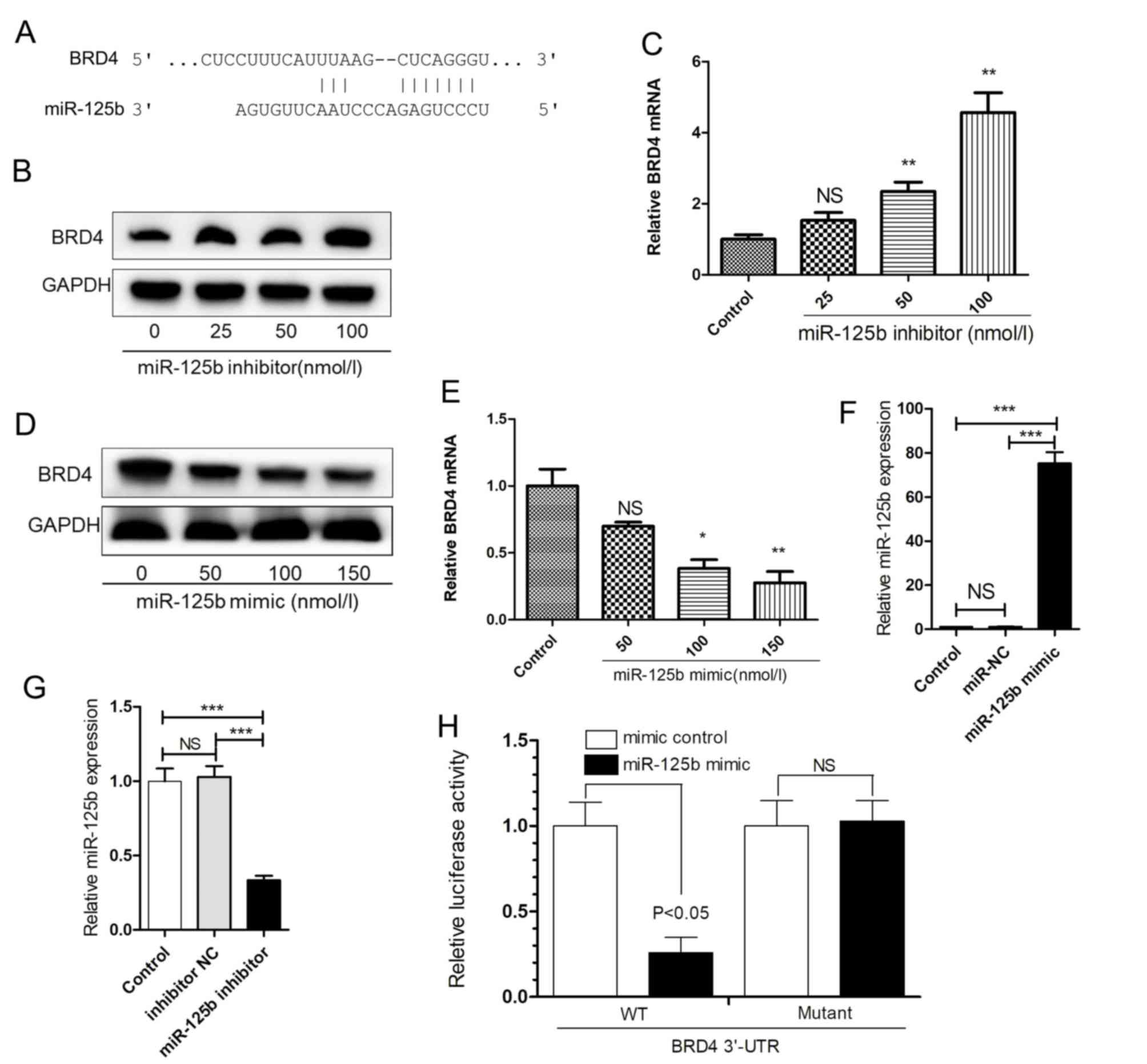 | Figure 4.miR-125b binds to the BRD4 3′-UTR.
(A) The binding site of miR-125b and the BRD4 3′-UTR was predicted.
(B and C) At 48 h after HaCaT cells were transfected with the
miR-125b inhibitor, the expression of BRD4 was detected by western
blotting and RT-qPCR. (D and E) At 48 h after HaCaT cells were
transfected with miR-125b mimic, the expression of BRD4 was
detected by western blotting and RT-qPCR. Following transfection of
293T cells for 48 h with (F) miR-125b mimic (50 nmol/l) or miR-NC
(50 nmol/l), or (G) miR-125b inhibitor (50 nmol/l) or inhibitor NC
(50 nmol/l), the levels of the miR-125b were detected by RT-qPCR to
verify the transfection. (H) Reporter constructs containing either
wild-type (WT) BRD4 3′-UTR, or BRD4 3′-UTR with mutation at the
predicted miR-125b target sequence were co-transfected into 293T
cells, along with miR-125b mimic (50 nmol/l) or mimic control (50
nmol/l). *P<0.05, **P<0.01 vs. control; ***P<0.001; NS,
not significant; BRD$, bromodomain-containing protein 4; miR,
microRNA; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction. |
To verify whether miR-125b directly acts on the
3′-UTR of BRD4, a luciferase assay was conducted in 293T cells
transfected with miR-125b mimic or inhibitor (Fig. 4F and G). The luciferase activity
was significantly decreased following co-transfection of
BRD4-3′-UTR-WT and miR-125b. When BRD4-3′-UTR-Mutant and miR-125b
were co-transfected, the inhibitory action of miR-125b was
eliminated (Fig. 4H).
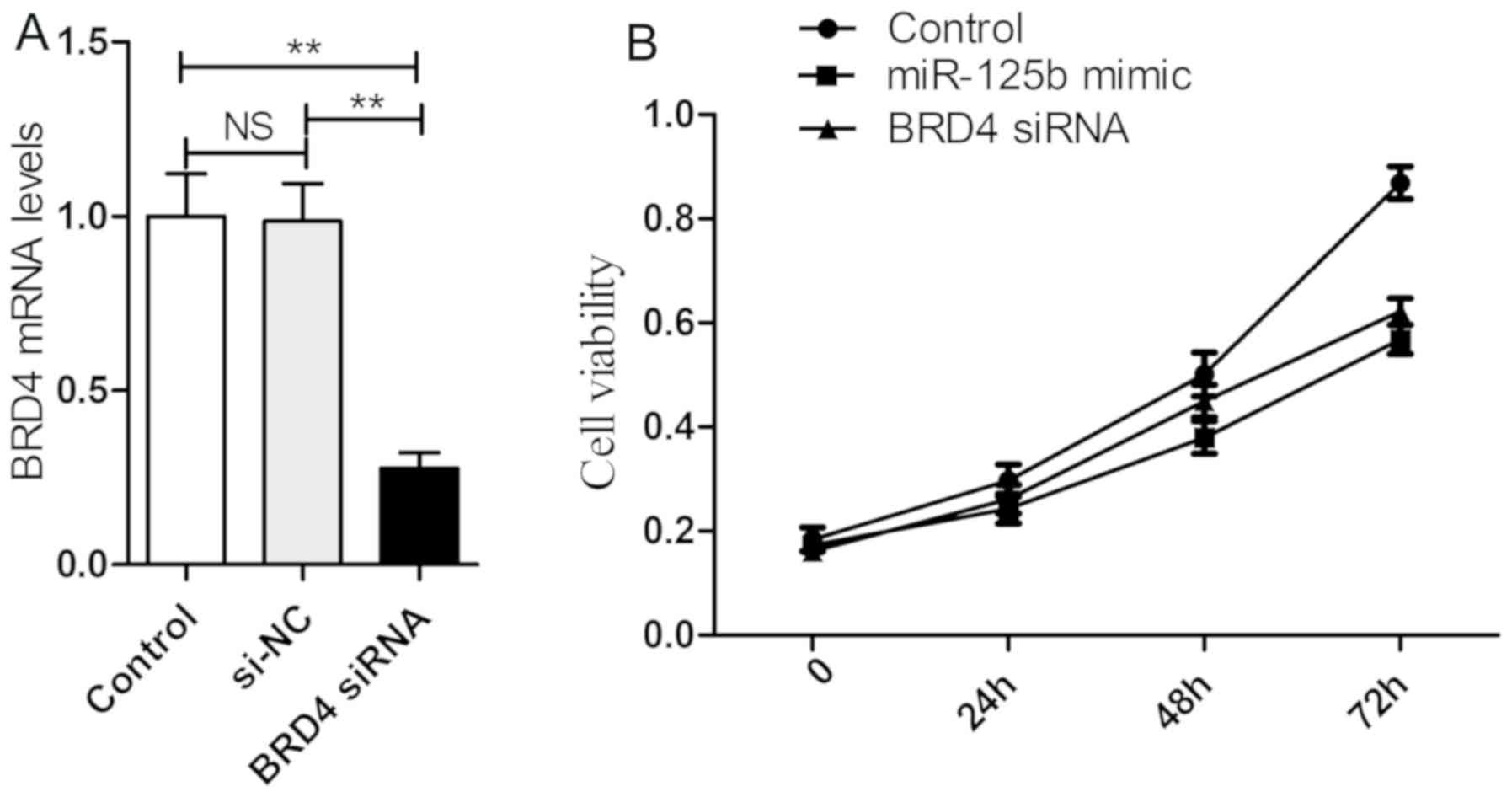
BRD4 siRNA and miR-125b mimic exhibit
similar inhibitory effects on HaCaT cell proliferation
The effects of BRD4 and miR-125b on HaCaT cell
proliferation were determined, and it was revealed that either the
transfection of BRD4 siRNA or miR-125b mimic markedly reduced cell
proliferation (Fig. 5).
Discussion
Psoriasis is a chronic inflammatory disease
characterized by the abnormal differentiation and
hyperproliferation of epidermal keratinocytes (3). It has several major histopathological
changes, including hyperproliferation of keratinocytes,
parakeratosis, inflammatory cell infiltration and
neovascularization, as well as scaly erythema (3). At present, the etiology and
pathogenesis of psoriatic skin lesions are too complex to be fully
elucidated (19). The
hyperproliferation of psoriatic epidermal cells is associated with
the Notch signaling pathway (20).
Furthermore, numerous miRNAs participate in the pathophysiological
process of psoriasis (21,22). In the present study, miR-125b
potentially regulates the activation of the Notch signaling pathway
by inhibiting the upstream protein BRD4 of Jagged-1 in
psoriasis.
Previous studies (14,15)
have demonstrated that miR-125b is mainly associated with
inflammation, cell proliferation and differentiation in psoriasis.
miR-125b acts on ubiquitin-specific peptidase 2 to induce psoriasis
(23). Injecting
lipopolysaccharide may decrease miR-125b levels in mice, and the
direct target gene of miR-125b is TNF-α. Furthermore, decreased
miR-125b levels may promote TNF-α secretion (24). miR-125b has also been demonstrated
to be associated with hyperproliferation. Furthermore, miR-125b
decreases keratinocyte proliferation by targeting fibroblast growth
factor receptor-2 (14). HaCaT
cells (25,26) are a non-tumor-immortalized human
normal skin keratinocyte strain characterized by continuous
differentiation and proliferation characteristics similar to normal
human keratinocytes, and are thus ideal as a cell model for
studying psoriasis. In the present study, adding miR-125b inhibitor
to HaCaT cell culture fluid markedly increased proliferation. The
Notch signaling pathway is associated with cell proliferation
(27). Next, the regulatory effect
of miR-125b on the proliferation of HaCaT cells via the Notch
signaling pathway was investigated. Previous studies have
demonstrated that Notch-1 is involved in the abnormal proliferation
and differentiation of vascular endothelial cells (28,29).
In patients with psoriasis, the mRNA and protein expression levels
of Notch-related proteins, including Notch-1, Notch-2, Jagged-1 and
Hes1, were markedly decreased following treatment with biological
agents. Therefore, the Notch signaling pathway is a key signaling
pathway involved in the onset and development of psoriasis
(30). In the Notch signaling
pathway, the cells are the ligands/receptors to each other, and the
ligand and receptor of the Notch signaling pathway are membrane
proteins (31). Following ligand
binding, the receptor is sheared by the γ-secretase in the
transmembrane region. The active intracellular domain (NICD) is
released into the nucleus, and the downstream pathway is activated
(32). The Notch signaling pathway
is specifically blocked by a γ-secretase inhibitor (33). To verify whether inhibiting
miR-125b results in HaCaT cell proliferation, the inhibitor DAPT of
the Notch signaling pathway was used along with the miR-125b
inhibitor. HaCaT cell proliferation was inhibited, and miR-125b
regulates such proliferation via the Notch signaling pathway.
Following miR-125b being inhibited, the mRNA and
protein expression of downstream genes (Hes1, Hey1 and
Hey2) of the Notch signaling pathway were upregulated,
indicating that the Notch signaling pathway was activated following
miR-125b inhibition. In order to detect which site of the Notch
signaling pathway was activated and in turn, which instigated the
pathway, four types of receptors and five types of ligands of the
Notch signaling pathway were investigated. The results demonstrated
that the mRNA expression of any of the four types of Notch receptor
was not upregulated (Notch-1, Notch-2, Notch-3 or Notch-4);
however, mRNA expression was upregulated for three (Jagged-1,
Jagged-2 and DLL1) of the five types of ligands, most notably
Jagged-1. Therefore, Jagged-1 was selected as the research target.
Following knockdown of endogenous miR-125b expression, Jagged-1 and
NICD1 were upregulated. Of note, NICD2 was also upregulated; NICD1
possesses the structural fragment(s) of the intracellular domains
of all the Notch receptors and is more highly expressed than NICD2.
Therefore, the changes in NICD1 expression were highlighted.
Transfection with miR-125b mimic revealed that Jagged-1 and NICD1
expression levels were downregulated, indicating that miR-125b
directly or indirectly acts on the Notch signaling pathway.
The results of the present study demonstrated that
BRD4 was a potential target gene of miR-125b; it was previously
reported to be an upstream signal molecule of Jagged-1 (34). BET proteins, including BRDT, BRD2,
BRD3 and BRD4, are novel ‘readers’ of epigenetic information. These
proteins regulate the activity of transcription factors by binding
to histone or non-histone lysine residues. Among these BET
proteins, BRD4 is the most widely studied. In the NF-κB signaling
pathway associated with tumorigenesis, BRD4 recruits positive
transcription elongation factor b proteins that activate the NF-κB
signaling pathway and are critical for nuclear signaling. The BRD4
and NF-κB enhancers also bind to each other and form a ‘super
enhancer’ complex that stimulates the NF-κB signaling pathway.
Luciferase assay results confirmed that miR-125b directly targets
the BRD4 3′-UTR.
In summary, miR-125b tightly binds to 3′-UTR of BRD4
with highly-marching sequences and restrains the translation
process of the Jagged-1 ligand. By further inhibiting the
activation of the Notch signaling pathway, miR-125b suppresses the
proliferation of psoriasis cells. Additionally, miR-125b is a
critical molecule in the progress of psoriasis.
The limitations of the present study were as
follows: The clinical sample size was small, and only few previous
studies had investigated BRD4 through clinical samples. Given the
extremely strong binding of BRD4 to histones and non-histone lysine
residues, the mechanisms of actions of BRD4 on any of the ligands
or receptors of the Notch signaling pathway have not been
investigated. Previous studies have focused on miR-125b and the
activation of T cells. Various activated T-cell subsets, including
Thl, Th2, Treg, Thl7 and CD8 T, under the action of different
adhesion factors, migrate to the skin and aggregate. Numerous
cytokines are then simultaneously released to stimulate the
hyperproliferation of keratinocytes, with an inflammatory reaction
after the epidermis considerably thickens (35,36).
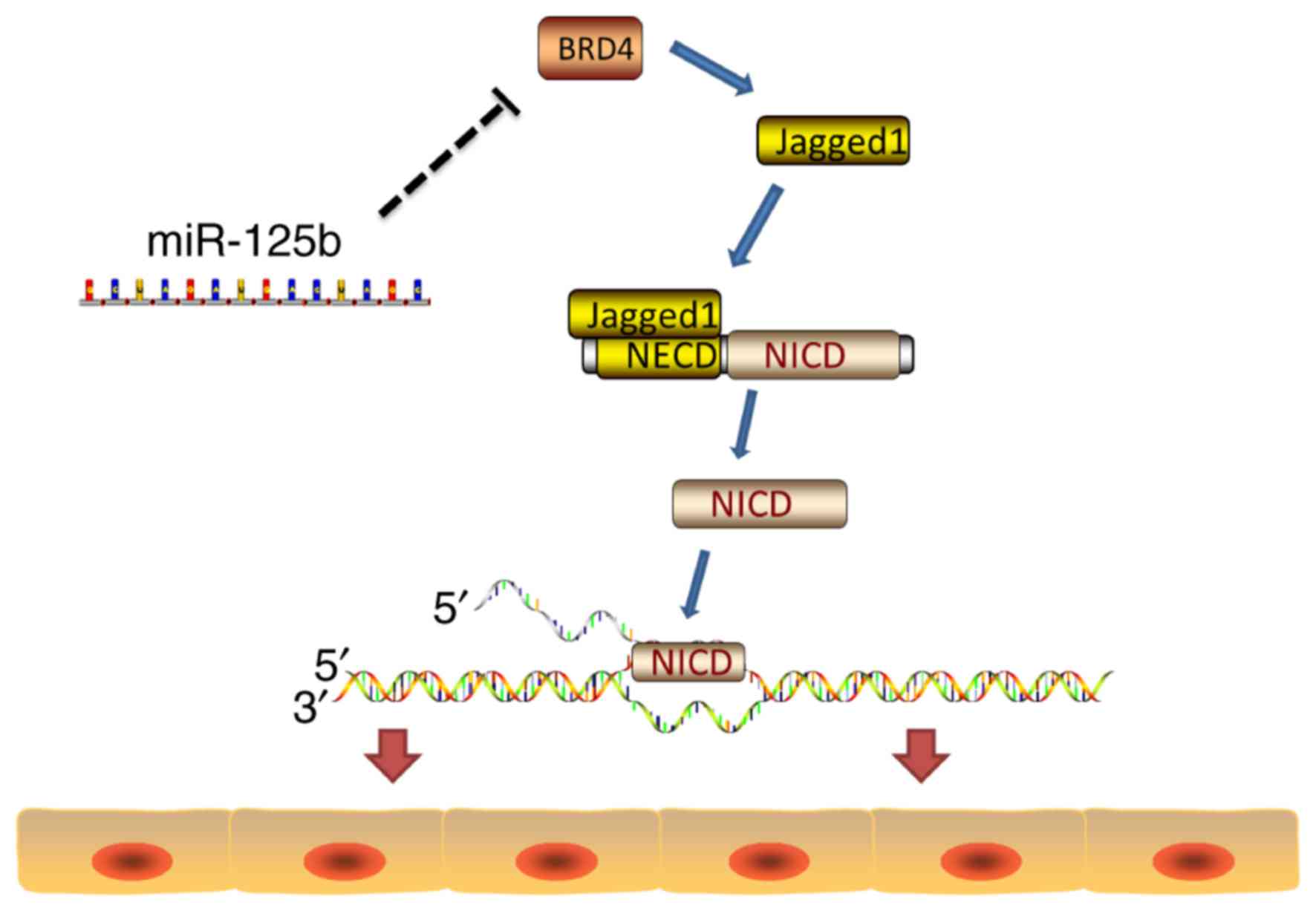
By investigating the differentiation/proliferation of psoriasis
cells, the significance of the present study lies in its discovery
that miR-125b does not directly act on the Notch signaling pathway
but on the upstream molecule BRD4 of the ligand Jagged-1 of the
Notch signaling pathway (Fig. 6).
Therefore, the miRNA indirectly regulates the Notch signaling
pathway. Furthermore, the Notch signaling pathway is associated
with the onset of psoriasis. Therefore, the present study is of
great importance in the regulation and treatment of psoriasis.
miR-125b may be a potential biomarker and therapeutic target for
the treatment of psoriasis.
The present study revealed that miR-125b inhibits
the expression of the upstream protein BRD4, and of the ligand
Jagged-1 of the Notch signaling pathway, thereby inhibiting the
HaCaT cell proliferation that is associated with the activation of
the Notch signaling pathway in psoriasis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
MP and DL conceived and designed the study. YH and
XZ designed and performed data analyses. MP and XL collected the
data and drafted the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study procedures were approved by the Ethics
Committee of the Northern Jiangsu Province Hospital. Written
informed consent was obtained from all subjects.
Patient consent for publication
Identifying information, including names, initials,
date of birth or hospital numbers, images or statements were not
included in this manuscript. The patients consented for the
publication of the associated data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Smith RL, Warren RB, Griffiths CE and
Worthington J: Genetic susceptibility to psoriasis: An emerging
picture. Genome Med. 1:722009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nestle FO, Kaplan DH and Barker J:
Psoriasis. N Engl J Med. 361:496–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lowes MA, Bowcock AM and Krueger JG:
Pathogenesis and therapy of psoriasis. Nature. 445:866–873. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miele L: Notch signaling. Clin Cancer Res.
12:1074–1079. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiao Z, Wang W, Guo M, Zhang T, Chen L,
Wang Y, You H and Li J: Expression analysis of Notch-related
molecules in peripheral blood T helper cells of patients with
rheumatoid arthritis. Scand J Rheumatol. 39:26–32. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Murea M, Park JK, Sharma S, Kato H,
Gruenwald A, Niranjan T, Si H, Thomas DB, Pullman JM, Melamed ML
and Susztak K: Expression of Notch pathway proteins correlates with
albuminuria, glomerulosclerosis, and renal function. Kidney Int.
78:514–522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nickoloff BJ and Nestle FO: Recent
insights into the immunopathogenesis of psoriasis provide new
therapeutic opportunities. J Clin Invest. 113:1664–1675. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lehar SM and Bevan MJ: Immunology:
Polarizing a T-cell response. Nature. 430:150–151. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Napolitani G, Rinaldi A, Bertoni F,
Sallusto F and Lanzavecchia A: Selected Toll-like receptor agonist
combinations synergistically trigger a T helper type 1-polarizing
program in dendritic cells. Nat Immunol. 6:769–776. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia J and Zhang W: MicroRNAs in normal and
psoriatic skin. Physiol Genomics. 46:113–122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan S, Xu Z, Lou F, Zhang L, Ke F, Bai J,
Liu Z, Liu J, Wang H, Zhu H, et al: NF-κB-induced microRNA-31
promotes epidermal hyperplasia by repressing protein phosphatase 6
in psoriasis. Nat Commun. 6:76522015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meisgen F, Xu N, Wei T, Janson PC, Obad S,
Broom O, Nagy N, Kauppinen S, Kemény L, Ståhle M, et al: MiR-21 is
up-regulated in psoriasis and suppresses T cell apoptosis. Exp
Dermatol. 21:312–314. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Stokes N, Polak L and Fuchs E:
Specific microRNAs are preferentially expressed by skin stem cells
to balance self-renewal and early lineage commitment. Cell Stem
Cell. 8:294–308. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu N, Brodin P, Wei T, Meisgen F, Eidsmo
L, Nagy N, Kemeny L, Ståhle M, Sonkoly E and Pivarcsi A: MiR-125b,
a microRNA downregulated in psoriasis, modulates keratinocyte
proliferation by targeting FGFR2. J Invest Dermatol. 131:1521–1529.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang RY, Li L, Wang MJ, Chen XM, Huang QC
and Lu CJ: An exploration of the role of MicroRNAs in psoriasis: A
systematic review of the literature. Medicine (Baltimore).
94:e20302015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Andrieu GP, Shafran JS, Deeney JT,
Bharadwaj KR, Rangarajan A and Denis GV: BET proteins in abnormal
metabolism, inflammation, and the breast cancer microenvironment. J
Leukoc Biol. 104:265–274. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang G, Liu R, Zhong Y, Plotnikov AN,
Zhang W, Zeng L, Rusinova E, Gerona-Nevarro G, Moshkina N, Joshua
J, et al: Down-regulation of NF-κB transcriptional activity in
HIV-associated kidney disease by BRD4 inhibition. J Biol Chem.
287:28840–28851. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Crow JM: Therapeutics: Silencing
psoriasis. Nature. 492:S58–S59. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ota T, Takekoshi S, Takagi T, Kitatani K,
Toriumi K, Kojima T, Kato M, Ikoma N, Mabuchi T and Ozawa A: Notch
signaling may be involved in the abnormal differentiation of
epidermal keratinocytes in psoriasis. Acta Histochem Cytochem.
47:175–183. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jinnin M: Recent progress in studies of
miRNA and skin diseases. J Dermatol. 42:551–558. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Q, Wu DH, Han L, Deng JW, Zhou L, He
R, Lu CJ and Mi QS: Roles of microRNAs in psoriasis: Immunological
functions and potential biomarkers. Exp Dermatol. 26:359–367. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei T, Folkersen L, Biskup E, Xu N, Manfe
V, Niazi O and Gniadecki R: Ubiquitin-specific peptidase 2 as a
potential link between microRNA-125b and psoriasis. Br J Dermatol.
176:723–731. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tili E, Michaille JJ, Cimino A, Costinean
S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA and
Croce CM: Modulation of miR-155 and miR-125b levels following
lipopolysaccharide/TNF-alpha stimulation and their possible roles
in regulating the response to endotoxin shock. J Immunol.
179:5082–5089. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boukamp P, Petrussevska RT, Breitkreutz D,
Hornung J, Markham A and Fusenig NE: Normal keratinization in a
spontaneously immortalized aneuploid human keratinocyte cell line.
J Cell Biol. 106:761–771. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schürer N, Köhne A, Schliep V, Barlag K
and Goerz G: Lipid composition and synthesis of HaCaT cells, an
immortalized human keratinocyte line, in comparison with normal
human adult keratinocytes. Exp Dermatol. 2:179–185. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thélu J, Rossio P and Favier B: Notch
signalling is linked to epidermal cell differentiation level in
basal cell carcinoma, psoriasis and wound healing. BMC Dermatol.
2:72002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abdou AG, Maraee AH, Sharaf A and
Elnaidany NF: Up-regulation of Notch-1 in psoriasis: An
immunohistochemical study. Ann Diagn Pathol. 16:177–184. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rooney P, Connolly M, Gao W, McCormick J,
Biniecka M, Sullivan O, Kirby B, Sweeney C, Molloy E, Markham T, et
al: Notch-1 mediates endothelial cell activation and invasion in
psoriasis. Exp Dermatol. 23:113–118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Skarmoutsou E, Trovato C, Granata M, Rossi
GA, Mosca A, Longo V, Gangemi P, Pettinato M, D'Amico F and
Mazzarino MC: Biological therapy induces expression changes in
Notch pathway in psoriasis. Arch Dermatol Res. 307:863–873. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kovall RA, Gebelein B, Sprinzak D and
Kopan R: The canonical Notch signaling pathway: Structural and
biochemical insights into shape, sugar, and force. Dev Cell.
41:228–241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bray SJ and Gomez-Lamarca M: Notch after
cleavage. Curr Opin Cell Biol. 51:103–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Locatelli M and Curigliano G: Notch
inhibitors and their role in the treatment of triple negative
breast cancer: Promises and failures. Curr Opin Oncol. 29:411–427.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Andrieu G, Tran AH, Strissel KJ and Denis
GV: BRD4 regulates breast cancer dissemination through
Jagged1/Notch1 signaling. Cancer Res. 76:6555–6567. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Diani M, Altomare G and Reali E: T cell
responses in psoriasis and psoriatic arthritis. Autoimmun Rev.
14:286–292. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen H, Liu H, Lu C, Wang M, Li X, Zhao H,
Yan Y, Yu W, Han L and Dai Z: PSORI-CM02 formula increases CD4+
Foxp3+ regulatory T cell frequency and ameliorates
imiquimod-induced psoriasis in mice. Front Immunol. 8:17672018.
View Article : Google Scholar : PubMed/NCBI
|