Introduction
Polycystic ovary syndrome (PCOS) is a common
endocrine disorder with an incidence of 6–21% (1,2).
PCOS affects women of reproductive age and symptoms include light
menstruation or even amenorrhea, hyperandrogenemia and polycystic
ovarian morphology in addition to metabolic disorders, including
insulin resistance, diabetes and obesity (3). PCOS is a primary cause of female
infertility due to a failure in oocyte-follicle maturation
(4).
Granulosa cells (GCs) serve an important role in
oocyte development and maintenance of the oocyte microenvironment
by secreting steroid hormones and producing growth hormones.
Additionally, GCs are the primary sites of estrogen synthesis and
provide endocrine signaling to other tissues (5). A previous study demonstrated that
dysfunction of GCs contributes to abnormal folliculogenesis in
patients with PCOS (6). GCs
provide nutrients and growth regulators to the oocyte. Therefore,
it is important to investigate the role of GCs in the development
of PCOS.
Previous studies have indicated that numerous genes
participate in the pathogenesis of PCOS (7–11). A
previous study revealed that co-activators of estrogen receptors,
including nuclear receptor co-activator (NCOA) 1, NCOA2, NCOA3,
CREB binding protein, histone acetyltransferase p300, lysince
acetyltransferase 2B and coactivator associated arginine
methyltransferase 1, as well as co-repressors such as nuclear
receptor corepressor 1 and nuclear receptor corepressor 2, were
deregulated in cumulus cells from patients with PCOS, which may
contribute to the etiology of PCOS (7). Jansen et al (8) demonstrated that androgen serves an
important role in the pathogenesis of PCOS through a microarray
analysis of PCOS compared with normal ovaries. In subcutaneous
adipose tissue from PCOS, C-C motif ligand 2 and heme oxygenase 1
were expressed at high levels, while adiponectin receptor 2,
lipoprotein lipase and twist-related protein 1 were expressed at
low levels. These molecules were associated with lipid metabolism,
insulin sensitivity, inflammation and oxidative stress (9). However, how these genes are
post-transcriptionally regulated is poorly understood.
An improved understanding regarding the regulation
of genes has resulted from the identification of microRNAs (miRNAs
or miRs). These small, non-coding RNAs of 20–22 nucleotides in
length are capable of inhibiting protein translation or degrading
target mRNAs in multiple biological and pathological processes,
including in cell proliferation, apoptosis, differentiation, cell
cycle control and metabolism (12,13).
Kim et al (14)
demonstrated that miR-27a, miR-let-7c and miR-322 can regulate the
meiotic competence of oocytes during the in vitro maturation
of mouse follicles. A previous study indicated that different
miRNAs are expressed in cumulus cells in patients with PCOS
compared with healthy controls (15). A number of previous studies have
also reported that miRNAs serve important roles in ovarian function
and GC apoptosis (16–18). Abnormal expression of miRNAs is
also associated with several reproductive disorders (19–22).
Murri et al (23)
identified that circulating miRNA-21, miRNA-27b, miRNA-103 and
miRNA-155 are differentially expressed between patients with PCOS
and healthy controls, and are involved in the pathogenesis of PCOS,
including inflammatory processes, hormone metabolism and insulin
signaling. Furthermore, Shi et al (24) indicated that miR-483-5p is
important to reduce insulin resistance, while miR-486-5p may
promote cumulus cell proliferation by activating PI3K/AKT signaling
in patients with PCOS. Jiang et al (25) reported that miRNA-93 could promote
ovarian GC proliferation by targeting cyclin-dependent kinase
inhibitor 1A in patients with PCOS. Zhu et al (26) indicated that miRNA-34a promoted
ovarian GC apoptosis by inhibiting B cell lymphoma-2 (Bcl-2) and
increasing the expression of Bcl-2-associated X protein and
caspase-3. However, to date, little is known about the miRNA
profiles in GCs in patients with PCOS.
The present study screened for differentially
expressed miRNAs (DE-miRNAs) from the dataset GSE84376. Target
genes of the DE-miRNAs were predicted and their potential functions
were analyzed by Gene Ontology (GO) enrichment analysis.
Furthermore, a protein-protein interaction (PPI) network was
constructed to identify hub genes. Finally, screening was conducted
to identify transcription factors that may regulate the target
genes. The present bioinformatics study on miRNAs may provide a
novel perspective into the pathological mechanism of PCOS.
Materials and methods
miRNA microarray analysis
The present study used ‘miRNA, granulosa cells,
PCOS, human’ as key words to extract expression data for GCs from
patients with PCOS and controls in the National Center for
Biotechnology Information Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84376;
accessed on December 2017). The dataset GSE84376, based on
Affymetrix Multispecies miRNA-3 Array platform (Affymetrix; Thermo
Fisher Scientific, Inc.) contained 15 cases of PCOS and 13 cases of
non-PCOS. The raw data were uploaded to the website of Gene-Cloud
of Biotechnology Information (GCBI; https://www.gcbi.com.cn/gclab/html/index; Genminix
Informatics Co., Ltd.) for analysis. GCBI, a good online
bioinformatics analysis platform, combines a variety of sample
information, research findings, genetic information, bioinformatics
and data algorithms to create a ‘gene knowledge base’. The GCBI
platform can be used to systematically analyze GEO dataset-derived
gene expression information. Cluster analysis was performed on
DE-miRNAs using a fold-change >2 and false discovery rate ≤0.05
as cut off values.
Prediction of target genes
Data mining tools, including three different target
prediction databases [TargetScan (http://www.targetscan.org/vert_71/), DIANA-microT-CDS
(http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=site/page&view=software)
and miRDB (http://mirdb.org/miRDB/)] and a
database containing validated targets (miRTarBase; http://mirtarbase.mbc.nctu.edu.tw/php/index.php) were
used to predict target genes of DE-miRNAs. According to the
parameters set for each bioinformatics tool, the predicted genes
were selected by ≥3 algorithms. Then, target genes were illustrated
from each tool using Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Gene ontology (GO) enrichment
analysis
GO was used for gene functional enrichment analysis
using Metascape software (http://metascape.org). The resulting GO terms with
P<0.05 were considered significantly enriched in the
differentially expressed genes.
PPI network
To further investigate the pathogenesis of PCOS, a
PPI network was constructed using Cytoscape software (version
3.6.1) (27). An integrated score
>0.4 [the default threshold in the STRING database; http://www.string-db.org/ (28)] was defined to construct the PPI
network. Then, the topological structure of the network was
analyzed and the degree for each gene was calculated. Hub genes
were defined as those with degree ≥10.
Screening of potential transcription
factors
The study identified the enriched transcription
factors regulating target genes of DE-miRNA using FunRich software
(version 3.1.3; http://www.funrich.org) (29), a functional enrichment and
interaction network analysis tool.
Enrollment of subjects
All the patients enrolled in the present study
underwent in vitro fertilization (IVF)/intracytoplasmic
sperm injection-embryo transfer at the Center of Reproductive
Medicine, Children's Hospital of Shanxi and Women Health Center of
Shanxi between January 2018 to July 2018. According to the 2003
Rotterdam Revised Criteria for PCOS (30), 38 patients with PCOS and 35 patient
controls with male or tubal infertility factors were enrolled into
the present study. Patients with prolactinoma, congenital adrenal
hyperplasia, thyroid disorder, Cushing syndrome and adrenal tumors
were excluded. The subjects did not receive any hormonal treatments
3 months prior to enrollment. The present study was approved by the
Ethics Committee of Children's Hospital of Shanxi and Women Health
Center of Shanxi. Written informed consent for inclusion was
obtained from each participant.
The demographic and clinical data, including age,
infertility duration, height, weight and serum levels of sex
hormones [luteinizing hormone (LH), follicle-stimulating hormone
(FSH), estrogen (E2), progesterone (P) and testosterone (T)] were
recorded (Table I). Reproductive
laboratory data were also obtained, including the number of
retrieved oocytes, metaphase II (MII) oocytes, fertilized oocytes,
cleavage rate and embryos.
 | Table I.Comparison of clinical
characteristics between the PCOS and control groups. |
Table I.
Comparison of clinical
characteristics between the PCOS and control groups.
| Variables | PCOS group
(n=38) | Control group
(n=35) | P-value |
|---|
| Age (year) | 29.60±0.66 | 29.66±0.82 | 0.960 |
| Infertility
duration (year) | 4.62±0.50 | 4.64±0.55 | 0.974 |
| Menarche age
(year) | 13.29±0.17 | 13.46±0.18 | 0.493 |
| Body mass index
(Kg/m2) | 25.25±0.59 | 22.63±0.53 | 0.002 |
| Basal hormone
levels |
|
|
|
| LH
(mIU/ml) | 8.10±0.69 | 5.50±0.59 | 0.005 |
| FSH
(mIU/ml) | 6.94±0.32 | 8.71±0.61 | 0.013 |
| E2
(pg/ml) | 90.43±11.85 | 88.80±12.95 | 0.926 |
| P
(ng/ml) | 0.62±0.09 | 0.50±0.06 | 0.275 |
| T
(ng/ml) | 45.09±3.66 | 41.58±3.60 | 0.498 |
|
LH/FSH | 1.21±0.10 | 0.66±0.07 | <0.001 |
| IVF outcomes | | |
|
| No. of
oocytes retrieved | 25.53±2.22 | 17.09±1.58 | 0.003 |
| MII
oocyte rate | 0.75±0.03 | 0.82±0.03 | 0.093 |
|
Fertilization rate | 0.72±0.03 | 0.81±0.02 | 0.012 |
|
Cleavage rate | 0.85±0.03 | 0.92±0.02 | 0.049 |
| Embryo
rate | 0.57±0.03 | 0.57±0.04 | 0.909 |
Isolation of GCs from follicular
fluid
GCs were collected from the follicular fluid during
oocyte collection 34–36 h after human chorionic gonadotrophin (hCG)
injection. The cumulus-oocyte complexes were used for IVF. The
remaining follicular fluid was transferred to 4°C within 1 h. The
follicular fluid was prepared by centrifugation at 400 × g for 5
min followed by layering on 40% Percoll® (Sigma-Aldrich;
Merck KGaA) at room temperature. Following this, three layers could
be distinguished; the middle ring-like layer was collected and
washed by centrifugation at 400 × g for 10 min. The pellet was
resuspended with 1X PBS and red blood cell lysate was added to a
volumetric ratio of 1:3 and kept at room temperature for 10 min.
Following centrifugation at 2,860 × g for 1 min at room
temperature, three wash steps were carried out using PBS and
centrifugation at 2,860 × g for 1 min at room temperature, and GCs
were collected.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
Total RNA was extracted from the purified human GCs
using an miRNeasy Mini kit (Qiagen GmbH) according to the
manufacturer's protocol. cDNA synthesis was performed using the
miScript II RT kit (Qiagen GmbH) following the manufacturer's
temperature protocol: 37°C for 60 min and 95°C for 5 min. PCR,
using miR-specific primers and universal adaptor PCR primers
(iGeneBio Co; GeneCopoeia, Inc.), was performed using a miScript
SYBR Green PCR kit (Qiagen GmbH) according to the manufacturer's
protocol using a CFX Connect Real-time System (Bio-Rad
Laboratories, Inc.). The miR-specific primers were as follows:
Hsa-miR-3135b AGCGAGTGCAGTGGTGAAA; hsa-miR-4433-3p
GAGTGGGGGTGGGACATAAA; hsa-miR-3188 GTGCGGATACGGGGAAAA; hsa-miR-1587
TGGGCTGGGTTGGGAAA; hsa-miR-1225-5p GGCCCAGTGGGGGGAA;
hsa-miR-4749-5p ACAGGCCAGGGCATCAAA; hsa-miR-4417 GGCTTCCCGGAGGGAAA.
The PCR conditions were as follows: 95°C pre-denaturation for 15
min, followed by 40 cycles of denaturation at 94°C for 15 sec,
annealing at 55°C for 30 sec and extension at 70°C for 30 sec.
Relative gene expression was analyzed using the 2−ΔΔCq
method (31). U6 small nuclear RNA
was used as a miRNA internal control. miRNA and U6 (cat. no.
HmiRQP9001) primers were purchased from iGeneBio (GeneCopoeia,
Inc.).
Statistical analysis
Statistical analyses were performed using SPSS
version 17.0 (SPSS, Inc.). Differences between groups were assessed
by Student's t-test (2-sided). Pearson's correlation coefficient
was used to assess correlations between variables. All quantitative
results from at least three independent experiments are presented
as the mean ± SEM. A two-tailed P<0.05 was considered to
indicate a statistically significant difference. All statistical
graphs were generated using GraphPad Prism 6.0 (GraphPad Software,
Inc.).
Results
Identification of DE-miRNAs and their
target genes
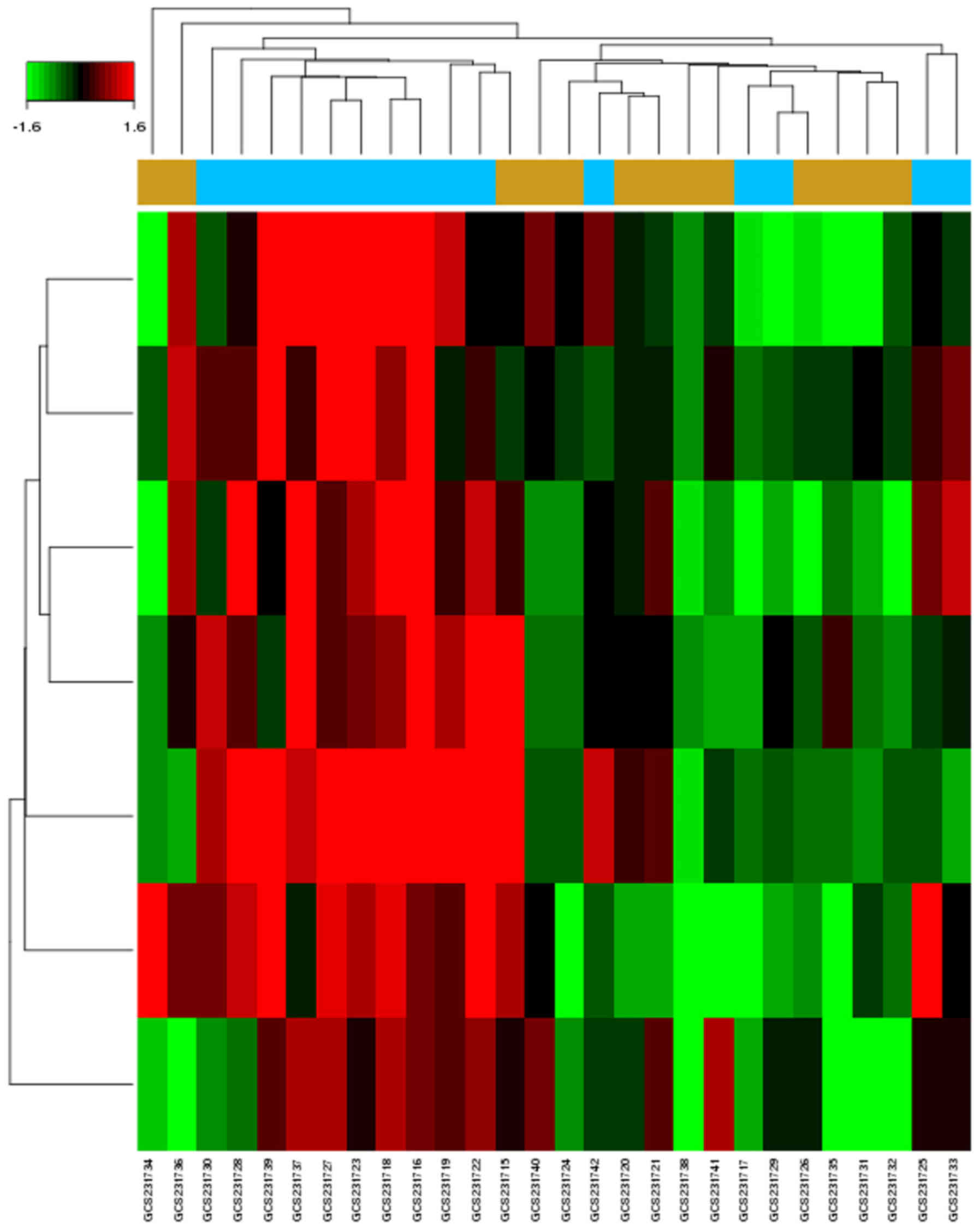
There were seven upregulated DE-miRNAs in GCs of
patients with PCOS identified from the microarray dataset
(GSE84376; Fig. 1). The dataset
contained 15 patients with PCOS and 13 control individuals with
male factor infertility. The computational analysis of DE-miRNA
target genes based on TargetScan, DIANA-microT-CDS, miRDB and
miRTarBase identified 935 genes (Table II). Using a combination of
algorithms, this approach provided a more accurate prediction of
target genes.
 | Table II.Differentially expressed miRNAs in
polycystic ovary syndrome and their target genes. |
Table II.
Differentially expressed miRNAs in
polycystic ovary syndrome and their target genes.
| miRNAs | Fold change
[Log(Fold Change)] | P-value | Target gene (number
of total target genes) |
|---|
| hsa-miR-3188 | 2.781063 | 0.003592 | HIF1AN ESPL1 SRCAP
ZSCAN25 TMBIM6 TFCP2L1 CSNK2A1 SDHAF2 MTMR3 RPLP1 (131) |
|
hsa-miR-4433-3p | 2.347565 | 0.003906 | TNRC6B BZW1 GPR63
DDX52 TMEM59 STK4 BICD2 TCHHL1 CYB561A3 OLR1 (205) |
| hsa-miR-3135b | 2.26603 | 0.006199 | YIPF4 ANKRD36 CUBN
METTL2B INTS3SIK2 FBXO27 CCDC142 DNAJC10 ST3GAL1 (313) |
| hsa-miR-1587 | 1.66782 | 0.007895 | PPM1N SLC2A8 DAO
RAD54L2 FBXL16 CD1B CHRFAM7A GIPC3 PLK2 ZNF609 (140) |
|
hsa-miR-1225-5p | 1.726874 | 0.008972 | PIFO ZNF37A KANSL1L
KIAA2022 COL8A1 FAM149B1 ERO1LB KIAA0196 PSME4 ZCCHC12 (55) |
| hsa-miR-4417 | 2.174213 | 0.009638 | ORAI2 MLF2 CHST2
ATP6V1B1 MTSS1 SMEK2 STK38 SYCE1 LSM12 KIR2DL4 (73) |
|
hsa-miR-4749-5p | 2.063519 | 0.009914 | NFIX SAP18 EMID1
GPRC5B ZNF558 FOXRED2 FOXE1 ABCE1 TMOD3 FAM84A (18) |
Functional characterization of target
genes
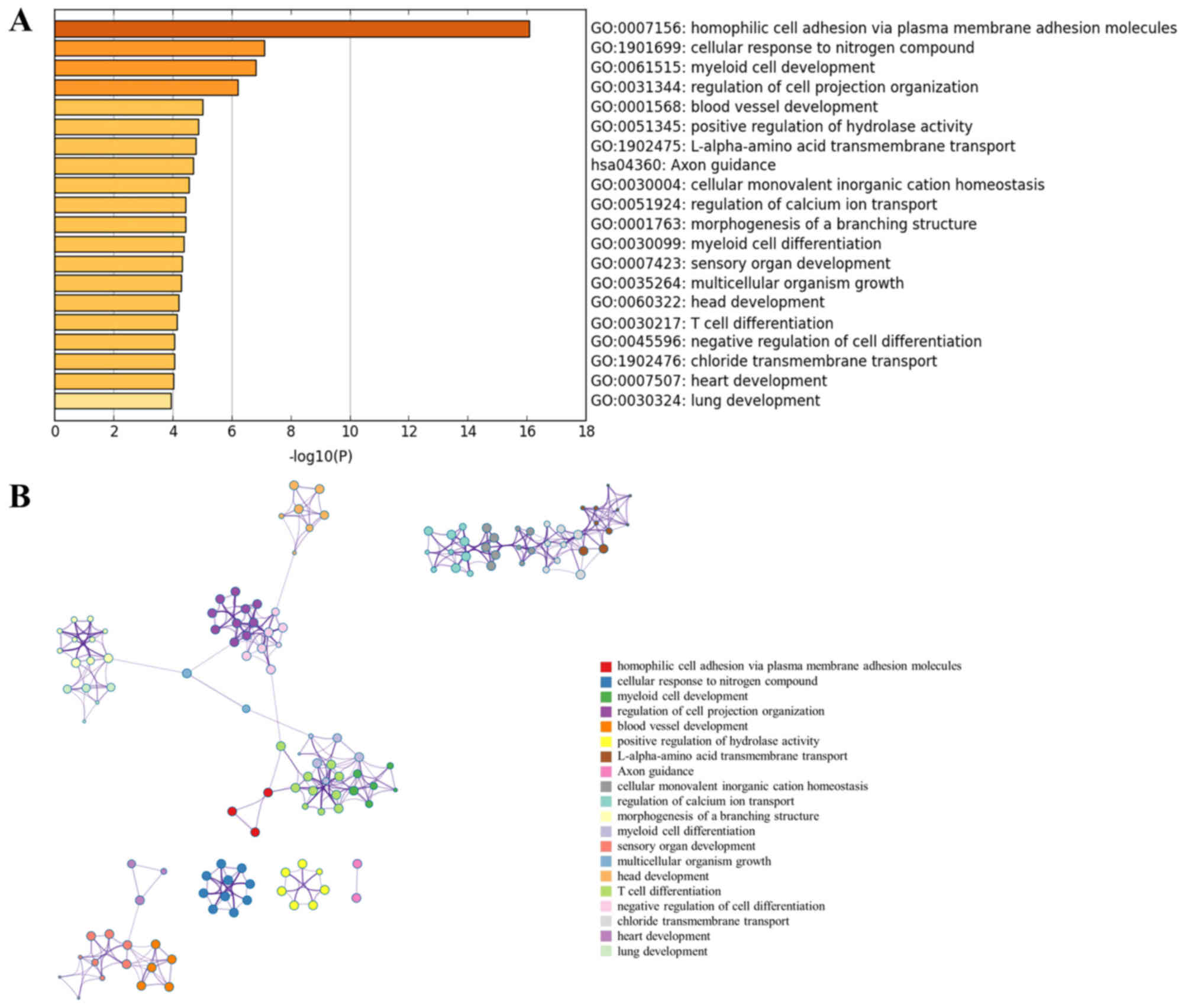
GO enrichment analysis was performed for the
DE-miRNA target genes using Metascape software. The most
significantly enriched gene set was ‘homophilic cell adhesion via
plasma membrane adhesion molecules’ (GO:0007156) (Fig. 2A). The analysis also revealed that
PCOS was associated with ‘cellular response to nitrogen compound’
(GO:1901699), ‘blood vessel development’ (GO:0001568), ‘positive
regulation of hydrolase activity’ (GO:0051345), ‘Axon guidance’
(hsa04360), ‘regulation of calcium ion transport’ (GO:0051924) and
‘chloride transmembrane transport’ (GO:1902476). Notably, all these
biological processes were closely interrelated (Fig. 2B).
PPI network analysis and miRNA-target
network
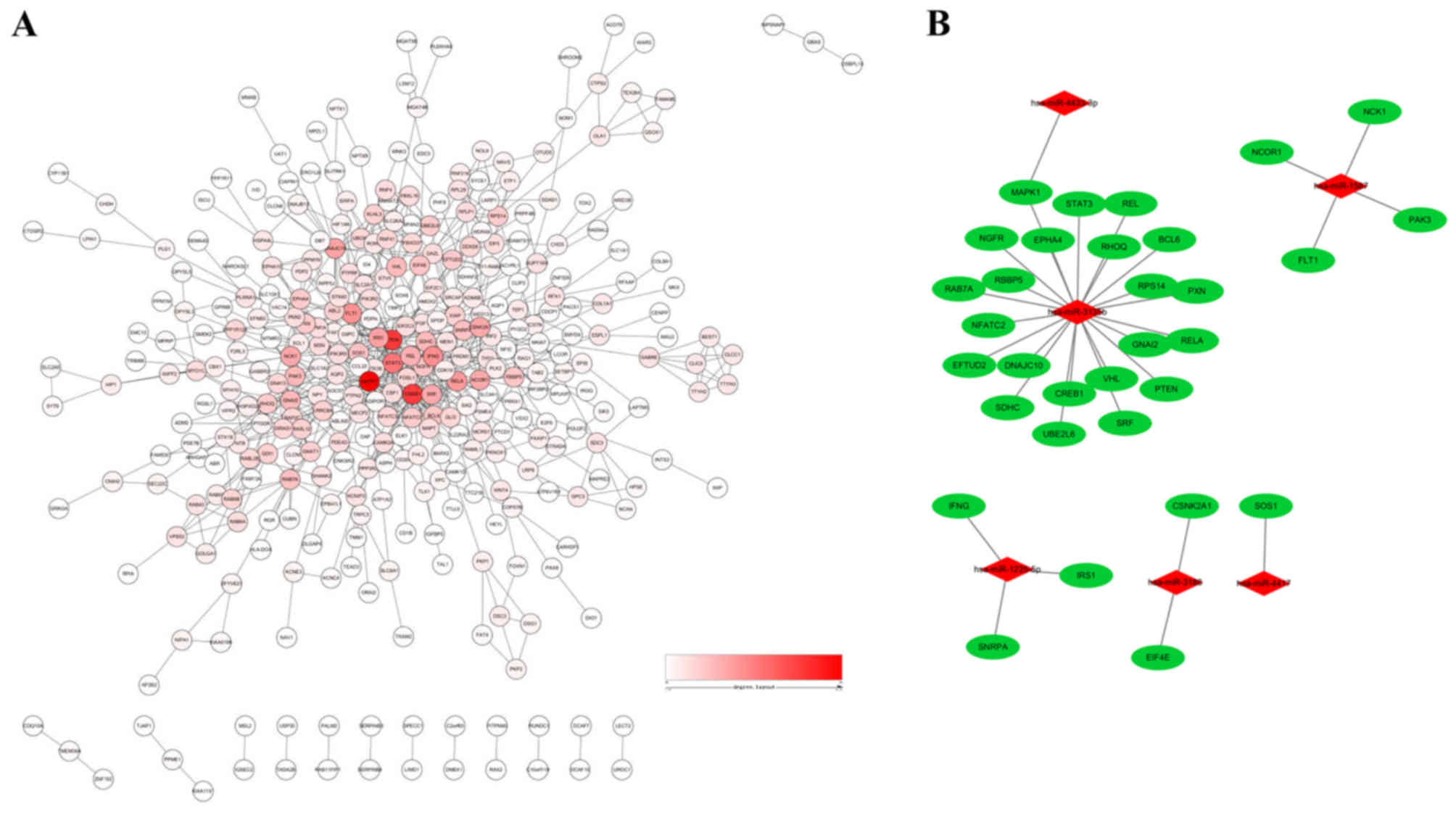
To reduce interference of unrelated genes and to
reduce the complexity of the list of PCOS-associated genes, a PPI
network was constructed. A total of 32 genes were identified as hub
genes with an interaction degree ≥10 (Fig. 3A and Table III). These hub genes were targets
of PCOS-associated miRNAs (Fig.
3B). Of these hub genes, mitogen-activated protein kinase 1
(MAPK1) had the highest degree of 46, followed by phosphatase and
tensin homolog (PTEN) and cAMP responsive element binding protein 1
(CREB1), with degrees of 36 and 35, respectively. The hub genes
identified by the PPI network analysis may serve as potential
targets for future research into PCOS treatment. As shown in
Fig. 3B, a miRNA-hub gene network
was constructed. The hub target genes may be regulated by Homo
sapiens (hsa)-miR-3135b, hsa-miR-1225-5p, hsa-miR-1587,
hsa-miR-3188, hsa-miR-4433-3p and hsa-miR-4417, with hsa-miR-3135b
having the highest correlation degree.
 | Table III.Hub genes identified in the protein
interaction network. |
Table III.
Hub genes identified in the protein
interaction network.
| Name | Degree | Betweenness | Closeness |
|---|
| MAPK1 | 46 | 0.23654846 | 0.39900249 |
| PTEN | 36 | 0.22614707 | 0.39653036 |
| CREB1 | 35 | 0.18022213 | 0.3874092 |
| STAT3 | 25 | 0.05874048 | 0.3567447 |
| IFNG | 21 | 0.07357852 | 0.33264033 |
| FLT1 | 20 | 0.06162387 | 0.35087719 |
| RELA | 19 | 0.03995804 | 0.34042553 |
| IRS1 | 17 | 0.02215209 | 0.34820457 |
| DNAJC10 | 17 | 0.06587252 | 0.29767442 |
| CSNK2A1 | 16 | 0.07117248 | 0.34408602 |
| NCOR1 | 16 | 0.05439941 | 0.31496063 |
| NCK1 | 16 | 0.04186164 | 0.31714569 |
| SRF | 15 | 0.01578066 | 0.33229491 |
| REL | 13 | 0.01440785 | 0.34042553 |
| PAK3 | 13 | 0.02088241 | 0.31037827 |
| VHL | 13 | 0.02583072 | 0.30798845 |
| GNAI2 | 13 | 0.0176689 | 0.30563515 |
| SDHC | 13 | 0.03731446 | 0.33970276 |
| SNRPA | 12 | 0.07384247 | 0.33826638 |
| SOS1 | 12 | 0.02049813 | 0.33862434 |
| RAB7A | 12 | 0.07246042 | 0.30828516 |
| RBBP5 | 11 | 0.01080627 | 0.2640264 |
| EIF4E | 11 | 0.06378144 | 0.33229491 |
| NGFR | 11 | 0.02044841 | 0.32753327 |
| BCL6 | 11 | 0.0410574 | 0.34115139 |
| RPS14 | 10 | 0.04376657 | 0.27633851 |
| EPHA4 | 10 | 0.01262707 | 0.30710173 |
| PXN | 10 | 0.01479177 | 0.32520325 |
| EFTUD2 | 10 | 0.02160464 | 0.28725314 |
| UBE2L6 | 10 | 0.00749018 | 0.26380874 |
| NFATC2 | 10 | 0.00103466 | 0.31840796 |
| RHOQ | 10 | 0.02818871 | 0.28193833 |
Screening of potential transcription
factors
The present study identified the top 10 most
significant transcription factors for target genes of miRNAs, based
on data from FunRich. These included transcription factor Sp1,
myogenic factor 5, homeobox protein Nkx-2.1, nuclear factor I
C-type, homeobox protein NOBOX, early growth response protein 1,
Ras-responsive element-binding protein 1, visual system homeobox 2,
T-cell acute lymphocytic leukemia protein 1 and central areolar
choroidal dystrophy (Fig. 4).
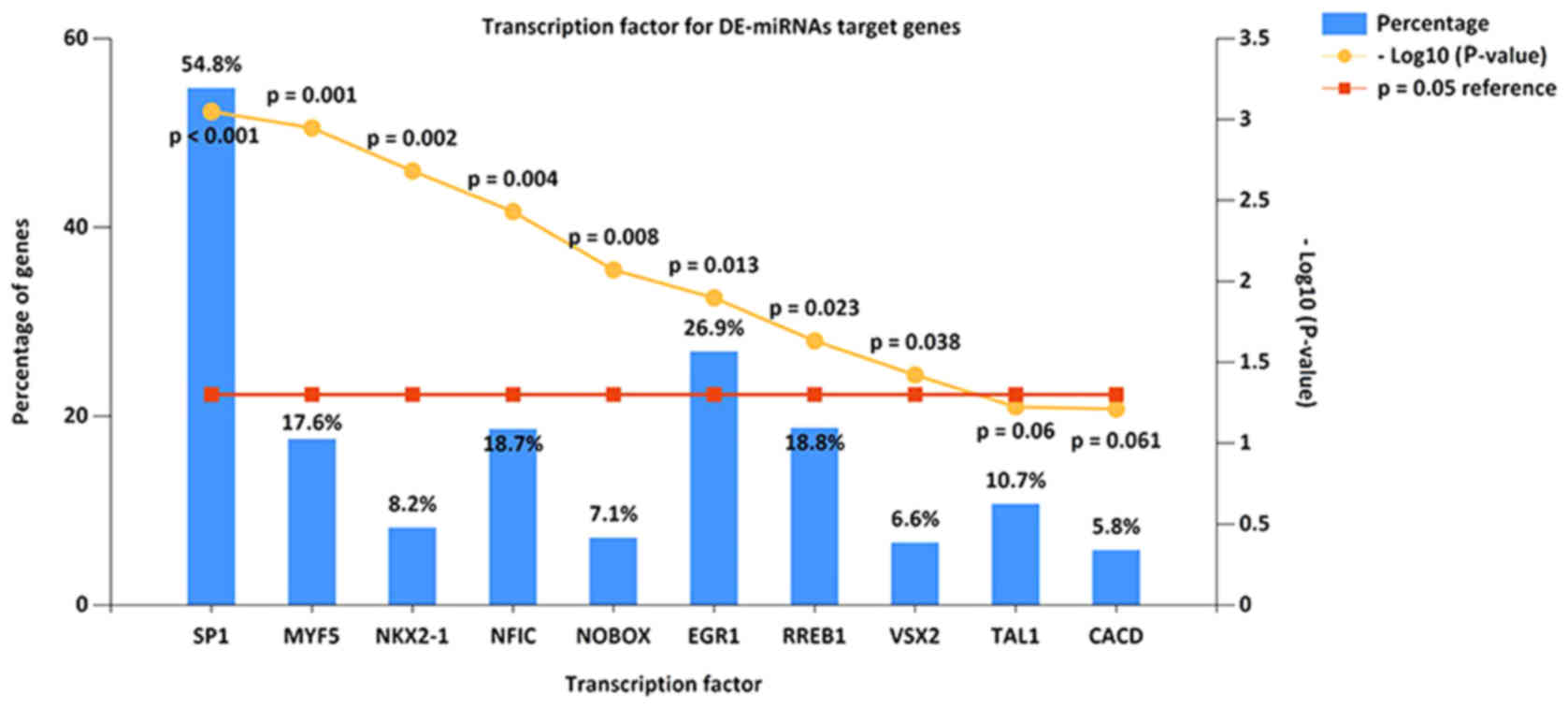 | Figure 4.Enriched transcription factors by
differentially expressed microRNAs target genes. The top 10 most
significant transcription factors include Sp1, MYF5, NKX2-1, NFIC,
NOBOX, EGR1, RREB1, VSX2, TAL1 and CACD. Sp1, transcription factor
Sp1; MYF5, myogenic factor 5; NKX2-1, homeobox protein Nkx-2.1;
NFIC, nuclear factor I C-type; NOBOX, homeobox protein NOBOX; EGR1,
early growth response protein 1; RREB1, Ras-responsive
element-binding protein 1; VSX2, visual system homeobox 2; TAL1,
T-cell acute lymphocytic leukemia protein 1; CACD, central areolar
choroidal dystrophy. |
Patient characteristics
The study population baseline descriptive parameters
are summarized in Table I. There
were no significant differences in age, infertility duration or
menarche age, with the exception of body mass index (P=0.002).
Patients with PCOS exhibited elevated LH levels (P=0.005) together
with significantly low levels of FSH (P=0.013) in patients with
PCOS on the third day of the menstrual cycle. Furthermore, LH/FSH
was significantly different (P<0.001) between the PCOS and
control groups. IVF outcomes revealed that the number of retrieved
oocytes in the PCOS group was higher than that in the control group
(P=0.003). Furthermore, the fertilization and cleavage rates in the
PCOS group were lower than that in control group (P<0.05).
Validation of candidate miRNAs using
RT-qPCR
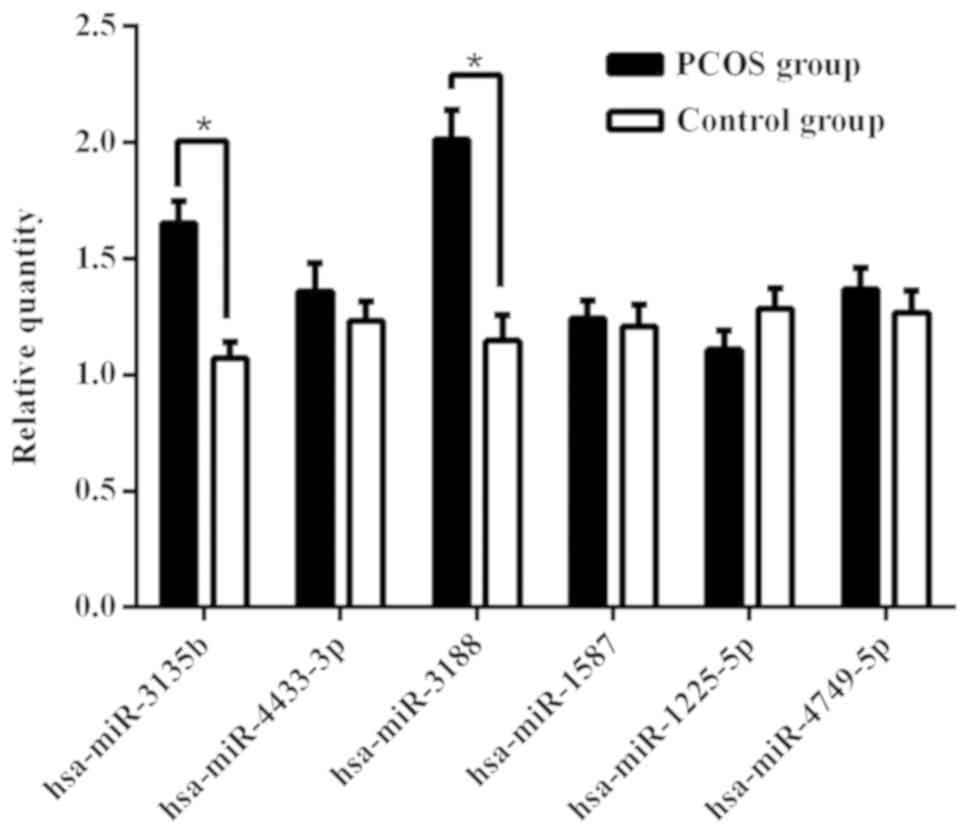
For confirmation purposes, the seven miRNAs
(hsa-miR-3188, hsa-miR-4433-3p, hsa-miR-3135b, hsa-miR-1587,
hsa-miR-1225-5p, hsa-miR-4417 and hsa-miR-4749-5p) identified were
re-examined using RT-qPCR in the 73 samples (38 patients with PCOS
and 35 from the control group) from the validation cohort. As shown
in Fig. 5, the levels of
hsa-miR-3188 and hsa-miR-3135b were significantly higher in GCs of
the PCOS group compared with those of the control group. However,
there was no significant difference between the two groups for the
levels of hsa-miR-4433-3p, hsa-miR-1587, hsa-miR-1225-5p or
hsa-miR-4749-5p. No hsa-miR-4417 expression was detected in GCs
(data not shown).
Correlations between validated miRNAs
and clinical parameters
Correlation analyses were used to examine the
association between miRNAs and clinical parameters of PCOS. The
results (data not shown) revealed that the levels of hsa-miR-3135b
were positively correlated with the number of oocytes retrieved
(r=0.450; P<0.001) and negatively correlated with fertilization
(r=−0.280; P=0.016) and cleavage rates (r=−0.232; P=0.049).
Discussion
PCOS is an endocrine disorder that affects 6–21% of
reproductive-aged women and 50% of sub-fertile women (32). PCOS is the leading cause of
menstrual complications in women, which involves a polygenic
multifactorial model with candidate genes involved in the
regulation of the biosynthesis and metabolism of folliculogenesis,
androgen, insulin and glucose metabolism (33). However, little is known about its
pathogenesis. Examining the molecular mechanism is of great
importance for the diagnosis and treatment of PCOS. The
identification of miRNAs involved in PCOS may provide novel insight
for detection, diagnosis, treatment and prognosis.
There is accumulating evidence that miRNAs serve
important roles in a variety of human diseases, including
development, proliferation, cellular differentiation, cell-cycle
control and cell death (34).
miRNAs participate in the formation of primordial follicles,
follicular recruitment and selection, follicular atresia,
oocyte-cumulus cell interaction and GC functions (35). Accumulating evidence suggested that
miRNAs contribute to the development and progression of PCOS
(36). miR-324-3p levels were
decreased in a PCOS rat model, while overexpression of miR-324-3p
can promote the apoptosis of GCs and inhibit cell proliferation by
targeting Wnt family member 2B (37). A previous study reported that a
series of miRNAs identified as clinically relevant markers of PCOS
are associated with obesity and metabolic dysfunction (38). miR-155 has been used as a biomarker
to monitor the efficacy of antiandrogen therapy in a
hyperandrogenic patient with PCOS (39). In addition, suppression of miR-19b
increased the proliferative ability of GCs by targeting
insulin-like growth factor 1 (IGF-1) in PCOS (40). Using Illumina deep sequencing
technology, Xue et al (36)
identified 263 DE-miRNAs in the follicular fluid of PCOS and
control groups, which were involved in the regulation of biological
functions and different signaling pathways. In a mouse PCOS model,
miR-27a-3p is involved in ovarian follicular development by
affecting estradiol and androgen imbalance (41). Previous studies have demonstrated
that several miRNAs are regulated by hormones, including LH, hCG
and FSH (42–45). These results suggested that miRNAs
may participate in the regulation of ovarian functions associated
with changes in hormone content. In the present study, based on the
GEO dataset (GSE84376), seven DE-miRNAs were identified using
bioinformatics analysis.
To date, there are a limited number of studies about
the role of miRNAs in patients with PCOS. In the present study,
hsa-miR-3188, hsa-miR-4433-3p, hsa-miR-3135b, hsa-miR-1587,
hsa-miR-1225-5p, hsa-miR-4417 and hsa-miR-4749-5p were found to be
differentially expressed compared with the control group. Zhou
et al (46) demonstrated
that miR-3188 knockout can inactivate the Notch signaling pathway
through upregulation of Zinc-fingers and homeoboxes 2 in
vivo and in vitro, thus inhibiting the growth and
metastasis of hepatocellular carcinoma cells. By contrast,
upregulation of miR-3188 regulates cancer cell proliferation,
apoptosis and migration in breast cancer by targeting
tumor-suppressor candidate 5 and activating the p38 MAPK signaling
pathway (47). In a previous study
on nasopharyngeal carcinoma, miR-3188 regulated cell proliferation
and chemosensitivity through a forkhead box O1 (FOXO1)-modulated
positive feedback loop with mammalian target of
rapamycin-p-PI3K/AKT-c-JUN (48).
Liu et al (49) reported a
high expression of miR-3135b in patients with coronary
calcification. Figueroa et al (50) identified miR-1587 as a mediator of
exosomes that can regulate tumor-initiating glioma stem-like cells
by suppressing nuclear receptor corepressor 1. In human laryngeal
carcinoma tissues, Sun et al (51) reported that miR-1225-5p promoted a
G1/S cell cycle arrest and enhanced cell death. In
addition, a previous study revealed that OTU deubiquitinase 5
suppression via hsa-miR-4417 and hsa-miR-6782 could maintain the
viability of tumor cells and suppress programmed cell death
(52). Song et al (53) demonstrated that, in hepatocellular
carcinoma cells, miR-4417 can promote proliferation and suppress
apoptosis by targeting tripartite motif-containing 35 and
regulating the phosphorylation of pyruvate kinase muscle.
Furthermore, the present study identified 935 target genes using
four databases. To better understand the functions of miRNA-target
genes, the present study performed GO analysis.
The GO enrichment analysis revealed that the most
significantly enriched gene set was ‘homophilic cell adhesion via
plasma membrane adhesion molecules’. The protocadherins (PCDHs),
the largest group of the cadherin superfamily, were identified as
the main target genes of the DE-miRNAs and were divided into two
major groups: ‘Clustered’ PCDHs that consisted of PCDH-α, PCDH-β
and PCDH-γ, and ‘non-clustered’ PCDHs (54). Previous studies have reported that
PCDHs are important for embryonic and neural development, and for
the etiology and progression of multiple types of cancer (55,56).
Inan et al (57) analyzed
the whole gene expression of GCs from a patient with recurrent
empty follicle syndrome (EFS) using an Affymetrix
GeneChip® and observed that PCDH17 was differentially
expressed in EFS compared with the control. Dickinson et al
(58) analyzed the effect of
pre-ovulatory follicle size on oocyte transcript abundance in beef
cows. A total of 106 transcripts were determined to be
differentially abundant, of which PCDH γ subfamily A 11 was in a
higher abundance in small follicle oocyte pools. In addition, the
present study identified a series of biological processes
associated with PCOS, including ‘cellular response to nitrogen
compound’, ‘blood vessel development’ and ‘regulation of calcium
ion and chloride transmembrane transport’. Previous studies have
revealed that follicular development disorder, ovulation inhibition
and GC apoptosis are associated with the downregulation of nitric
oxide (59,60). Liu et al (61) indicated that the abnormal
expression patterns of angiopoietin-like (ANGPT) 1 and ANGPTL2
mRNAs in cumulus cells are associated with impaired oocyte
developmental competence in PCOS, via pathological angiogenesis and
metabolism. Chen et al (62) demonstrated that impaired cystic
fibrosis transmembrane conductance regulator (a chloride ion
channel) resulted in ovarian disorders by amplifying FSH-stimulated
estrogen production in cystic fibrosis and PCOS. In addition,
transmembrane member 16A, a calcium-activated chloride channel,
regulates the morphology of oocytes and affects the synthesis of
estrogen through the MAPK kinase/ERK signaling cascade (63). These findings provide novel
insights into the molecular mechanisms of the regulation of
folliculogenesis and ovulation in PCOS.
Furthermore, a PPI network was constructed in the
present study, and MAPK1 was identified as the hub gene with the
highest connectivity degree (=46), followed by PTEN (=36) and CREB1
(=35). In 2017, Li et al (64) reported that MAPK1, epidermal growth
factor receptor, receptor tyrosine-protein kinase ERBB2, FOXO1,
NF-κβ1, IGF-1, cellular tumor antigen P53 and MAPK9 may activate
autophagy in the ovarian tissue of patients with PCOS. Cui et
al (65) indicated that the
MAPK signaling pathway promoted androgen receptor (AR) nuclear
translocation, thus influencing AR activity. In a PCOS-prone
metabolic syndrome rodent model, metformin appeared to decrease
fasting plasma glucose, insulin and homeostatic model
assessment-insulin resistance via the MAPK1, AKT2 and
5′-AMP-activated protein kinase catalytic subunit α2 signaling
pathways (66). These data
supported the hypothesis that MAPK1 may be a candidate target gene
associated with PCOS. Based on the hub genes identified, the
miRNA-gene regulatory network in the present study revealed that
hsa-miR-3135b had the highest correlation with the hub genes. In
summary, hsa-miR-3135b may participate in the development of PCOS
by regulating the expression of hub genes.
In order to better understand the mechanisms of
target genes in DE-miRNAs, the present study screened possible
transcription factors. Specificity Protein 1 (SP1) is the most
common transcription factor, a recent study indicated that SP1
serves an important role in regulating vascular endothelial growth
factor (VEGF) production by binding to specific sites in the VEGF
promoter (67). Previous studies
suggested that SP1 may be involved in the mechanism of insulin
resistance in patients with PCOS by mediating the gene expression
of insulin (68,69). Anjali et al (69) revealed that FSH can stimulate the
expression of insulin receptor substrate 2 in human GCs through
cAMP/SP1 in patients with PCOS. Enrichment of SP1 by the target
genes of DE-miRNAs suggests their potential involvement in the
development and progression of PCOS.
Based on the findings of this present study, only
hsa-miR-3188 and hsa-miR-3135b were found to display significantly
high levels in GCs of patients with PCOS compared with controls
with male factor infertility. An association was found between
hsa-miR-3135b and the hub genes, the expression level of
hsa-miR-3135b was observed to be significantly correlated with the
number of oocytes retrieved, fertilization and cleavage rate
(P<0.05). Overall, these results demonstrate that hsa-miR-3135b
may participate in the development and maturation of oocytes by the
regulation of target genes.
There are several limitations to the present study.
First, the study is based on microarray data from GSE84376. The
sample size is small; thus, further studies are necessary to
investigate additional samples from multiple centers. Second,
various enriched functions and hub genes were identified in the
present study; however, their associations were not fully
elucidated. In conclusion, the data presented provide a
comprehensive analysis of DE-miRNAs that may be involved in the
development of PCOS. The DE-miRNAs and hub target genes identified
in the present study may be used in the future as targets for PCOS
prevention and treatment. However, their biological functions and
mechanism of action in PCOS require further investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the project
sponsored by the Research Fund of National Key Research and
Development Program (grant no. 2018YFC1002103) and the Research
Fund of National Health and Family Planning Commission of China
(grant no. RFNHFPCC, 201402004).
Availability of data and materials
The dataset used and/or analyzed in the present
study is available from the corresponding author on reasonable
request.
Authors' contributions
XW conceived and supervised the project. YH and SX
performed the experiments. YW and GQ analyzed the data. YH, YW and
XW wrote the manuscript. All the authors reviewed the
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Children's Hospital of Shanxi and Women Health Center of Shanxi.
Written informed consent for inclusion was obtained from each
participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Franks S: Polycystic ovary syndrome. N
Engl J Med. 333:853–861. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lizneva D, Suturina L, Walker W, Brakta S,
Gavrilova-Jordan L and Azziz R: Criteria, prevalence, and
phenotypes of polycystic ovary syndrome. Fertil Steril. 106:6–15.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao Y and Qiao J: Ethnic differences in
the phenotypic expression of polycystic ovary syndrome. Steroids.
78:755–760. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dumesic DA, Padmanabhan V and Abbott DH:
Polycystic ovary syndrome and oocyte developmental competence.
Obstet Gynecol Surv. 63:39–48. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nelson LR and Bulun SE: Estrogen
production and action. J Am Acad Dermatol 45 (3 Suppl). S116–S124.
2001. View Article : Google Scholar
|
|
6
|
Artimani T, Saidijam M, Aflatoonian R,
Amiri I, Ashrafi M, Shabab N, Mohammadpour N and Mehdizadeh M:
Estrogen and progesterone receptor subtype expression in granulosa
cells from women with polycystic ovary syndrome. Gynecol
Endocrinol. 31:379–383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haouzi D, Assou S, Monzo C, Vincens C,
Dechaud H and Hamamah S: Altered gene expression profile in cumulus
cells of mature MII oocytes from patients with polycystic ovary
syndrome. Hum Reprod. 27:3523–3530. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jansen E, Laven JS, Dommerholt HB, Polman
J, van Rijt C, van den Hurk C, Westland J, Mosselman S and Fauser
BC: Abnormal gene expression profiles in human ovaries from
polycystic ovary syndrome patients. Mol Endocrinol. 18:3050–3063.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Manneras-Holm L, Benrick A and
Stener-Victorin E: Gene expression in subcutaneous adipose tissue
differs in women with polycystic ovary syndrome and controls
matched pair-wise for age, body weight, and body mass index.
Adipocyte. 3:190–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sorensen AE, Wissing ML, Salö S, Englund
AL and Dalgaard LT: MicroRNAs related to polycystic ovary syndrome
(PCOS). Genes (Basel). 5:684–708. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wood JR, Ho CK, Nelson-Degrave VL,
McAllister JM and Strauss JF III: The molecular signature of
polycystic ovary syndrome (PCOS) theca cells defined by gene
expression profiling. J Reprod Immunol. 63:51–60. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rossi JJ: New hope for a microRNA therapy
for liver cancer. Cell. 137:990–992. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim YJ, Ku SY, Kim YY, Liu HC, Chi SW, Kim
SH, Choi YM, Kim JG and Moon SY: MicroRNAs transfected into
granulosa cells may regulate oocyte meiotic competence during in
vitro maturation of mouse follicles. Hum Reprod. 28:3050–3061.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu B, Zhang YW, Tong XH and Liu YS:
Characterization of microRNA profile in human cumulus granulosa
cells: Identification of microRNAs that regulate Notch signaling
and are associated with PCOS. Mol Cell Endocrinol. 404:26–36. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Imbar T and Eisenberg I: Regulatory role
of microRNAs in ovarian function. Fertil Steril. 101:1524–1530.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Tu F, Yao W, Li X, Xie Z, Liu H, Li
Q and Pan Z: Conserved miR-26b enhances ovarian granulosa cell
apoptosis through HAS2-HA-CD44-Caspase-3 pathway by targeting HAS2.
Sci Rep. 6:211972016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou J, Liu J, Pan Z, Du X, Li X, Ma B,
Yao W, Li Q and Liu H: The let-7g microRNA promotes follicular
granulosa cell apoptosis by targeting transforming growth
factor-beta type 1 receptor. Mol Cell Endocrinol. 409:103–112.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Carletti MZ and Christenson LK: MicroRNA
in the ovary and female reproductive tract. J Anim Sci 87 (14
Suppl). E29–E38. 2009. View Article : Google Scholar
|
|
20
|
Chen YH, Heneidi S, Lee JM, Layman LC,
Stepp DW, Gamboa GM, Chen BS, Chazenbalk G and Azziz R: miRNA-93
inhibits GLUT4 and is overexpressed in adipose tissue of polycystic
ovary syndrome patients and women with insulin resistance.
Diabetes. 62:2278–2286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Long W, Zhao C, Ji C, Ding H, Cui Y, Guo
X, Shen R and Liu J: Characterization of serum microRNAs profile of
PCOS and identification of novel non-invasive biomarkers. Cell
Physiol Biochem. 33:1304–1315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roth LW, McCallie B, Alvero R, Schoolcraft
WB, Minjarez D and Katz-Jaffe MG: Altered microRNA and gene
expression in the follicular fluid of women with polycystic ovary
syndrome. J Assist Reprod Genet. 31:355–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murri M, Insenser M, Fernandez-Duran E,
San-Millan JL and Escobar-Morreale HF: Effects of polycystic ovary
syndrome (PCOS), sex hormones, and obesity on circulating miRNA-21,
miRNA-27b, miRNA-103, and miRNA-155 expression. J Clin Endocrinol
Metab. 98:E1835–E1844. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi L, Liu S, Zhao W and Shi J: miR-483-5p
and miR-486-5p are down-regulated in cumulus cells of metaphase II
oocytes from women with polycystic ovary syndrome. Reprod Biomed
Online. 31:565–572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang L, Huang J, Li L, Chen Y, Chen X,
Zhao X and Yang D: MicroRNA-93 promotes ovarian granulosa cells
proliferation through targeting CDKN1A in polycystic ovarian
syndrome. J Clin Endocrinol Metab. 100:E729–E738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu XL, Chen XF and Xu WL: miR-34a
regulates apoptosis of human ovarian granulosa cells. J Jiangsu
Univ (Med Ed). 26:470–474. 2016.(In Chinese).
|
|
27
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res 41
(Database Issue). D808–D815. 2013.
|
|
29
|
Pathan M, Keerthikumar S, Ang CS, Gangoda
L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim
A, et al: FunRich: An open access standalone functional enrichment
and interaction network analysis tool. Proteomics. 15:2597–2601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rotterdam ESHRE/ASRM-Sponsored PCOS
consensus workshop Group, : Revised 2003 consensus on diagnostic
criteria and long-term health risks related to polycystic ovary
syndrome (PCOS). Hum Reprod. 19:41–47. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Azziz R, Carmina E, Dewailly D,
Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen
OE, Legro RS, Norman RJ, Taylor AE, et al: Positions statement:
Criteria for defining polycystic ovary syndrome as a predominantly
hyperandrogenic syndrome: An Androgen Excess Society guideline. J
Clin Endocrinol Metab. 91:4237–4245. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Escobar-Morreale HF, Luque-Ramirez M and
San Millan JL: The molecular-genetic basis of functional
hyperandrogenism and the polycystic ovary syndrome. Endocr Rev.
26:251–282. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miska EA: How microRNAs control cell
division, differentiation and death. Curr Opin Genet Dev.
15:563–568. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maalouf SW, Liu WS and Pate JL: MicroRNA
in ovarian function. Cell Tissue Res. 363:7–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xue Y, Lv J, Xu P, Gu L, Cao J, Xu L, Xue
K and Li Q: Identification of microRNAs and genes associated with
hyperandrogenism in the follicular fluid of women with polycystic
ovary syndrome. J Cell Biochem. 119:3913–3921. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang YC and Ma JX: The role of MiR-324-3p
in polycystic ovary syndrome (PCOS) via targeting WNT2B. Eur Rev
Med Pharmacol Sci. 22:3286–3293. 2018.PubMed/NCBI
|
|
38
|
Murri M, Insenser M, Fernandez-Duran E,
San-Millan JL, Luque-Ramirez M and Escobar-Morreale HF:
Non-targeted profiling of circulating microRNAs in women with
polycystic ovary syndrome (PCOS): Effects of obesity and sex
hormones. Metabolism. 86:49–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Arancio W, Calogero Amato M, Magliozzo M,
Pizzolanti G, Vesco R and Giordano C: Serum miRNAs in women
affected by hyperandrogenic polycystic ovary syndrome: The
potential role of miR-155 as a biomarker for monitoring the
estroprogestinic treatment. Gynecol Endocrinol. 34:704–708. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhong Z, Li F, Li Y, Qin S, Wen C, Fu Y
and Xiao Q: Inhibition of microRNA-19b promotes ovarian granulosa
cell proliferation by targeting IGF-1 in polycystic ovary syndrome.
Mol Med Rep. 17:4889–4898. 2018.PubMed/NCBI
|
|
41
|
Wang M, Liu M, Sun J, Jia L, Ma S, Gao J,
Xu Y, Zhang H, Tsang SY and Li X: MicroRNA-27a-3p affects estradiol
and androgen imbalance by targeting Creb1 in the granulosa cells in
mouse polycytic ovary syndrome model. Reprod Biol. 17:295–304.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yao G, Yin M, Lian J, Tian H, Liu L, Li X
and Sun F: MicroRNA-224 is involved in transforming growth
factor-beta-mediated mouse granulosa cell proliferation and
granulosa cell function by targeting Smad4. Mol Endocrinol.
24:540–551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Carletti MZ, Fiedler SD and Christenson
LK: MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa
cells. Biol Reprod. 83:286–295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yin M, Wang X, Yao G, Lü M, Liang M, Sun Y
and Sun F: Transactivation of micrornA-320 by microRNA-383
regulates granulosa cell functions by targeting E2F1 and SF-1
proteins. J Biol Chem. 289:18239–18257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fiedler SD, Carletti MZ, Hong X and
Christenson LK: Hormonal regulation of MicroRNA expression in
periovulatory mouse mural granulosa cells. Biol Reprod.
79:1030–1037. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou SJ, Deng YL, Liang HF, Jaoude JC and
Liu FY: Hepatitis B virus X protein promotes CREB-mediated
activation of miR-3188 and Notch signaling in hepatocellular
carcinoma. Cell Death Differ. 24:1577–1587. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen X and Chen J: MiR-3188 regulates cell
proliferation, apoptosis, and migration in breast cancer by
targeting TUSC5 and regulating the p38 MAPK signaling pathway.
Oncol Res. 26:363–372. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao M, Luo R, Liu Y, Gao L, Fu Z, Fu Q,
Luo X, Chen Y, Deng X, Liang Z, et al: miR-3188 regulates
nasopharyngeal carcinoma proliferation and chemosensitivity through
a FOXO1-modulated positive feedback loop with
mTOR-p-PI3K/AKT-c-JUN. Nat Commun. 7:113092016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu W, Ling S, Sun W, Liu T, Li Y, Zhong
G, Zhao D, Zhang P, Song J, Jin X, et al: Circulating microRNAs
correlated with the level of coronary artery calcification in
symptomatic patients. Sci Rep. 5:160992015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Figueroa J, Phillips LM, Shahar T, Hossain
A, Gumin J, Kim H, Bean AJ, Calin GA, Fueyo J, Walters ET, et al:
Exosomes from Glioma-associated mesenchymal stem cells increase the
tumorigenicity of glioma stem-like cells via transfer of miR-1587.
Cancer Res. 77:5808–5819. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sun P, Zhang D, Huang H, Yu Y, Yang Z, Niu
Y and Liu J: MicroRNA-1225-5p acts as a tumor-suppressor in
laryngeal cancer via targeting CDC14B. Biol Chem. 400:237–246.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Fomicheva KA, Osip'yants AI, Knyazev EN,
Samatov TR and Shkurnikov MY: Detection of potential metastatic
prostate cancer circulating biomarkers by comparison of miRNA
profiles in DU145 cells and culture medium. Bull Exp Biol Med.
162:792–796. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Song L, Zhang W, Chang Z, Pan Y, Zong H,
Fan Q and Wang L: miR-4417 targets tripartite motif-containing 35
(TRIM35) and regulates pyruvate kinase muscle 2 (PKM2)
phosphorylation to promote proliferation and suppress apoptosis in
hepatocellular carcinoma cells. Med Sci Monit. 23:1741–1750. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Schreiner D and Weiner JA: Combinatorial
homophilic interaction between gamma-protocadherin multimers
greatly expands the molecular diversity of cell adhesion. Proc Natl
Acad Sci USA. 107:14893–14898. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yagi T and Takeichi M: Cadherin
superfamily genes: Functions, genomic organization, and neurologic
diversity. Genes Dev. 14:1169–1180. 2000.PubMed/NCBI
|
|
56
|
Mah KM and Weiner JA: Regulation of Wnt
signaling by protocadherins. Semin Cell Dev Biol. 69:158–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Inan MS, Al-Hassan S, Ozand P and Coskun
S: Transcriptional profiling of granulosa cells from a patient with
recurrent empty follicle syndrome. Reprod Biomed Online.
13:481–491. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dickinson SE: Effect of pre-ovulatory
follicle size on oocyte transcript abundance in beef cows. Master's
Thesis University of Missouri-Columbia. 2016.
|
|
59
|
Mitchell LM, Kennedy CR and Hartshorne GM:
Expression of nitric oxide synthase and effect of substrate
manipulation of the nitric oxide pathway in mouse ovarian
follicles. Hum Reprod. 19:30–40. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Guo YX, Zhang GM, Yao XL, Tong R, Cheng
CY, Zhang TT, Wang ST, Yang H and Wang F: Effects of nitric oxide
on steroidogenesis and apoptosis in goat luteinized granulosa
cells. Theriogenology. 126:55–62. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu Z, Liu C, Hao C, Xue Q, Huang X, Zhang
N, Bao H and Qu Q: Aberrant expression of angiopoietin-like
proteins 1 and 2 in cumulus cells is potentially associated with
impaired oocyte developmental competence in polycystic ovary
syndrome. Gynecol Endocrinol. 32:557–561. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen H, Guo JH, Lu YC, Ding GL, Yu MK,
Tsang LL, Fok KL, Liu XM, Zhang XH, Chung YW, et al: Impaired
CFTR-dependent amplification of FSH-stimulated estrogen production
in cystic fibrosis and PCOS. J Clin Endocrinol Metab. 97:923–932.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sun M, Sui Y, Li L, Su W, Hao F, Zhu Q, Di
W, Gao H and Ma T: Anoctamin 1 calcium-activated chloride channel
downregulates estrogen production in mouse ovarian granulosa cells.
Endocrinology. 155:2787–2796. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li D, You Y, Bi FF, Zhang TN, Jiao J, Wang
TR, Zhou YM, Shen ZQ, Wang XX and Yang Q: Autophagy is activated in
the ovarian tissue of polycystic ovary syndrome. Reproduction.
155:85–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Cui Y, Sun Y, Hu S, Luo J, Li L, Li X, Yeh
S, Jin J and Chang C: Neuroendocrine prostate cancer (NEPCa)
increased the neighboring PCa chemoresistance via altering the
PTHrP/p38/Hsp27/androgen receptor (AR)/p21 signals. Oncogene.
35:6065–6076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kupreeva M, Diane A, Lehner R, Watts R,
Ghosh M, Proctor S and Vine D: Effect of metformin and flutamide on
insulin, lipogenic and androgen-estrogen signaling and
cardiometabolic risk in a PCOS-prone rodent model. Am J Physiol
Endocrinol Metab. 316:E16–E33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu N, Ding D, Hao W, Yang F, Wu X, Wang
M, Xu X, Ju Z, Liu JP, Song Z, et al: hTERT promotes tumor
angiogenesis by activating VEGF via interactions with the Sp1
transcription factor. Nucleic Acids Res. 44:8693–8703. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Solomon SS, Majumdar G, Martinez-Hernandez
A and Raghow R: A critical role of Sp1 transcription factor in
regulating gene expression in response to insulin and other
hormones. Life Sci. 83:305–312. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Anjali G, Kaur S, Lakra R, Taneja J,
Kalsey GS, Nagendra A, Shrivastav TG, Devi MG, Malhotra N, Kriplani
A and Singh R: FSH stimulates IRS-2 expression in human granulosa
cells through cAMP/SP1, an inoperative FSH action in PCOS patients.
Cell Signal. 27:2452–2466. 2015. View Article : Google Scholar : PubMed/NCBI
|