Introduction
Glioblastoma multiforme (GBM) is one of the most
common malignant primary brain tumours in adults (1). Technological advances in surgery,
radiotherapy and chemotherapy have been made in previous decades;
however, the average overall survival time of patients with GBM is
14.6 months (2). As a result of
the invasive growth of tumour cells, challenges include complete
resection and low sensitivity to standard therapy (3,4).
Temozolomide (TMZ) is the standard first-line chemotherapeutic
treatment for glioma; however, the overall clinical prognosis
remains unsatisfactory due to the resistance of glioma cells to TMZ
(5). Therefore, determination of
the mechanisms underlying drug resistance is important for
improving chemotherapeutic efficacy in GBM.
MicroRNAs (miRNAs/miRs) are a class of small
noncoding RNAs, 20–22 nucleotides in length, which negatively
regulate their target genes by binding to the 3′-untranslated
region (3′UTR) of mRNA (6). miRNAs
possess important roles in tumourigenesis, such as in the invasion
and drug resistance of tumour cells (7,8).
miR-186 has been reported to affect the proliferation,
infiltration, metastasis and drug resistance of various types of
cancer, including ovarian, prostate and lung cancer (9–11).
Downregulation of miR-186 results in the increased invasion and
proliferation of non-small cell lung cancer (NSCLC) cells, and is
associated with poor patient prognosis (11). Decreased miR-186 expression has
also been detected in cisplatin-resistant ovarian cancer tissues
and cell lines, and may act as a potent epithelial-mesenchymal
transition (EMT) regulator that is involved in the drug resistance
of ovarian cancer cells (9).
however, the role of miR-186 in the chemoresistance of glioma cells
remains unknown.
The highly conserved basic helix-loop-helix
transcription factor Twist-related protein 1 (Twist1) has been
proposed to act as an oncogenic transcription factor; increased
expression of Twist proteins promotes tumour initiation,
progression, metastasis, EMT, and protection from
chemotherapy-induced apoptosis and senescence (12–15).
Evidence has also suggested that Twist1 is upregulated in malignant
glioma and promotes glioma cell invasion via the induction of
mesenchymal molecular and cellular alterations (15). However, the role of Twist1 in the
drug resistance of human glioma cells to TMZ is unclear.
In the present study, it was reported that the
expression levels of miR-186-5p were decreased, whereas Twist1
expression was upregulated during the development of drug
resistance in glioma cells. Furthermore, it was revealed that
decreased expression of miR-186-5p contributed to the proliferation
and drug resistance of glioma cells via the direct targeting of
Twist1. The results provided novel insight into the role of
miR-186-5p and its downstream target, Twist1, in the drug
resistance of glioma.
Materials and methods
Cell lines and tissue samples
Human U87 (U87 MG HTB-14, likely glioblastoma of
unknown origin) and T98G glioblastoma cell lines were purchased
from the American Type Culture Collection (ATCC). The human normal
glial cell line HEB (ATCC0459) and 293T cells were purchased from
Shanghai Beinuo Biotechnology Co., Ltd. Cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
foetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100
mg/ml streptomycin and 100 IU/ml penicillin (Thermo Fisher
Scientific, Inc.). Cells were maintained at 37°C in a 5%
CO2 humidified atmosphere. Human GBM (n=11) and normal
brain specimens (n=20) used in the present study were obtained from
patients operated for glioma and epilepsy, respectively. The
samples were collected from the Department of Neurosurgery, The
First Affiliated Hospital of Nanchang University between January
2011 and December 2017. Patients with primary tumours that had not
received radiotherapy or chemotherapy prior to surgery were
included in the study. Detailed clinical data and surgical records
were available. The age and sex distributions of patients are
presented in Table I. Diagnosis of
GBM was confirmed by histopathological examination. This study was
approved by the Ethics Committee of The First Affiliated Hospital
of Nanchang University and protocols were conducted according to
the principles of the Declaration of Helsinki. Written informed
consent was obtained from all patients in the study.
 | Table I.Comparison of clinicopathological
features between the control and glioma groups. |
Table I.
Comparison of clinicopathological
features between the control and glioma groups.
| Clinicopathological
features | Control group
(n=11) | Glioma group
(n=20) | P-value |
|---|
| Sex |
|
| 0.76 |
|
Male | 6 | 11 |
|
|
Female | 5 | 9 |
|
| Age (years) | 46 (30–65) | 48 (32–68) | 0.86 |
| Tumour site |
| Frontal
lobe | – | 4 |
|
|
Temporal lobe | – | 4 |
|
|
Parietal lobe | – | 2 |
|
|
Occipital lobe | – | 3 |
|
| Insular
lobe | – | 2 |
|
|
Multiple lobes | – | 5 |
|
| Infiltration
depth |
|
T1+T2 | – | 4 |
|
|
T3+T4 | – | 16 |
|
| Tumour diameter
(cm) | – | 3.18±0.45 |
|
Reverse transcription-quantitative PCR
(RT-qPCR)
Cells were treated with TMZ (0, 50, 100 and 200 µM;
Sigma-Aldrich; Merck KGaA) for 24 h at 37°C. Twist1 expression was
determined via RT-qPCR analysis. Total RNA was extracted from cell
lines and tissues using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). cDNA was synthesised from total
RNA using the PrimeScript® RT reagent kit (Takara
Biotechnology Co., Ltd.), according to the manufacturer's protocol.
PCR reactions were performed using the SYBR® Premix
Dimer Eraser kit (Takara Biotechnology Co., Ltd.) with the ABI
Prism 7500 system (Applied Biosystems; Thermo Fisher Scientific,
Inc.), under the following conditions: 4 min at 94°C, then 40
cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 60 sec. The
expression of Twist1 was normalized to the expression of β-actin.
For analysis of the expression of miR-186-5p, the TaqMan MicroRNA
Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) was
used, and qPCR was conducted as follows: 4 min at 94°C, then 40
cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 60 sec. U6
was used as an endogenous control. The primers used were as
follows: Twist1, forward 5′-AGCTACGCCTTCTCCGTCT-3′, reverse,
5′-TCCTTCTCTGGAAACAATGACA-3′; β-actin, forward
5′-GGCGGCACCACCATGTACCCT-3′, reverse, 5′-AGGGGCCGGACTCGTCATACT-3′;
U6, forward 5′-CTCGCTTCGGCAGCACA-3′, reverse,
5′-AACGCTTCACGAATTTGCGT-3′; and miR-186-5p, forward
5′-GCGGCGCAAAGAATTCTCCT-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′. The
2ΔΔCq method (16) was
used to quantify the relative expression levels of Twist1 and
miR-186-5p.
Cell transfection
miR-186-5p mimic (5′-CAAAGAAUUCUCCUUUUGGGCU-3′,
5′-CCCAAAAGGAGAAUUCUUUGUU-3′), inhibitor
(5′-AGCCCAAAAGGAGAAUUCUUUG-3′), mimic negative control (NC;
5′-UUCUCCGAACGUGUCACGUTT-3′, 5′-ACGUGACACGUUCGGAGAATT-3′) and
inhibitor NC (5′-CAGUACUUUUGUGUAGUACAA-3′) were synthesised by
Guangzhou RiboBio Co., Ltd. The cell lines (U87, T98G) were seeded
in 6-well plates at a density of 2×105 cells/ml, and
then maintained for 24 h at 37°C in a 5% CO2 humidified
atmosphere. Cells were transfected with miR-186-5p mimics,
miR-186-5p inhibitors, mimic NC or inhibitor NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), at a final concentration of 100 nM for 24 or 48
h at 37°C. Twist1 small interfering RNA [siRNA (siTwist1-1:
5′-CCTCTGCATTCTGATAGAA-3′; siTwist1-2: 5′-TGAGCAACAGCGAGGAAGA-3′)]
and siRNA NC (5′-UUCUCCGAACGUGUCACGUTT-3′) were obtained from
Guangzhou RiboBio Co., Ltd. siTwist1-2 (20 nM) induced notably
increased effects on Twist1 expression compared with siTwist1-1 (20
nM), and was therefore selected for use as the si-Twist1 presented
in this study (data not shown). The Twist1 expression vector
(pcDNA3.1-Twist1) was constructed by Shanghai GenePharma Co., Ltd.
The pcDNA3.1-Twist1 vector (2 µg/ml) and empty vector control were
transfected into cells using Lipofectamine 2000, according to the
manufacturer's protocols. At 24 h following transfection, RT-qPCR
was performed. At 48 h following transfection, western blot,
5-ethynyl-2′-deoxyuridine (EdU) and flow cytometry assays were
performed.
Western blot analysis
Western blot analysis was performed as previously
described (17). Cells or tissues
were harvested in lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl,
1% Triton X-100) containing protease and phosphatase inhibitors
(Roche Applied Science). The protein concentration was determined
using a BCA kit (Pierce; Thermo Fisher Scientific, Inc.). Then,
proteins (20 µg/lane) were separated by 10% SDS-PAGE and
transferred onto polyvinylidene difluoride membranes (EMD
Millipore). Membranes were incubated for 1 h at room temperature in
PBS-0.1% Tween-20 and 5% non-fat skim milk, and then probed at 37°C
for 1 h with antibodies against Twist1 (1:500; cat. no. ab50581;
Abcam) and β-actin (1:2,000; cat. no. ab8227; Abcam). After
washing, the blots were incubated at 37°C for 1 h with horseradish
peroxidase-conjugated secondary antibodies (1:30,000; cat. no.
AP187P; EMD Millipore), and the signals were visualised using
enhanced chemiluminescence substrate (Bio-Rad Laboratories, Inc.)
by a luminescent image analyser (Fujifilm).
Luciferase reporter assays
To identify putative targets involved in the effects
of miR-186 on the proliferation and drug resistance of glioblastoma
cells, bioinformatics was employed using miRanda version 3.3a
(http://www.microrna.org/microrna/home.do). The
wild-type (WT) 3′-UTR of Twist1 that contains the predicted
miR-186-5p target sequence was amplified using Pyrobest DNA
polymerase (Fermentas; Thermo Fisher Scientific, Inc.) and cloned
into the miRNA Expression Reporter Vector (pGL3 vector; Promega
Corporation) at the XbaI site. A mutation from GTA to CAT
was introduced in the potential miR-186-5p binding sites (named
pGL3-Twist1-Mut) by using the QuickChange Stratagene method. 293T
cells were seeded in a 24-well plate 24 h prior to transfection at
a density of 2.5×104 cells/well, and co-transfected with
0.15 µg of pGL3-Twist1-WT or pGL3-Twist1-Mut, 0.05 µg of the
control vector containing Renilla luciferase (pRL-TK;
Promega Corporation) and 40 pmol of miR-186-5p mimics or NC using
Lipofectamine® 2000. At 24 h following transfection,
firefly and Renilla luciferase activity were measured using
the Dual-Luciferase Reporter Assay system (Promega Corporation),
and firefly luciferase activity was normalized to Renilla
luciferase activity.
Evaluation of apoptosis
Post-transfection, cells were treated with TMZ (100
µM) for 24 h at 37°C. Cell death was detected using an Annexin
V-phycoerythrin/7-aminoactinomycin D (7-AAD) apoptosis kit (BD
Biosciences), according to the manufacturer's protocol. Cells were
resuspended in 1X binding buffer (BD Biosciences) at a
concentration of 1×106 cells/ml. Then, 5 µl PE-Annexin V
and 5 µl 7-AAD were added. Cells were incubated for 15 min at room
temperature in the dark. Then, 400 µl of 1X binding buffer was
added to each tube, and samples were analysed using a flow
cytometer (BD Biosciences). The data were analysed using FlowJo
version 10 (FlowJo LLC).
5-ethynyl-2′-deoxyuridine (EdU)
assay
Cells were seeded at a density of 2.5×104
cells/well in a 24-well plate 24 h prior to transfection.
Subsequently, cells were transfected with mimics, mimic NC,
inhibitors and inhibitor NC as previously described. At 48 h
following transfection, the cells were then fixed with 4%
formaldehyde for 15 min at room temperature, and stained with an
EdU DNA Proliferation In vitro Detection kit (Guangzhou
RiboBio Co., Ltd.), according to the manufacturer's protocols.
Images of three randomly selected fields were acquired to visualise
the number of EdU-stained cells using a fluorescence microscope
(Nikon Corporation), and the data were analysed using ImageJ
version 1 (National Institutes of Health).
Analysis of cell proliferation and
IC50
A Cell Counting Kit-8 (CCK-8) Assay kit
(Sigma-Aldrich; Merck KGaA) was used to measure cell proliferation
and drug resistance. Cells were seeded in 96-well plates at a
density of 5,000 cells/well and incubated for 24 h. Then, the cells
were treated with various concentrations (0, 5, 10, 20, 40, 80, 160
and 320 µM) of TMZ and incubated for 24, 48 and 72 h in 37°C. CCK-8
solution (10 µl) was added to each well and cells were incubated
for 2 h at 37°C. The absorbance was detected at 450 nm using a
Multi-mode Plate Reader (BioTek Instruments, Inc.). IC50
values were calculated from dose-response curves generated using a
polynomial dose-response approximation using SPSS version 17.0
software (SPSS, Inc.).
Statistical analysis
Statistical analysis was performed using SPSS
version 13.0 software (SPSS, Inc.). All results are expressed as
the means ± standard deviation of three independent experiments.
Comparison of means between two groups was performed using
Student's t-test. Comparisons of means between multiple groups were
conducted using one-way analyses of variance followed by post hoc
pairwise comparisons using Tukey's test. Differences in miRNA
expression levels between normal and GBM brain tissues were
evaluated using Mann-Whitney U tests. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-186-5p is downregulated in
glioblastoma and contributes to the proliferation of glioblastoma
cells
To determine whether miR-186-5p is involved in human
glioblastoma, the expression levels of miR-186-5p were detected in
human glioblastoma tissues and two human glioblastoma cell lines
[U87 (likely glioblastoma of unknown origin) and T98G] via RT-qPCR
analysis. The results revealed that miR-186-5p levels in human
glioblastoma tissues were significantly reduced compared with in
normal brain tissues (Fig. 1A). It
was also demonstrated that miR-186-5p was significantly
downregulated in U87 and T98G cells compared with in the human
normal glial cell line, HEB (Fig.
1B). To investigate the effects of miR-186-5p on the
proliferation of glioblastoma cells, proliferation was measured
using CCK-8 and EdU assays 24 h post-transfection of U87 and T98G
cells with miR-186-5p mimics, mimic NC, miR-186-5p inhibitors and
inhibitor NC. It was revealed that upregulated miR-186-5p
expression significantly decreased the proliferation of U87 and
T98G cells compared with the NC, whereas inhibition of miR-186-5p
significantly increased the proliferation of U87 and T98G cells
(Fig. 1C-H). These results
indicated that miR-186-5p was downregulated in GBM tissues and
cells and suppressed the proliferation of glioblastoma cells.
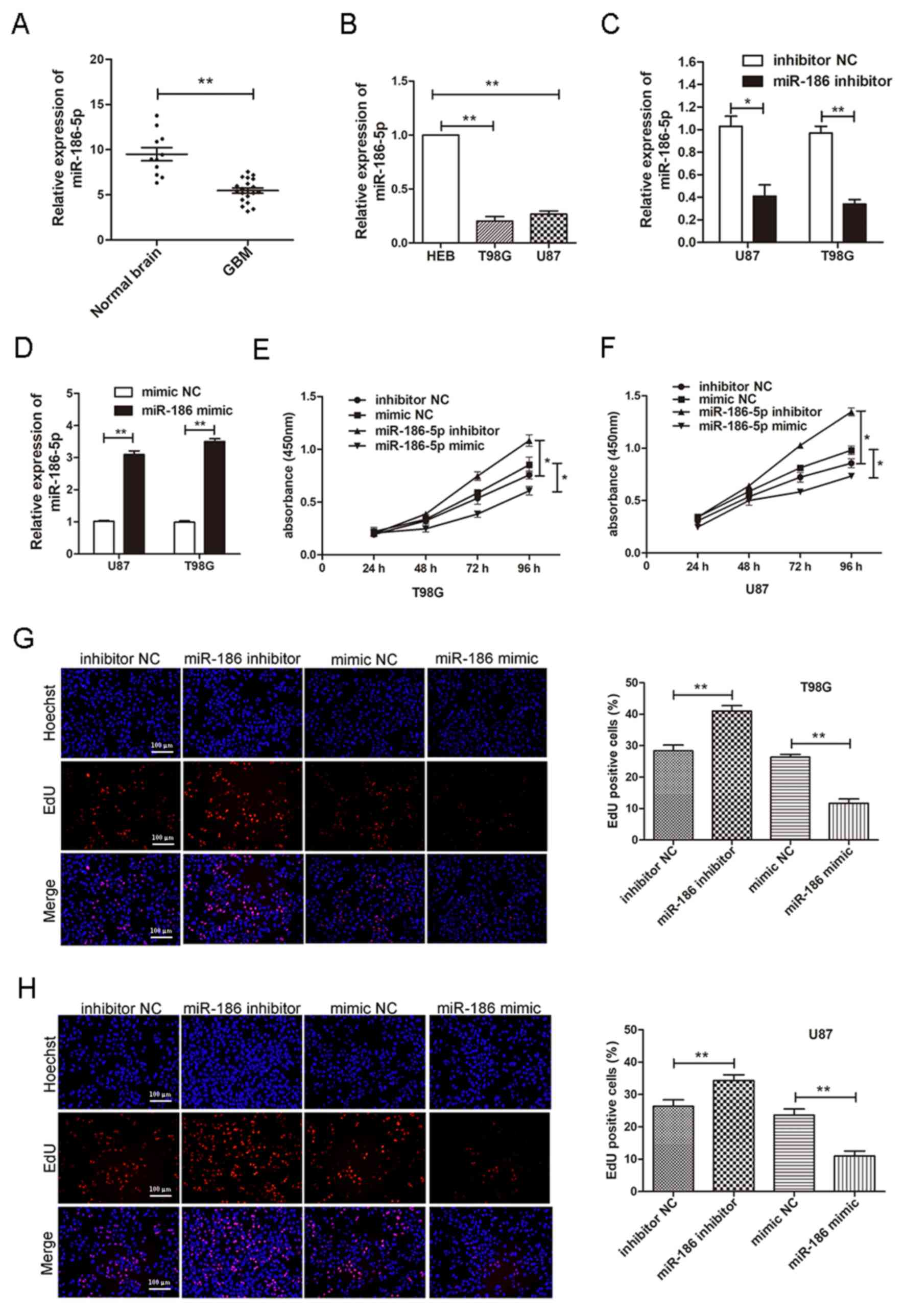 | Figure 1.miR-186-5p is downregulated in
glioblastoma. (A) Expression of miR-186-5p in GBM and normal
non-neoplastic tissues, as determined by RT-qPCR analysis. (B)
Expression of miR-186-5p in HEB, T98G and U87 cells, as determined
by RT-qPCR analysis. miR-186-5p expression in T98G and U87 cells
transfected with (C) miR-186-5p inhibitors or inhibitor NC, and (D)
miR-186-5p mimics or mimic NC. Proliferation of (E) T98G and (F)
U87 cells transfected with miR-186-5p inhibitors, mimics or their
NC, as determined via a Cell Counting kit-8 assay. Proliferation of
(G) T98G and (H) U87 cells transfected with miR-186-5p inhibitors,
mimics or their NC, as determined by an EdU assay (scale bar=100
µm). All data are presented as the means ± standard deviation.
*P<0.05, **P<0.01. EdU, 5-ethynyl-2′-deoxyuridine; GBM,
glioblastoma multiforme; miR-186-5p, microRNA-186-5p; NC, negative
control; RT-qPCR, reverse transcription-quantitative PCR. |
miR-186-5p decreases TMZ resistance in
glioblastoma cells
TMZ has been widely used as a treatment for
glioblastoma. U87 and T98G cells were treated with TMZ (50, 100 and
200 µM) for 24 h; it was demonstrated that miR-186-5p was
significantly downregulated in U87 and T98G cells treated with TMZ
(Fig. 2A and B). Subsequently, the
IC50 values of TMZ in U87 and T98G cells transfected
with miR-186-5p mimics or NC were investigated, and the results
revealed that miR-186-5p significantly decreased the
IC50 values of TMZ in glioblastoma cells (Fig. 2C and D). Furthermore, the apoptosis
of U87 and T98G cells was measured following treatment with 100 µM
TMZ via flow cytometry, and it was observed that miR-186-5p
overexpression significantly increased the number of apoptotic
cells, whereas miR-186-5p inhibition reduced the number of
apoptotic cells (Fig. 2E and F).
The results suggested that downregulation of miR-186-5p contributed
to the development of TMZ resistance in glioblastoma cells.
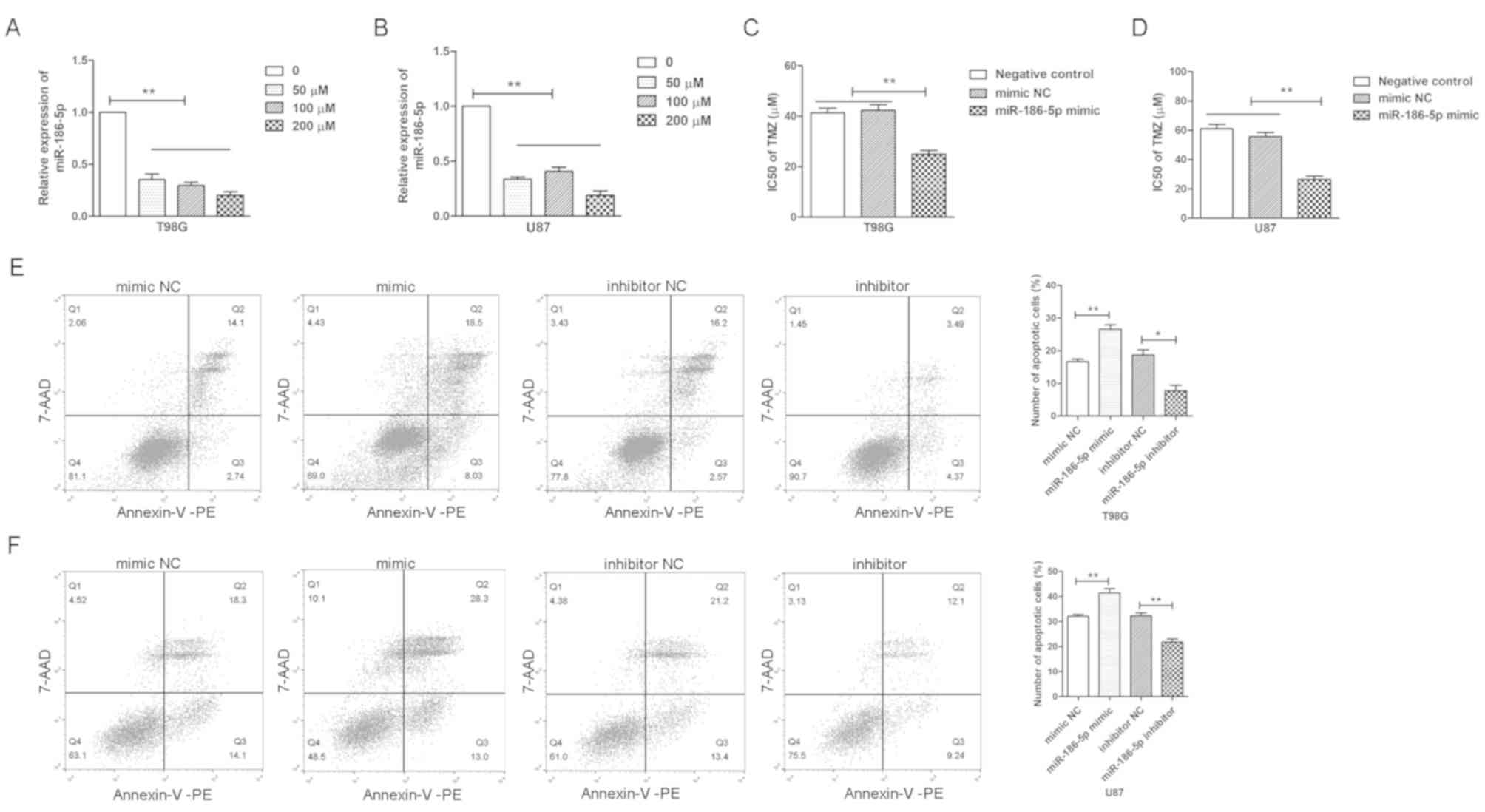 | Figure 2.miR-186-5p reduces the TMZ resistance
of glioblastoma cells. Reverse transcription-quantitative PCR
analysis of the relative expression levels of miR-186-5p in (A)
T98G and (B) U87 cells treated with TMZ (0, 50, 100 and 200 µM) for
24 h. IC50 values of TMZ in (C) T98G and (D) U87 cells
transfected with miR-186-5p mimics, mimic NC or blank control, as
determined via a Cell Counting Kit-8 assay. Flow cytometric
analysis of the apoptosis of (E) T98G and (F) U87 cells transfected
with miR-186-5p inhibitors, mimics or their NC and treated with 100
µM TMZ for 24 h. All data are presented as the means ± standard
deviation. *P<0.05, **P<0.01. 7-AAD, 7-aminoactinomycin; PE,
phycoerythrin; miR-186-5p, microRNA-186-5p; NC, negative control;
TMZ, temozolomide. |
Twist1 is associated with the drug
resistance of glioblastoma cells
It has been reported that Twist1 is upregulated in
glioblastoma and promotes the invasion of glioblastoma cells
(15). In the present study, it
was observed that the expression levels of Twist1 were
significantly higher in U87 and T98G cell lines compared with in
HEB cells (Fig. 3A and B). In
addition, it was demonstrated that Twist mRNA and protein levels
were increased in U87 and T98G cells treated with TMZ compared with
the control (Fig. 3C-F). To
investigate whether Twist1 was involved in the drug resistance of
glioblastoma cells, U87 and T98G cells were transfected with
si-Twist1 or Twist1 overexpression vector. Si-Twist1 significantly
downregulated the expression of Twist1 in U87 and T98G cells,
whereas Twist1 vector upregulated the expression of Twist1
(Fig. 3G and H). Subsequently,
transfected T98G and U87 cells were treated with 100 µM TMZ; it was
observed that the overexpression of Twist1 decreased TMZ-induced
apoptosis, whereas the inhibition of Twist1 increased the number of
apoptotic T98G and U87 cells (Fig. 3I
and J). Twist1 overexpression and knockdown most notably
affected the number of early apoptotic U87 cells (Fig. 3I and J). These data suggested that
Twist1 was involved in the drug resistance of glioblastoma cells,
and that Twist1 may primarily affect the early apoptosis of U87
cells. In addition, it was observed that when cells were treated
with 100 µM TMZ (>IC50; Fig. 2B), the percentage of apoptotic
cells was <50% (Fig. 3J); it
may be that the percentage of dead cells was >50% cells, but
that these dead cells presented as cell debris that were not
detected as apoptotic cells.
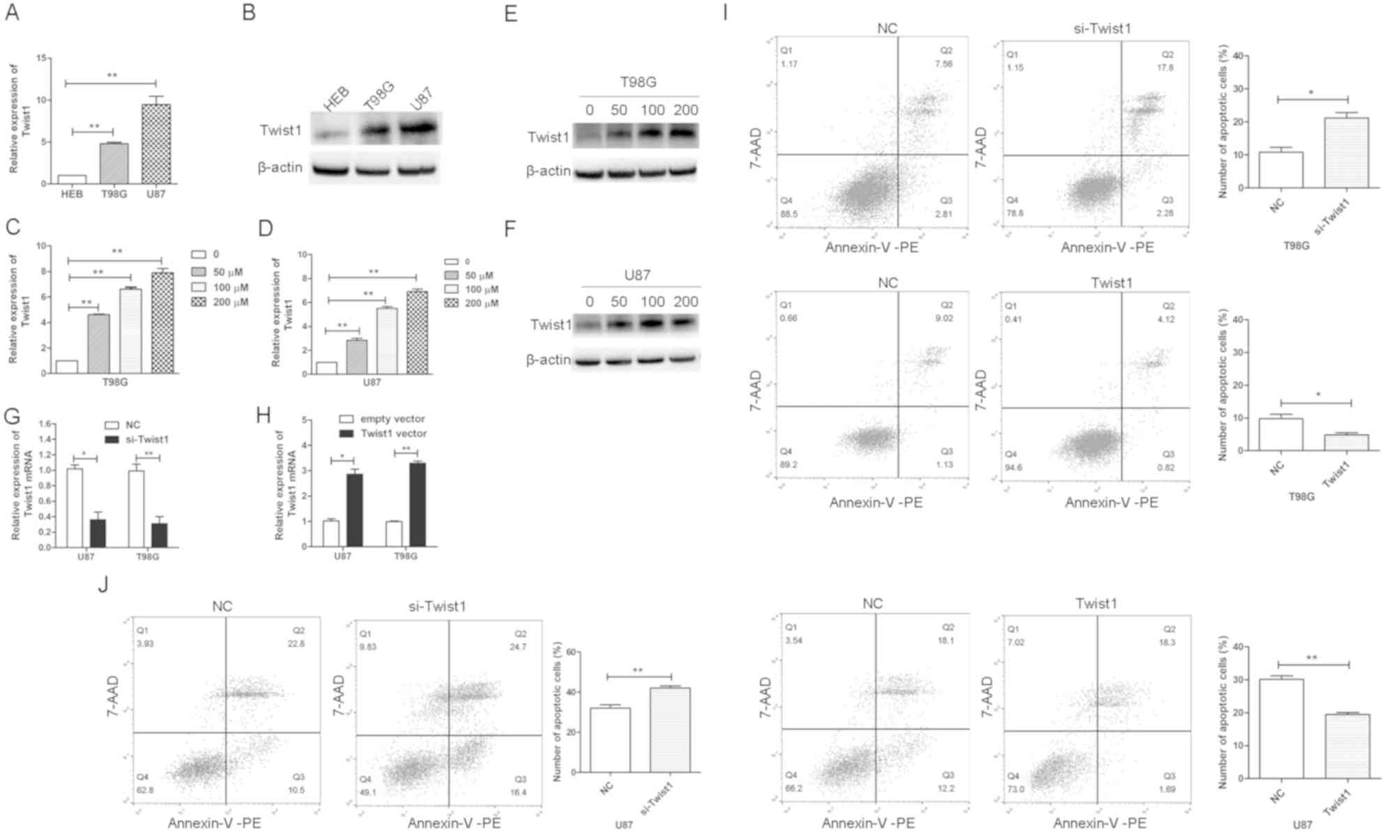 | Figure 3.Twist1 is associated with the TMZ
resistance of glioblastoma cells. (A) Twist1 mRNA and (B) protein
levels in HEB, T98G and U87 cells, as determined by RT-qPCR and
western blot analysis, respectively. (C and D) Twist1 mRNA and (E
and F) protein levels in (C and E) T98G and (D and F) U87 cells
treated with TMZ (0, 50, 100, and 200 µM) for 24 h, as determined
by RT-qPCR and western blot analysis, respectively. Expression of
Twist1 mRNA in U87 and T98G cells transfected with (G) si-Twist1 or
siRNA NC, and (H) Twist1 vector or empty vector. Flow cytometric
analysis of the apoptosis of (I) T98G and (J) U87 cells transfected
with si-Twist1, Twist1 vector or NC, and treated with 100 µM TMZ
for 24 h. All data are presented as the means ± standard deviation.
*P<0.05, **P<0.01. 7-AAD, 7-aminoactinomycin; PE,
phycoerythrin; miR-186-5p, microRNA-186-5p; NC, negative control;
RT-qPCR, reverse transcription-quantitative PCR; si/siRNA, small
interfering RNA; TMZ, temozolomide; Twist1, Twist-related protein
1. |
miR-186-5p contributes to the
proliferation and drug resistance of glioblastoma cells by
targeting Twist1
It was predicted that Twist1 was a potential target
of miR-186-5p. The binding sites of miR-186-5p and Twist1 are
presented in (Fig. 4A). To verify
that Twist1 was a direct target of miR-186-5p, luciferase reporter
assays were conducted, and it was revealed that miR-186-5p
significantly inhibited the luciferase activity of reporters
containing the WT, but not the MUT 3′UTR of Twist1 (Fig. 4B). To determine whether miR-186-5p
regulates the expression of Twist1, glioblastoma cells were
transfected with miR-186-5p mimics with or without Twist1 vector.
The results demonstrated that miR-186-5p transfection significantly
suppressed the expression of Twist1 mRNA and protein, whereas
overexpression of Twist1 significantly reversed the effects of
miR-186-5p mimic (Fig. 4C-F). To
investigate whether miR-186-5p affected the proliferation and drug
resistance of glioblastoma cells by targeting Twist1, the
IC50 of TMZ in, and proliferation of U87 and T98G cells
following transfection with miR-186-5p mimics with or without Twist
vector was determined. Overexpression of miR-186-5p significantly
decreased cell proliferation and the IC50 of TMZ,
whereas the overexpression of Twist1 reversed the effects of
miR-186-5p on cell proliferation and the IC50 of TMZ in
glioblastoma cells (Fig. 4G-J).
The results indicated that Twist1 may be a functional target of
miR-186-5p.
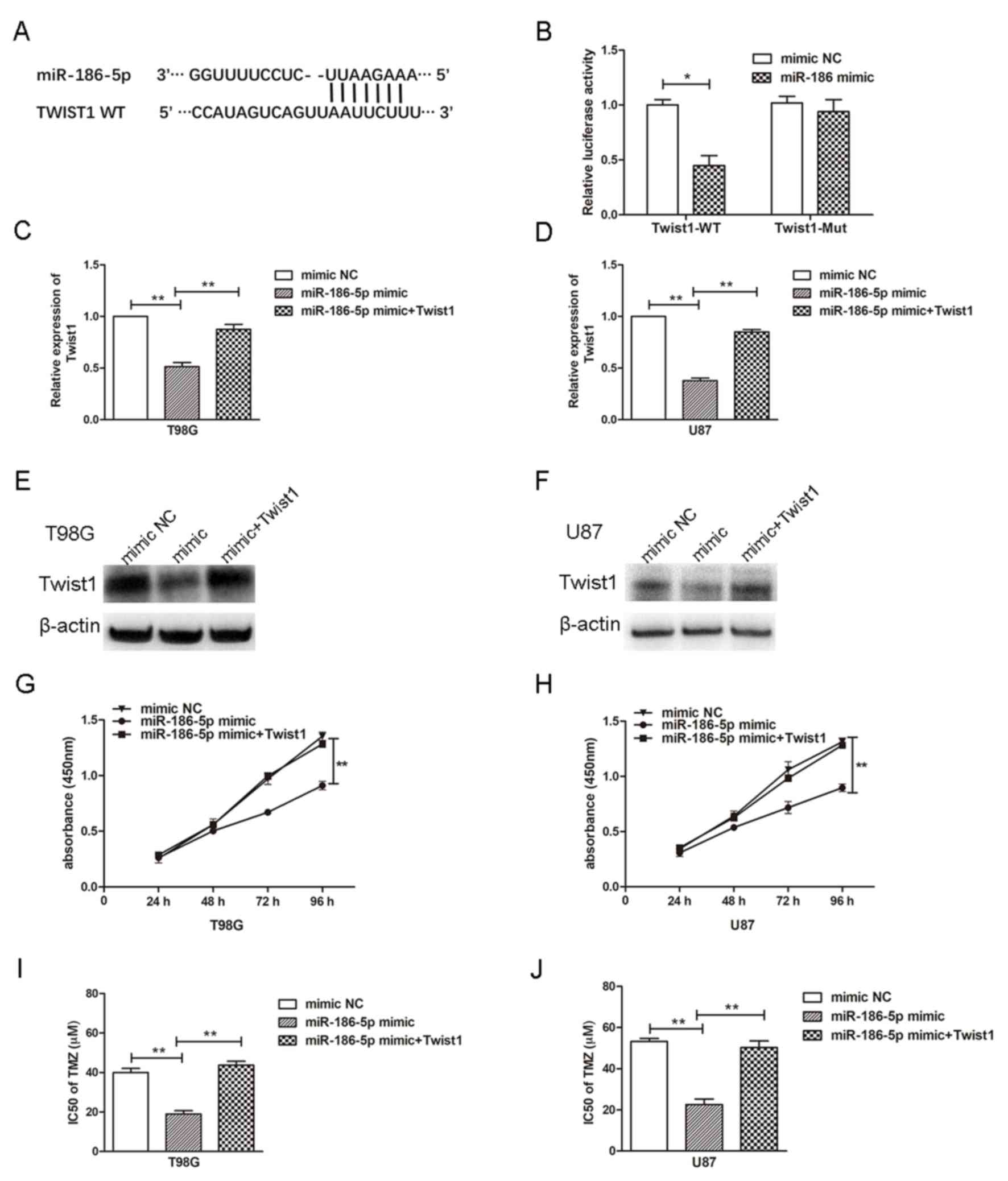 | Figure 4.miR-186-5p contributes to the
proliferation and drug resistance of glioblastoma cells by
targeting Twist1. (A) Potential binding sites of miR-186-5p in the
3′UTR of Twist1. (B) 293T cells were co-transfected with miR-186-5p
and reporter plasmids containing the WT or MUT 3′UTR of Twist1.
Expression of Twist1 mRNA in (C) T98G and (D) U87 cells transfected
with miR-186-5p mimic with or without Twist1 vector, as determined
via reverse transcription-quantitative PCR. Expression of Twist1
protein in (E) T98G and (F) U87 cells transfected with miR-186-5p
mimic with or without Twist1 vector, as determined by western blot
analysis. Proliferation of (G) T98G and (H) U87 cells transfected
with miR-186-5p mimic with or without Twist1 vector, as determined
via a CCK-8 assay. IC50 values of TMZ in (I) T98G and
(J) U87 cells transfected with miR-186-5p mimic with or without
Twist1 vector as determined via a CCK-8 assay. All data are
presented as the means ± standard deviation. *P<0.05,
**P<0.01. CCK-8, Cell Counting kit-8; miR-186-5p,
microRNA-186-5p; MUT, mutant; NC, negative control; Twist1,
Twist-related protein 1; 3′-UTR, 3′-untranslated region; WT,
wild-type. |
Discussion
In the present study, it was demonstrated that
miR-186-5p was downregulated in glioblastoma cell lines and
tissues. Silencing of miR-186-5p increased the proliferation and
decreased the apoptosis of glioblastoma cells, whereas ectopic
overexpression of miR-186-5p induced opposing effects. Recently,
Jiang et al (18) also
reported that miR-186 is downregulated in glioblastoma tissue, and
that upregulation of miR-186 decreases cell proliferation. In
addition, it was demonstrated that miR-186-5p may be involved in
the TMZ resistance of glioblastoma cells. The present findings
indicated that miR-186-5p served a role in the proliferation and
TMZ resistance of glioma cells via the direct targeting of Twist1.
Conversely, Jiang et al (18) reported insulin-like growth factor 1
receptor to be a target of miR-186; therefore, Twist1 may not be
the only functionally relevant target of miR-186-5p in
glioblastoma.
miR-186 has previously been investigated in various
tumours. In NSCLC, miR-186 expression is downregulated compared
with in normal lung tissues or epithelial cell lines, and it is
associated with patient survival (11). Further studies have indicated that
overexpression of miR-186 in NSCLC cells inhibits proliferation by
inducing G1/S checkpoint arrest (11). Conversely, it has been reported
that miR-186 is a tumour-promoting miRNA in endometrial cancer and
hepatic carcinoma by targeting the tumour suppressors forkhead box
protein O1 and A-kinase anchoring protein 12, respectively
(19,20). The present study suggested that
miR-186-5p acts as a tumour suppressor in glioblastoma, and that
Twist1 was the functional target of miR-186-5p.
Drug resistance is the main reason for the failure
of chemotherapeutic treatment of glioma. Numerous studies have
reported that certain miRNAs, including miR-634, miR-205 and
miR-497, contribute to drug resistance in glioma (17,21,22).
For example, miR-497 is upregulated in glioma and hypoxia induces
the expression of miR-497. Ectopic overexpression of miR-497
decreases the sensitivity of glioma cells to TMZ by targeting
programmed cell death protein 4 (17). To the best of our knowledge, the
present study was the first to report that downregulation of
miR-186-5p in GBM promoted the proliferation and drug resistance of
glioma cells.
It has been demonstrated that miR-186 serves a role
in cell biological processes by inhibiting the expression of its
downstream targets, including β-secretase 1, collagen type V α1,
casein kinase 2α and Twist1 (9,23–25).
In the present study, Twist1 was identified as a direct downstream
target of miR-186-5p. It was demonstrated that miR-186-5p
overexpression downregulated the expression of Twist1 in glioma
cells. Ectopic expression of miR-186-5p in glioma cells reduced
cell proliferation, and the IC50 of TMZ and the levels
of Twist1 in U87 and T98G cells. Conversely, overexpression of
Twist1 rescued the miR-186-5p-induced decrease in cell
proliferation and the IC50 of TMZ. These data suggested
that miR-186-5p participated in the proliferation and drug
resistance of glioblastoma cells by targeting Twist1. The results
further indicated that Twist1 may primarily affect the early
apoptosis of U87 cells; however, it was observed that miR-186-5p
inhibition or overexpression notably affected the number of early
and late apoptotic U87 cells. This may be due to miR-186-5p
influencing the apoptosis of U87 cells by regulating the expression
of other genes aside from Twist1.
It has been reported that Twist1, a
basic-helix-loop-helix transcription factor, serves a role in the
regulation of EMT, cell proliferation, cell migration, stem cell
self-renewal, tumour initiation, senescence and drug resistance
(12,26,27).
Twist1 is overexpressed in various types of tumour, including
glioma, skin tumours, breast and ovarian cancer, and melanoma
(15,28,29).
Mechanistically, Twist1 has been revealed to prevent apoptosis or
senescence by inhibiting key regulators of the p16 and/or p53
pathways (14,26); however, in the study, the
mechanisms by which Twist1 regulated apoptosis and proliferation
were not investigated.
In conclusion, the findings of the present study
revealed that miR-186-5p was downregulated in glioblastoma cells
and tissues, and functioned as a tumour suppressor. Biological or
pharmacological intervention based on miR-186-5p may have
therapeutic potential in reversing drug resistance and improving
chemotherapeutic efficacy in human glioblastoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Jiangxi Province (grant no. 20161BAB215230)
and the Science and Technology Research Project of Jiangxi Province
Education Department (grant no. GJJ150119).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CW conceived and designed the experiments; YX and RC
performed the experiments; LW and SW analyzed and interpreted the
data, YT and LZ contributed to the analysis of data and drafted the
manuscript. All authors gave final approval of the version to be
published.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
The First Affiliated Hospital of Nanchang University and protocols
were conducted according to the principles of the Declaration of
Helsinki. Written informed consent was obtained from all patients
in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lathia JD, Mack SC, Mulkearns-Hubert EE,
Valentim CL and Rich JN: Cancer stem cells in glioblastoma. Genes
Dev. 29:1203–1217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim SS, Harford JB, Pirollo KF and Chang
EH: Effective treatment of glioblastoma requires crossing the
blood-brain barrier and targeting tumors including cancer stem
cells: The promise of nanomedicine. Biochem Biophys Res Commun.
468:485–489. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mirimanoff RO: High-grade gliomas: Reality
and hopes. Chin J Cancer. 33:1–3. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grauer OM, Wesseling P and Adema GJ:
Immunotherapy of diffuse gliomas: Biological background, current
status and future developments. Brain Pathol. 19:674–693. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Patil SA, Hosni-Ahmed A, Jones TS, Patil
R, Pfeffer LM and Miller DD: Novel approaches to glioma drug design
and drug screening. Expert Opin Drug Discov. 8:1135–1151. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sarkar FH, Li Y, Wang Z, Kong D and Ali S:
Implication of microRNAs in drug resistance for designing novel
cancer therapy. Drug Resist Updat. 13:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu X, Shen H, Yin X, Long L, Xie C, Liu
Y, Hui L, Lin X, Fang Y, Cao Y, et al: miR-186 regulation of Twist1
and ovarian cancer sensitivity to cisplatin. Oncogene. 35:323–332.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Erdmann K, Kaulke K, Thomae C, Huebner D,
Sergon M, Froehner M, Wirth MP and Fuessel S: Elevated expression
of prostate cancer-associated genes is linked to down-regulation of
microRNAs. BMC Cancer. 14:822014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai J, Wu J, Zhang H, Fang L, Huang Y,
Yang Y, Zhu X, Li R and Li M: miR-186 downregulation correlates
with poor survival in lung adenocarcinoma, where it interferes with
cell-cycle regulation. Cancer Res. 73:756–766. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X, Ling MT, Guan XY, Tsao SW, Cheung
HW, Lee DT and Wong YC: Identification of a novel function of
TWIST, a bHLH protein, in the development of acquired taxol
resistance in human cancer cells. Oncogene. 23:474–482. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maestro R, Dei Tos AP, Hamamori Y,
Krasnokutsky S, Sartorelli V, Kedes L, Doglioni C, Beach DH and
Hannon GJ: Twist is a potential oncogene that inhibits apoptosis.
Genes Dev. 13:2207–2217. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mikheeva SA, Mikheev AM, Petit A, Beyer R,
Oxford RG, Khorasani L, Maxwell JP, Glackin CA, Wakimoto H,
González-Herrero I, et al: TWIST1 promotes invasion through
mesenchymal change in human glioblastoma. Mol Cancer. 9:1942010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lan J, Xue Y, Chen H, Zhao S, Wu Z, Fang
J, Han C and Lou M: Hypoxia-induced miR-497 decreases glioma cell
sensitivity to TMZ by inhibiting apoptosis. FEBS Lett.
588:3333–3339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang J, Wang W, Fang D, Jin X, Ding L and
Sun X: MicroRNA186 targets IGF-1R and exerts tumor-suppressing
functions in glioma. Mol Med Rep. 16:7821–7828. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Myatt SS, Wang J, Monteiro LJ, Christian
M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S and Lam EW:
Definition of microRNAs that repress expression of the tumor
suppressor gene FOXO1 in endometrial cancer. Cancer Res.
70:367–377. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goeppert B, Schmezer P, Dutruel C, Oakes
C, Renner M, Breinig M, Warth A, Vogel MN, Mittelbronn M, Mehrabi
A, et al: Down-regulation of tumor suppressor A kinase anchor
protein 12 in human hepatocarcinogenesis by epigenetic mechanisms.
Hepatology. 52:2023–2033. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tan Z, Zhao J and Jiang Y: MiR-634
sensitizes glioma cells to temozolomide by targeting CYR61 through
Raf-ERK signaling pathway. Cancer Med. 7:913–921. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li FF, Xing C, Wu LL and Xue F: MiR-205
enhances cisplatin sensitivity of glioma cells by targeting E2F1.
Eur Rev Med Pharmacol Sci. 22:299–306. 2018.PubMed/NCBI
|
|
23
|
Kim J, Yoon H, Chung DE, Brown JL,
Belmonte KC and Kim J: miR-186 is decreased in aged brain and
suppresses BACE1 expression. J Neurochem. 137:436–445. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lei GS, Kline HL, Lee CH, Wilkes DS and
Zhang C: Regulation of Collagen V expression and
epithelial-mesenchymal transition by miR-185 and miR-186 during
idiopathic pulmonary fibrosis. Am J Pathol. 186:2310–2316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee YH, Kim SY and Bae YS: Upregulation of
miR-760 and miR-186 is associated with replicative senescence in
human lung fibroblast cells. Mol Cells. 37:620–627. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee KE and Bar-Sagi D: Oncogenic KRas
suppresses inflammation-associated senescence of pancreatic ductal
cells. Cancer Cell. 18:448–458. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Beck B, Lapouge G, Rorive S, Drogat B,
Desaedelaere K, Delafaille S, Dubois C, Salmon I, Willekens K,
Marine JC and Blanpain C: Different levels of Twist1 regulate skin
tumor initiation, stemness, and progression. Cell Stem Cell.
16:67–79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ansieau S, Morel AP, Hinkal G, Bastid J
and Puisieux A: TWISTing an embryonic transcription factor into an
oncoprotein. Oncogene. 29:3173–3184. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nordfors K, Haapasalo J, Makela K,
Granberg KJ, Nykter M, Korja M, Paavonen T, Haapasalo H and Soini
Y: Twist predicts poor outcome of patients with astrocytic glioma.
J Clin Pathol. 68:905–912. 2015. View Article : Google Scholar : PubMed/NCBI
|


















