Introduction
Intrahepatic cholestasis leads to systemic and
intrahepatic retention of toxic bile acids that initially cause
liver injury and subsequently result in biliary fibrosis and
cirrhosis (1). Estrogens and their
metabolites are well-known causes of estrogen-induced intrahepatic
cholestasis (EIC) during pregnancy and in women taking oral
contraception and postmenopausal hormone replacement therapy
(2). Its pathogenesis arises
primarily through genetic predisposition but also to a lesser
extent by environment and immunology factors (3). Intrahepatic cholestasis of pregnancy
(ICP) is the most common type of EIC, with an estimated prevalence
of 0.4–1.5% worldwide (4). Delayed
diagnosis of ICP may increase the risks of stillbirth and early
delivery (5). However, the
pathogenic and physiopathological mechanisms of EIC are not well
understood. EIC is currently treated with ursodeoxycholic acid
(UDCA), but 30–50% of patients do not respond to UDCA treatment
(6). Therefore, identification of
key pathways and candidate genes for EIC may provide a greater
understanding of this disease and aid in the development of novel
drug targets.
Estrogen levels increase significantly during
pregnancy and following administration of oral contraception and
postmenopausal replacement therapy, particularly in patients with
EIC (7). High estrogen levels
induce acute cholestasis by impairing both bile flow and bile acid
metabolism, causing downstream dysfunction of bile acid homeostasis
(8). Accordingly, toxic bile acids
accumulate in the liver, inducing oxidative stress and inflammatory
reactions and the onset of liver injury (9). Clinical studies have indicated that
oxidative stress responses are amplified in ICP patients compared
to healthy pregnant women (10,11).
In addition, pro-inflammatory cytokines, including interleukin
(IL)-6 and IL-17, are markedly increased in patients with ICP
compared with healthy pregnant women (12,13).
Animal models of EIC have also demonstrated high expression levels
of pro-inflammatory cytokines and an increased oxidative stress
response (14,15).
The development of bioinformatics has provided new
insights into the diagnosis and treatment of cholestatic liver
diseases (16–18). Using integral microRNA (miRNA) and
mRNA microarrays, Nakagawa et al (16) identified that the miR-425-mediated
T cell receptor signaling pathway had an important role in
regulating the synthesis of inflammatory cytokines in
CD4+ T cells of patients with primary biliary
cholangitis (PBC). Suppression of the T cell receptor
signaling-associated regulator or miR-425 has therefore been
suggested to be a promising immunotherapeutic strategy against PBC
(16). Wang et al (17) identified serpin family E member 1
(PAI-1) as the key gene in mice with extrahepatic
cholestasis following bile duct ligation, and demonstrated that
inhibiting PAI-1 may attenuate cholestatic liver diseases.
These studies suggest that bioinformatics is a useful strategy to
identify key pathways and candidate genes involved in disease as a
foundation to develop new therapies. However, to the best of our
knowledge, a comprehensive bioinformatical analyses of the cellular
and molecular mechanisms underlying EIC has not been performed.
Cholestasis induced by 17α-ethinylestradiol (EE) is
a rodent model widely used to investigate the molecular mechanisms
of EIC (19). The present study
aimed to identify key pathways and candidate genes using a
whole-genome microarray (4×44K) to analyze an EE-induced
cholestatic rat model. The most significant differentially
expressed genes (DEGs) involved in the metabolism of estrogen and
bile acids and the regulation of inflammation and oxidative stress
were shortlisted as candidate genes. In addition, Gene Ontology
(GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment and protein-protein interaction (PPI) network analyses
were performed to identify key pathways involved in EIC. A total of
5 candidate genes and several key pathways were identified, which
may be candidate diagnostic biomarkers or therapeutic targets for
EIC.
Materials and methods
Animals and treatments
A total of 12 male Sprague Dawley rats weighing
200±20 g were obtained from the Center of Experimental Animals of
Hubei Province. All animals acclimated to the laboratory conditions
for 1 week prior to the experiment and were kept at 25±2°C with a
12:12 h light: dark cycle. Animals had ad libitum access to
standard laboratory chow and tap water. The present study was
approved by the Ethical Committee on Animal Experimentation of
Tongji Medical College, Huazhong University of Science and
Technology (Wuhan, China). Animals were randomly divided into
control and model groups (n=6 per group). Control and model rats
were administered subcutaneous (s.c.) propylene glycol (solvent of
EE, 0.25 ml/100 g, s.c.) or EE (5 mg/kg, s.c.) for 5 consecutive
days, respectively. Rat conditions and body weight were recorded
every day. Rats were fasted overnight following the last treatment
prior to sacrifice. Bile flow was measured according to a
previously described method (14).
Blood samples and liver tissues were collected for subsequent
analysis.
Biochemical determinations and
histological analyses
Serum aspartate aminotransferase (ALT), alanine
aminotransferase (AST), alkaline phosphatase (ALP) and total bile
acids (TBA) were analyzed using commercial kits (Nanjing Jiancheng
Bioengineering Institute) according to the manufacturer's protocol.
A section of the same liver parts from each rat was dissected and
fixed in 10% formalin for 24 h in a room temperature, washed,
dehydrated in alcohol gradients (100, 100, 95, 80 and 75%, 2 min
each) and paraffin-embedded. Each slide was subsequently stained
with hematoxylin (0.2%, 5 min, room temperature) and eosin (0.5%,
30 sec, room temperature), and then assessed histologically with a
light microscope (magnification, ×200, EVOS FL Auto; Thermo Fisher
Scientific, Inc.).
Gene expression analyses
Total RNA of 3 randomly selected rat liver tissues
were extracted using RNAiso Plus (Takara Bio, Inc.) and purified
using an RNeasy mini kit (Qiagen GmbH) following the manufacturer's
instructions. Total RNA was amplified and labeled with an Agilent
Quick Amp Labeling kit (Agilent Technologies, Inc.). Each slide
containing rat liver total RNA was hybridized with an Agilent Whole
Rat Genome Oligo Microarray (4×44K) for 17 h. Slides were then
washed and scanned using an Agilent DNA Microarray Scanner (Agilent
Technologies, Inc.). Data was extracted with Agilent Feature
Extraction software v10.7 (Agilent Technologies, Inc.) and
subsequently filtered for detection of normalization and
significant changes, using P<0.05 as the cut-off value, and DEGs
compared to the control rats, using fold change >2 or <0.5 as
the cut-off. The heatmap of DEGs was analyzed by SBC analysis
system (Shanghai Biotechnology Corporation). Technical support was
provided by the Shanghai Biotechnology Corporation.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA of all rat liver samples was isolated
using TRIzol® reagent (Thermo Fisher Scientific, Inc.)
and then reverse transcribed into cDNA using the PrimeScript™ RT
Master Mix (Takara Biotechnology Co., Ltd.). The RT steps were:
37°C, 15 min and 85°C, 5 sec. Each cDNA was used as a template for
RT-qPCR amplification, performed using SYBR Premix Ex Taq™ (Takara
Biotechnology Co., Ltd.) and pairs of forward/reverse primers for
the candidate genes and rat β-actin gene. The qPCR steps were:
initial denaturation, 95°C, 30 sec; PCR reaction, 95°C, 5 sec,
60°C, 34 sec, 40 cycles. Independent reactions were performed in
triplicate using an Applied Biosystems StepOnePlus Real-time PCR
system (Applied Biosystems). Threshold cycle (Cq) values were
normalized to those for β-actin (ΔCq). The 2−ΔΔCq method
was used to calculate the relative changes of gene expression
(20). The primer sequences of
candidate genes and β-actin are presented in Table I.
 | Table I.Sequence of primers for 6 candidate
genes and β-actin for reverse transcription quantitative polymerase
chain reaction. |
Table I.
Sequence of primers for 6 candidate
genes and β-actin for reverse transcription quantitative polymerase
chain reaction.
| Gene name | Direction | Sequence
(5′-3′) |
|---|
| Sult1e1 | Forward |
ATTCAATGCTGCCAGAGACC |
|
| Reverse |
TCATTTGCTGCTGGTAGTGC |
| Cyp3a2 | Forward |
TGACTGCTCTTGATGCATGGTT |
|
| Reverse |
ATCACAGACCTTGCCAACTCCT |
| Car3 | Forward |
GCTCCTTTTAATCACTTCGACC |
|
| Reverse |
AGCCACACAATGCACTCCTC |
| Ltc4s | Forward |
GAAGAACTTTCCACGTGTCG |
|
| Reverse |
GTGCAGCCATTGCCACTAGC |
| Adam8 | Forward |
GCCTCGGACCTTAGAAAT |
|
| Reverse |
GGCATAACGGCTGATGTA |
| β-actin | Forward |
CACCCGCGAGTACAACCTTC |
|
| Reverse |
CCCATACCCACCATCACACC |
Functional enrichment analyses of the
DEGs
Functional annotation of DEGs was performed using
the online Database for Annotation, Visualization, and Integrated
Discovery (DAVID; http://david.ncifcrf.gov/). DAVID provides
comprehensive functional annotations tools for the analysis of the
genes and proteins included in the database, for users to obtain
biological information. The functions of the DEGs was determined
via GO terms and KEGG pathway enrichment using DAVID, with
P<0.05 used as the cut-off for statistical significance.
Construction of PPI networks and
analysis of modules
The Search Tool for Retrieval of Interacting Genes
and Proteins (STRING) database (v10.0; http://string-db.org/) was used for PPI network
construction of the DEGs. Genes without associations with other
genes were removed. Interactions with a combined score >0.4 were
considered significant. PPI networks were used for module screening
using the Molecular Complex Detection (MCODE) in Cytoscape (v3.6.0)
bioinformatics integration platform (21). Significant modules were those with
scores >3 and nodes >4. KEGG pathway enrichment analysis for
DEGs was performed in each module using ClueGO software (version
2.5.0, Institute for Genomics and Bioinformatics Graz University of
Technology, Graz, Austria).
Statistical analyses
Statistical analyses were performed using SPSS
software v13.0 (SPSS Software, Inc.). All data are presented as the
mean ± standard deviation. A Student's t-test was used to compare
the differences between two groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
EE-induced cholestasis and liver
injury
The body weight of the control rats gradually
increased, while that of model rats initially increased, but then
fell. A significant difference (P<0.01) in body weight between
control and model rats was observed at days 5 and 6 (Fig. 1A). Bile flow was suppressed
(P<0.01) in the EE-treated rats (Fig. 1B). Serum ALT, AST, ALP and TBA
levels were significantly increased in the EE-induced rats compared
with the control rats (Fig. 1C).
Histological evaluation of the hepatic tissues indicated that
control rats exhibited normal structures, while there was evidence
of neutrophil infiltration, edema and hepatic necrosis in model
rats (Fig. 1D). These results were
in concordance with the previously published data (22) and thereby suggest that EE induced
cholestasis and liver injury in rats.
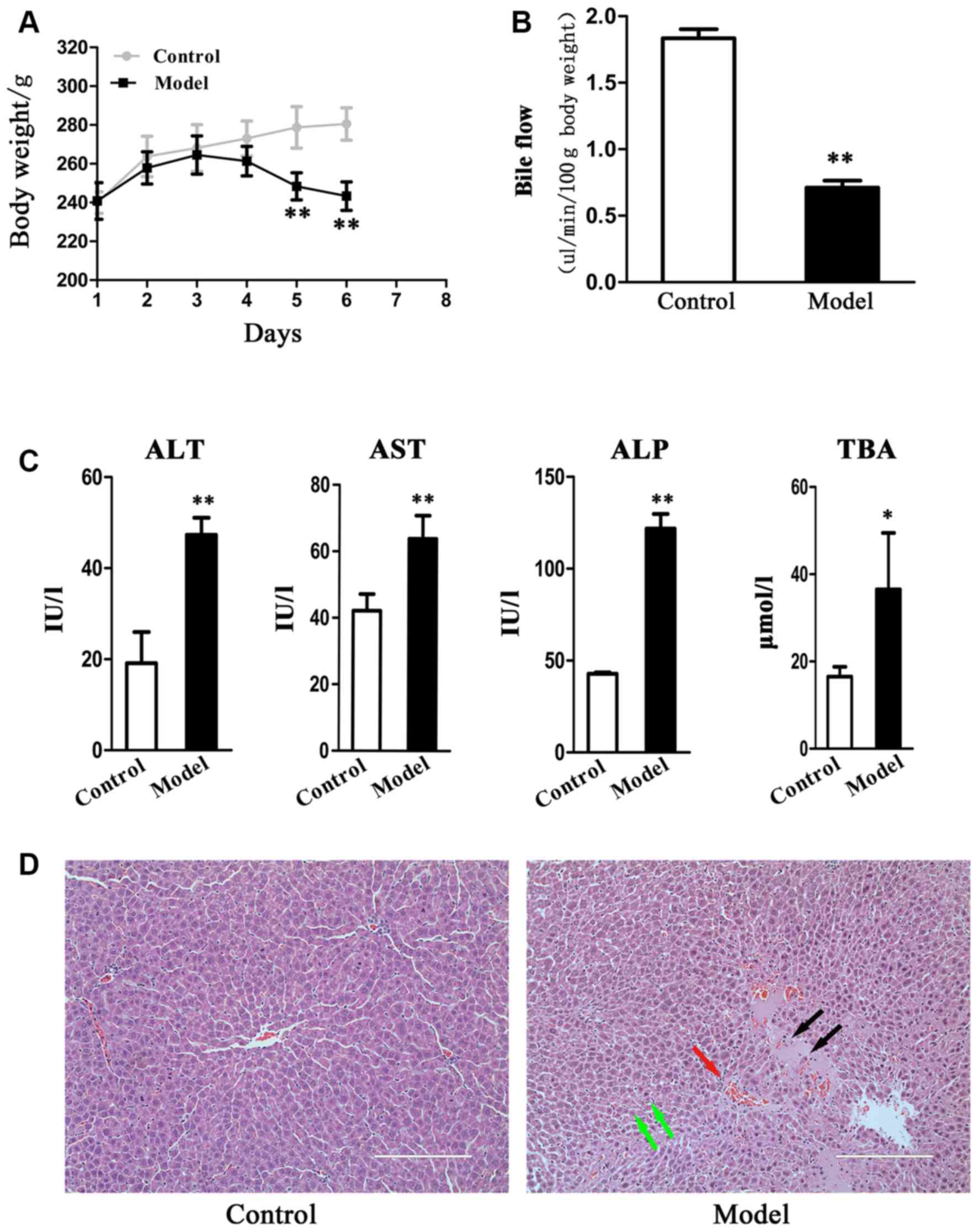 | Figure 1.17α-ethinylestradiol-induced
cholestasis and liver injury. (A) Changes in body weight and (B)
bile flow in EIC rats. Bile was collected in a 10-minute period to
monitor the flow. (C) Serum biochemical index of AST, ALT, ALP and
TBA. (D) The images of representative hematoxylin and eosin stained
liver sections (magnification, ×200). Liver necrosis, neutrophil
infiltration and edema are marked by black, red, and green arrows,
respectively. Data are presented as the mean ± standard deviation
(n=6). *P<0.05 and **P<0.01 vs. control. EIC,
estrogen-induced intrahepatic cholestasis; AST, aspartate
aminotransferase; ALT, alanine aminotransferase; ALP, alkaline
phosphatase; TBA, total bile acids. |
Identification of DEGs and candidate
genes
Following preprocessing of the microarray data, the
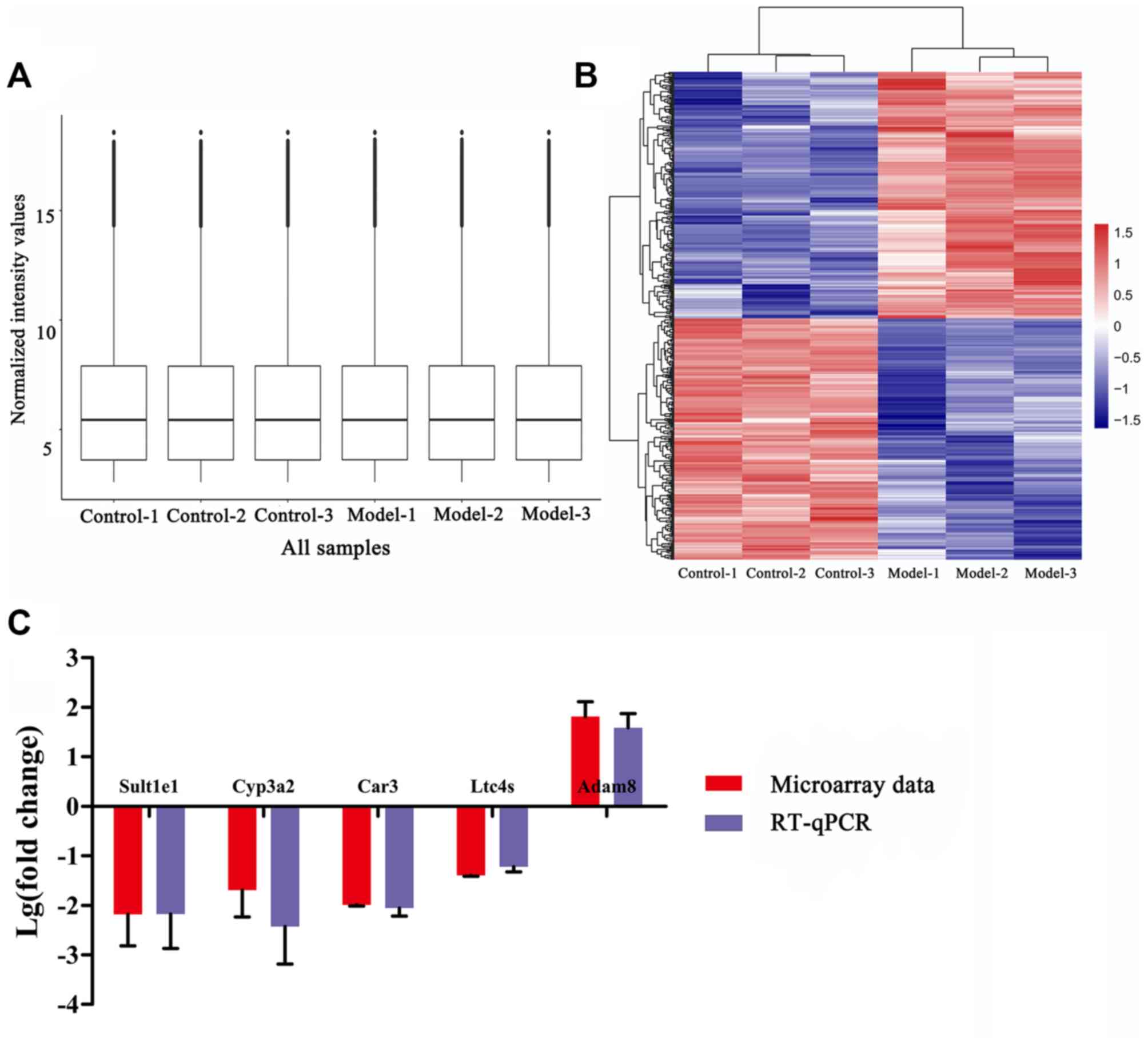
gene expression data were normalized and visualized with a box
plot. A good normalization performance was obtained (Fig. 2A). In total, 455 DEGs were
identified, including 225 downregulated genes and 230 upregulated
genes. The heatmap function was used to observe DEGs in different
samples (Fig. 2B). The top 5
downregulated and upregulated DEGs are listed in Table II. Genes were thereafter
shortlisted as candidate genes based on evidence of their
involvement in the metabolism of estrogen and bile acids or in the
regulation of inflammatory reaction and oxidative stress, either
from the literature or the data from the present study. These were
sulfotransferase family 1E member 1 (Sult1e1), cytochrome
P450 family 3 subfamily A member 2 (Cyp3a2), carbonic
anhydrase 3 (Car3), leukotriene C4 synthase (Ltc4s)
and ADAM metallopeptidase domain 8 (Adam8). To confirm the
reliability of DNA microarray data, the mRNA expression levels of
these candidate genes were investigated by RT-qPCR. The results
were consistent with those from the DNA microarray data (Fig. 2C).
 | Table II.Top 5 downregulated and upregulated
differentially expressed genes (n=3). |
Table II.
Top 5 downregulated and upregulated
differentially expressed genes (n=3).
| Direction | Symbol | Log10
FC | P-values |
|---|
| Downregulated | Car3 | −1.988 | 0.0019 |
|
| Sult1e1 | −1.966 | 0.0272 |
|
| Cyp3a2 | −1.531 | 0.0322 |
|
| LOC100912610 | −1.444 | 0.0277 |
|
| Ltc4s | −1.396 | 0.0042 |
| Upregulated | Rasd2 | 1.894 | 0.0034 |
|
| Adam8 | 1.882 | 0.0074 |
|
| Pvalb | 1.859 | 0.0153 |
|
| Slc5a1 | 1.705 | 0.0010 |
|
| Cited4 | 1.638 | 0.0009 |
Functional enrichment analysis for
gene expression
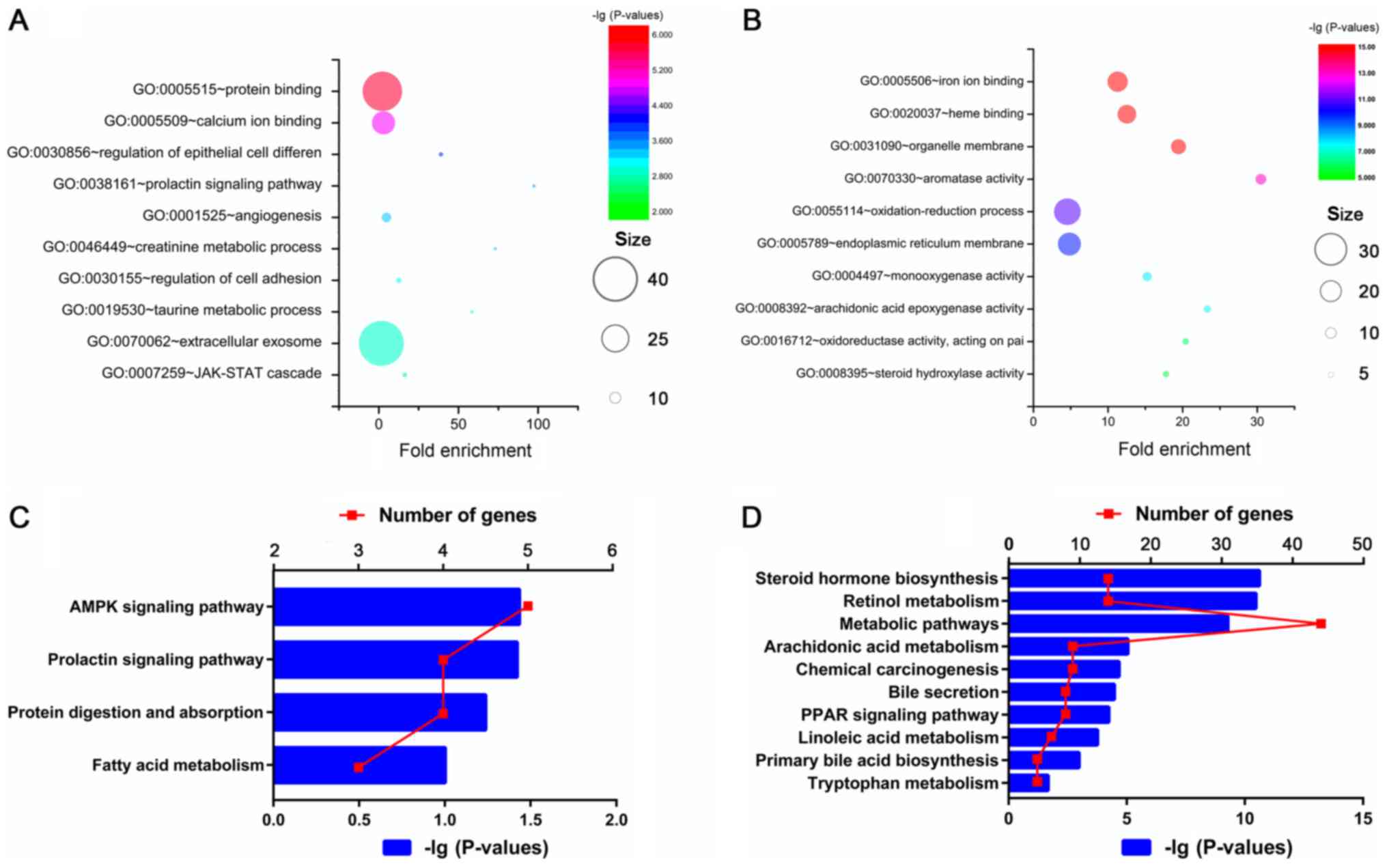
To explore the functions of DEGs, GO and KEGG
pathway enrichment analyses were performed using the DAVID. As
demonstrated in Fig. 3, analysis
of GO indicated that the upregulated genes were primarily enriched
in the regulation of protein binding, calcium ion binding,
epithelial cell differentiation and extracellular exosome, while
the downregulated genes were primarily enriched in regulators of
iron ion binding, heme binding, aromatase activity and
oxidation-reduction.
KEGG analysis was used for the identification of
significantly enriched pathways of the DEGs. The upregulated DEGs
were primarily enriched in the pathways of 5′AMP-activated protein
kinase (AMPK) and prolactin and those involved in protein
digestion, protein absorption and fatty acid metabolism. The
downregulated DEGs were primarily enriched in steroid hormone
biosynthesis, retinol metabolism, metabolic pathways and
arachidonic acid metabolism. A total of 3 pathways also enriched in
the downregulated DEGs were involved in regulating bile acid
homeostasis, including bile secretion, peroxisome
proliferator-activated receptor (PPAR) signaling and primary bile
acid biosynthesis.
PPI network construction and module
selection
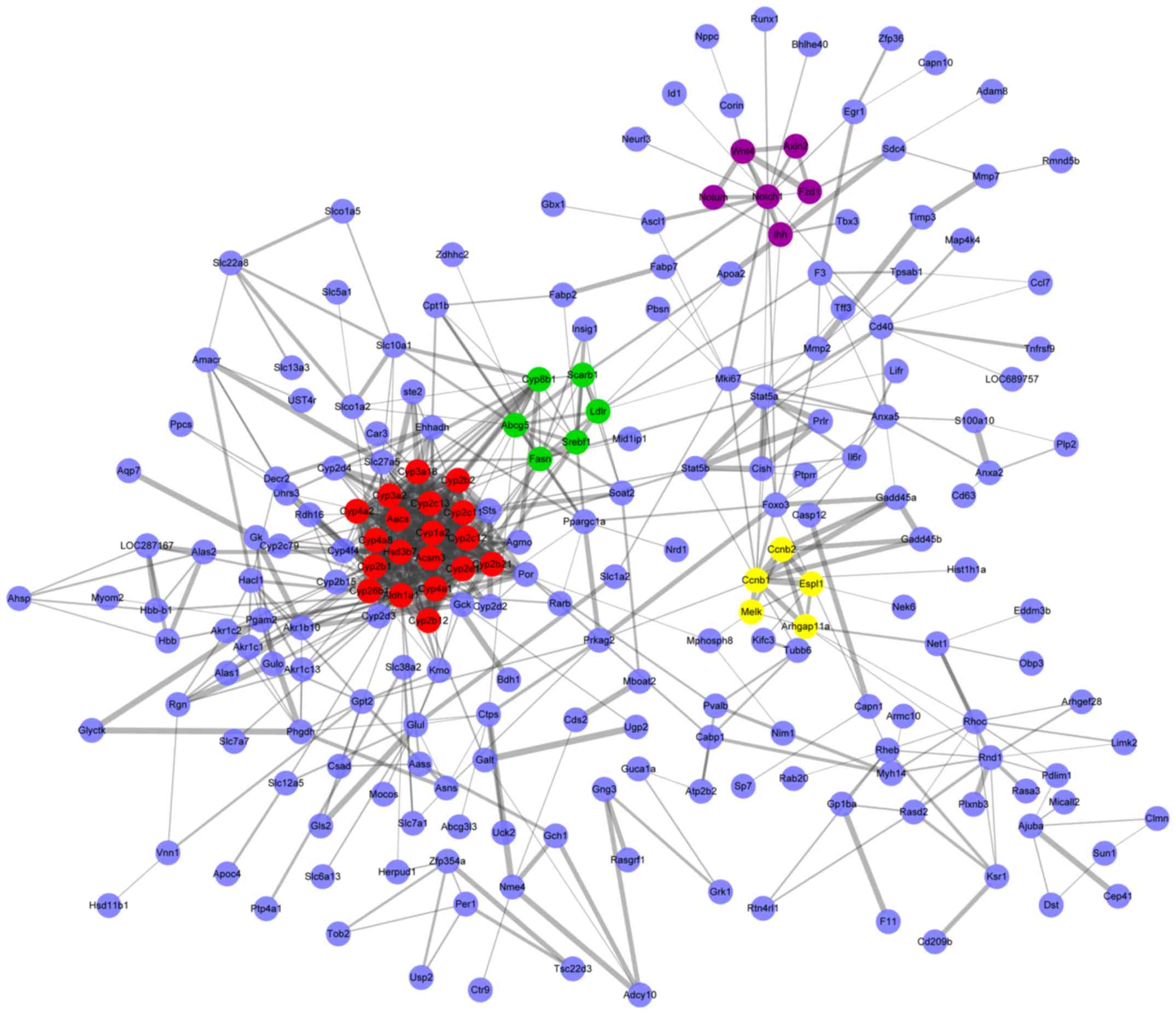
Protein interactions of DEGs were predicted by the
STRING database and then the PPI networks were constructed for
those with a combined protein pair score >0.4 (Fig. 4). Modules of genes in the PPI
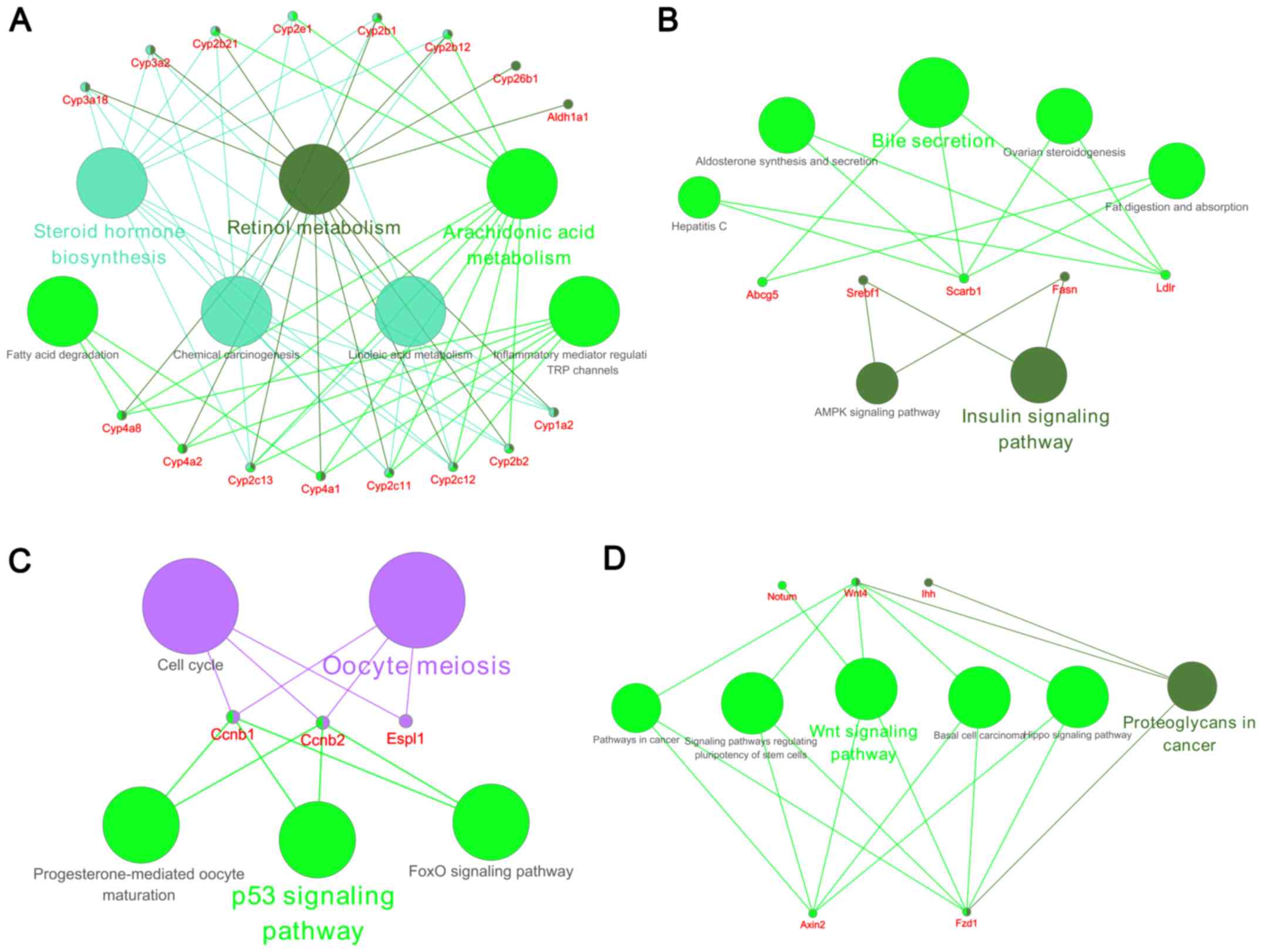
network were identified by the MCODE plugin in Cytoscape. The top 4
significant modules were selected, and the cellular pathways of the
genes involved in these modules were analyzed using ClueGO. As
demonstrated in Fig. 5, the
cellular pathways were involved in steroid hormone biosynthesis,
retinol metabolism, arachidonic acid metabolism, bile secretion,
and p53 and Wnt signaling.
Discussion
EIC is characterized by high estrogen levels,
accumulation of toxic bile acids, hepatic inflammation and
oxidative stress, which leads to the fibrosis, cirrhosis and
eventually failure of the liver (23). Liver transplantation remains the
only strategy for patients with the end-stage EIC (24). For EICs detected at earlier stages,
UDCA and obeticholic acid, a nuclear receptor agonist, are the
first- and second-line drugs; however, their curative effects are
limited (25). In addition, the
mechanisms involved in the pathogenesis and physiopathology of EIC
are not well understood. Understanding the molecular mechanisms
involved in the development and progression of EIC is therefore
crucial for developing and evaluating diagnostic and management
strategies.
In the present study, a whole-genome microarray was
used to identify key pathways and candidate genes within EE-induced
cholestatic rats. The results identified a total of 455 DEGs,
including 225 downregulated genes and 230 upregulated genes. Among
the top upregulated and downregulated genes, those involved in the
metabolism of estrogens and bile acids, and the regulation of
inflammation and oxidative stress, were identified and validated as
candidate genes. These were Sult1e1, Cyp3a2, Car3, Ltc4s and
Adam8. The elevated estrogen levels that are typically
observed in patients with EIC and animal models induce dysfunction
of bile acid hemostasis through activating estrogen-responsive
receptors, leading to the accumulation of toxic bile acids in the
liver and the development of liver injury (1). A phase II drug-metabolizing enzyme,
Sult1e1, is known to catalyze the sulfoconjugation of estrogens and
their metabolites (26). It has
been demonstrated Sult1e1 is downregulated during several liver
diseases, including cholestasis, and this is associated with
elevated serum estrogen (27,28).
An additional study investigating livers in a cystic fibrosis mouse
model indicated that elevated hepatic Sult1e1 levels may result in
decreased levels of estrogens and decreased expression of
estrogen-responsive receptors (29). Cyp3a2, a human ortholog of
cytochrome P450 family 3 subfamily A member 4 (CYP3A4), is a phase
I detoxifying cytochrome P450 enzyme that catalyzes the elimination
of bile acid (30) and thereby
decreases toxic bile acid levels in the liver (31). Numerous studies have demonstrated
that approved drugs including rifampicin, phenobarbital and
bezafibrate protect against cholestasis through inducing CYP3A4
activity and detoxifying bile acid levels (32–34).
In the present study, the extremely low expression of
Sult1e1 and Cyp3a2 may limit the metabolism of
estrogens and bile acids and lead to their accumulation in the
liver. Therefore, reversing the expression of Sult1e1 and
Cyp3a2 may be a target for EIC therapy.
Accumulation of hepatic bile acids often results in
oxidative stress and inflammation, which then leads to
hepatocellular injury (9).
Car3, a human ortholog of CA3, is responsible for
catalyzing the hydration of carbon dioxide in response to oxidative
stress, but is nonetheless expressed at low levels in EIC. It has
been demonstrated that Car3 serves an important role in
glutathionylation during cellular oxidative stress (35). Compared with parental cells,
Car3-transfected cells exhibited lower levels of the
intracellular reactive oxygen species (36). More importantly, Car3 decreases the
toxicity of hydrophobic bile acids on biliary epithelium in
bicarbonate-rich hydrocholeresis (37). Additionally, Car3 is involved in
mediating oxidative stress in various tissues and pathological
processes, including in skeletal muscle and during alcoholic liver
disease and aging (38–40). As a nuclear-membrane enzyme, Ltc4s
catalyzes the conjugation of leukotriene A4 to glutathione,
generating leukotriene C4, which is the first reaction in the
synthesis of cysteinyl leukotrienes (LTs) (41). The previously described role of LTs
strongly implicates them in the pathogenesis of inflammatory
diseases (41–42). In addition, increasing evidence has
demonstrated that LTs are associated with cholestasis, hepatic
inflammation during metabolic disease and fulminant hepatic failure
(42). Adam8 is a member of the
ADAM protein family, which serves critical roles in certain liver
diseases. Li et al (43),
for example, demonstrated that Adam8 promoted liver injury by
inhibiting the proliferation of hepatocytes and promoting
angiogenesis, and by affecting the metabolic function of the liver
during acute liver injury induced by CCl4. Accordingly,
neutralization of Adam8 ameliorated liver injury and accelerated
liver repair. Concomitantly, Higuchi et al (44) identified that oxazolone-induced
contact hypersensitivity reactions were more severe in Adam8
transgenic mice compared with wild-type mice. Adam8, directly or
indirectly, regulates leukocyte infiltration in mice (44). The microarray data and RT-qPCR
results from the present study indicated that Car3, Ltc4s
and Adam8 were the most significantly altered DEGs in the
EE-induced rat model. Therefore, these 3 candidate genes may have
crucial roles in the regulation of oxidative stress and
inflammation during the pathogenesis of EIC.
Key pathways underlying EIC were investigated
through KEGG and PPI network-associated module analyses in the
present study. The KEGG analysis indicated that the AMPK signaling
pathway was primarily enriched in the upregulated DEGs. It has
previously been demonstrated that the activation of AMPK signaling
is integral to the EE-mediated disruption of the expression of bile
acid transporters and promoting the EIC process (45). In addition, inhibition of AMPK
activation attenuated EE-induced cholestasis in vitro and
in vivo (45). AMPK
signaling is also crucial for the pathogenesis of cholestatic liver
injury involved in regulating hepatic polarity, inflammation and
fibrosis (46). Fatty acid
metabolism was enriched in the upregulated DEGs, whereas steroid
hormone biosynthesis and PPAR signaling pathway were enriched in
the downregulated DEGs. These 3 pathways are all involved in lipid
metabolism. It has been demonstrated previously that alterations in
lipid metabolism may promote inflammation, fibrosis and
proliferation in a mouse model of chronic cholestatic liver injury
(47). Fenofibrate is a PPARα
agonist that protects against EE-induced cholestasis in mice
(48). In addition, it is well
recognized that bile acid accumulation contributes to cholestasis.
The results of the present study indicated that bile secretion and
primary bile acid biosynthesis were primarily enriched in the
downregulated DEGs, which suggests that bile acid homeostasis is
disrupted during EE-induced cholestasis. In previous decades,
numerous studies have struggled to regulate the expression of bile
acid transporters, metabolic enzymes and nuclear receptors to
improve bile acid homeostasis in cholestatic liver diseases
(49). Accordingly, the inhibition
of AMPK signaling or maintenance of lipid and bile acid homeostasis
may provide potential strategies for EIC treatment.
The p53 and Wnt signaling pathways were identified
as key pathways based on the data from the PPI network
associated-modules. The role of p53 signaling on cholestasis is
controversial. Chen et al (50) identified that the p53 activator
doxorubicin attenuated cholic acid-induced cholestasis in mice
through promoting the deposition of bile acid and alleviation of
cholestatic syndrome, and that cholic acid-induced cholestatic
liver injury was aggravated in p53-knockout mice. Concomitantly,
other studies hypothesized that p53 signaling served an important
role in promoting apoptosis during cholestatic liver injury
(51,52). Future studies are required to
determine the role of p53 signaling on EIC and other cholestatic
diseases. In addition, it has been demonstrated that Wnt signaling
is a critical regulator of the pathophysiology of cholestasis
(53,54). Wnt signaling regulates
hepatobiliary repair in cholestatic mice (54) and controls intrahepatic biliary
network formation in zebrafish (53). Therefore, the regulation of p53 and
Wnt signaling pathways may also provide novel insights into EIC
treatment.
There are several limitations of the present study.
Due to the fact that there are limited data available on the
pathways and genes involved in EIC, the identified key pathways and
candidate genes were not discussed in detail. Additionally, these
results were generated from rat liver samples, which may cause
tissue or sample heterogeneity in independent studies. Finally,
only bioinformatics-based evidence and part of RT-qPCR data were
presented, and additional studies are therefore required to confirm
the functions of these candidate genes and key pathways in EIC, and
to verify the results.
In summary, the present study identified 5 candidate
genes and several key pathways that were demonstrated to be
important in the pathogenesis of EIC. These pathways were involved
in the homeostasis of lipids and bile acids and in AMPK, p53 and
Wnt signaling. The results of the present study indicated that
these candidate genes and key pathways may serve critical roles in
the development and progression of EIC. Reversing the abnormal
expression of candidate genes and dysfunction of key pathways may
provide opportunities for novel EIC therapies.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from
National Natural Science Foundation of China (grant nos. 81670521
and 81803798).
Availability of data and materials
The microarray data and other data sets in the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
DL and CZ designed, supervised and revised the
study. DX and CZ collected and analyzed the data, and wrote the
manuscript. YX and WH contributed to microarray data collection and
analysis. JY assisted in performing experiments and analysis of the
data. All authors reviewed and approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee on Animal Experimentation of Tongji Medical College,
Huazhong University of Science and Technology (Wuhan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Marrone J, Soria LR, Danielli M, Lehmann
GL, Larocca MC and Marinelli RA: Hepatic gene transfer of human
aquaporin-1 improves bile salt secretory failure in rats with
estrogen-induced cholestasis. Hepatology. 64:535–548. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schreiber AJ and Simon FR:
Estrogen-induced cholestasis: Clues to pathogenesis and treatment.
Hepatology. 3:607–613. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stieger B, Fattinger K, Madon J,
Kullak-Ublick GA and Meier PJ: Drug- and estrogen-induced
cholestasis through inhibition of the hepatocellular bile salt
export pump (Bsep) of rat liver. Gastroenterology. 118:422–430.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hillman SC, Stokes-Lampard H and Kilby MD:
Intrahepatic cholestasis of pregnancy. BMJ. 353:i12362016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kondrackiene J and Kupcinskas L:
Intrahepatic cholestasis of pregnancy-current achievements and
unsolved problems. World J Gastroenterol. 14:5781–5788. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Lu L, Victor DW, Xin Y and Xuan
S: Ursodeoxycholic acid and S-adenosylmethionine for the treatment
of intrahepatic cholestasis of pregnancy: A meta-analysis. Hepat
Mon. 16:e385582016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zucchetti AE, Barosso IR, Boaglio AC,
Basiglio CL, Miszczuk G, Larocca MC, Ruiz ML, Davio CA, Roma MG,
Crocenzi FA and Pozzi EJ: G-protein-coupled receptor 30/adenylyl
cyclase/protein kinase A pathway is involved in estradiol
17ss-D-glucuronide-induced cholestasis. Hepatology. 59:1016–1029.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reyes H and Simon FR: Intrahepatic
cholestasis of pregnancy: An estrogen-related disease. Semin Liver
Dis. 13:289–301. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Copple BL, Jaeschke H and Klaassen CD:
Oxidative stress and the pathogenesis of cholestasis. Semin Liver
Dis. 30:195–204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ozler A, Ucmak D, Evsen MS, Kaplan I,
Elbey B, Arica M and Kaya M: Immune mechanisms and the role of
oxidative stress in intrahepatic cholestasis of pregnancy. Cent Eur
J Immunol. 39:198–202. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sanhal CY, Daglar K, Kara O, Yilmaz ZV,
Turkmen GG, Erel O, Uygur D and Yucel A: An alternative method for
measuring oxidative stress in intrahepatic cholestasis of
pregnancy: Thiol/disulphide homeostasis. J Matern Fetal Neonatal
Med. 31:1477–1482. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Biberoglu E, Kirbas A, Daglar K, Kara O,
Karabulut E, Yakut HI and Danisman N: Role of inflammation in
intrahepatic cholestasis of pregnancy. J Obstet Gynaecol Res.
42:252–257. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kirbas A, Biberoglu E, Ersoy AO, Dikmen
AU, Koca C, Erdinc S, Uygur D, Caglar T and Biberoglu K: The role
of interleukin-17 in intrahepatic cholestasis of pregnancy. J
Matern Fetal Neonatal Med. 29:977–981. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu D, Wu T, Zhang CL, Xu YJ, Chang MJ, Li
XP and Cai HJ: Beneficial effect of Calculus Bovis Sativus on
17α-ethynylestradiol-induced cholestasis in the rat. Life Sci.
113:22–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamamoto Y, Moore R, Hess HA, Guo GL,
Gonzalez FJ, Korach KS, Maronpot RR and Negishi M: Estrogen
receptor alpha mediates 17alpha-ethynylestradiol causing
hepatotoxicity. J Biol Chem. 281:16625–16631. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakagawa R, Muroyama R, Saeki C, Goto K,
Kaise Y, Koike K, Nakano M, Matsubara Y, Takano K, Ito S, et al:
miR-425 regulates inflammatory cytokine production in CD4(+) T
cells via N-Ras upregulation in primary biliary cholangitis. J
Hepatol. 66:1223–1230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Vohra BP, Zhang Y and Heuckeroth
RO: Transcriptional profiling after bile duct ligation identifies
PAI-1 as a contributor to cholestatic injury in mice. Hepatology.
42:1099–1108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sakamoto T, Morishita A, Nomura T, Tani J,
Miyoshi H, Yoneyama H, Iwama H, Himoto T and Masaki T:
Identification of microRNA profiles associated with refractory
primary biliary cirrhosis. Mol Med Rep. 14:3350–3356. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Carreras FI, Lehmann GL, Ferri D, Tioni
MF, Calamita G and Marinelli RA: Defective hepatocyte aquaporin-8
expression and reduced canalicular membrane water permeability in
estrogen-induced cholestasis. Am J Physiol Gastrointest Liver
Physiol. 292:G905–G912. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Crocenzi FA, Sánchez PE, Pellegrino JM,
Favre CO, Rodríguez GE, Mottino AD, Coleman R and Roma MG:
Beneficial effects of silymarin on estrogen-induced cholestasis in
the rat: A study in vivo and in isolated hepatocyte couplets.
Hepatology. 34:329–339. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak JK and Schmittgen DT: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen J, Zhao KN and Liu GB:
Estrogen-induced cholestasis: Pathogenesis and
therapeuticimplications. Hepatogastroenterology. 60:1289–1296.
2013.PubMed/NCBI
|
|
24
|
Glantz A, Marschall HU and Mattsson LA:
Intrahepatic cholestasis of pregnancy: Relationships between bile
acid levels and fetal complication rates. Hepatology. 40:467–474.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lammert F, Marschall HU, Glantz A and
Matern S: Intrahepatic cholestasis of pregnancy: Molecular
pathogenesis, diagnosis and management. J Hepatol. 33:1012–1021.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Y, Yang X, Wang Z, Li M, Ning Y, Chen
S, Yin L and Li X: Estrogen sulfotransferase (SULT1E1) regulates
inflammatory response and lipid metabolism of human endothelial
cells via PPARgamma. Mol Cell Endocrinol. 369:140–149. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu X, Xue R, Yang C, Gu J, Chen S and
Zhang S: Cholestasis-induced bile acid elevates estrogen level via
farnesoid X receptor-mediated suppression of the estrogen
sulfotransferase SULT1E1. J Biol Chem. 293:12759–12769. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yalcin EB, More V, Neira KL, Lu ZJ,
Cherrington NJ, Slitt AL and King RS: Downregulation of
sulfotransferase expression and activity in diseased human livers.
Drug Metab Dispos. 41:1642–1650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li L and Falany CN: Elevated hepatic
SULT1E1 activity in mouse models of cystic fibrosis alters the
regulation of estrogen responsive proteins. J Cyst Fibros. 6:23–30.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deo AK and Bandiera SM: Identification of
human hepatic cytochrome p450 enzymes involved in the
biotransformation of cholic and chenodeoxycholic acid. Drug Metab
Dispos. 36:1983–1991. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saini SP, Sonoda J, Xu L, Toma D, Uppal H,
Mu Y, Ren S, Moore DD, Evans RM and Xie W: A novel constitutive
androstane receptor-mediated and CYP3A-independent pathway of bile
acid detoxification. Mol Pharmacol. 65:292–300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li T and Chiang JY: Rifampicin induction
of CYP3A4 requires pregnane X receptor cross talk with hepatocyte
nuclear factor 4alpha and coactivators, and suppression of small
heterodimer partner gene expression. Drug Metab Dispos. 34:756–764.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Honda A, Ikegami T, Nakamuta M, Miyazaki
T, Iwamoto J, Hirayama T, Saito Y, Takikawa H, Imawari M and
Matsuzaki Y: Anticholestatic effects of bezafibrate in patients
with primary biliary cirrhosis treated with ursodeoxycholic acid.
Hepatology. 57:1931–1941. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Back P: Therapeutic use of phenobarbital
in intrahepatic cholestasis. Inductions in bile acid metabolism.
Pharmacol Ther. 33:153–155. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao X, Sheng L, Wang L, Hong J, Yu X,
Sang X, Sun Q, Ze Y and Hong F: Mechanisms of nanosized titanium
dioxide-induced testicular oxidative stress and apoptosis in male
mice. Part Fibre Toxicol. 11:472014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Räisänen SR, Lehenkari P, Tasanen M,
Rahkila P, Harkonen PL and Väänänen HK: Carbonic anhydrase III
protects cells from hydrogen peroxide-induced apoptosis. FASEB J.
13:513–522. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Miethke AG, Zhang W, Simmons J, Taylor AE,
Shi T, Shanmukhappa SK, Karns R, White S, Jegga AG, Lages CS, et
al: Pharmacological inhibition of apical sodium-dependent bile acid
transporter changes bile composition and blocks progression of
sclerosing cholangitis in multidrug resistance 2 knockout mice.
Hepatology. 63:512–523. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Parkkila S, Halsted CH, Villanueva JA,
Väänänen HK and Niemelä O: Expression of testosterone-dependent
enzyme, carbonic anhydrase III, and oxidative stress in
experimental alcoholic liver disease. Dig Dis Sci. 44:2205–2213.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zimmerman UJ, Wang P, Zhang X, Bogdanovich
S and Forster R: Anti-oxidative response of carbonic anhydrase III
in skeletal muscle. IUBMB Life. 56:343–347. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cabiscol E and Levine RL: Carbonic
anhydrase III. Oxidative modification in vivo and loss of
phosphatase activity during aging. J Biol Chem. 270:14742–14747.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hong F and Yang S: Ischemic
preconditioning decreased leukotriene C4 formation by depressing
leukotriene C4 synthase expression and activity during hepatic I/R
injury in rats. J Surg Res. 178:1015–1021. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Martínez-Clemente M, Ferré N,
González-Périz A, López-Parra M, Horrillo R, Titos E,
Morán-Salvador E, Miquel R, Arroyo V, Funk CD and Clària J:
5-lipoxygenase deficiency reduces hepatic inflammation and tumor
necrosis factor α-induced hepatocyte damage in hyperlipidemia-prone
ApoE-null mice. Hepatology. 51:817–827. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li SQ, Zhu S, Wan XD, Xu ZS and Ma Z:
Neutralization of ADAM8 ameliorates liver injury and accelerates
liver repair in carbon tetrachloride-induced acute liver injury. J
Toxicol Sci. 39:339–351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Higuchi Y, Yasui A, Matsuura K and
Yamamoto S: CD156 transgenic mice. Different responses between
inflammatory types. Pathobiology. 70:47–54. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li X, Liu R, Luo L, Yu L, Chen X, Sun L,
Wang T, Hylemon PB, Zhou H, Jiang Z and Zhang L: Role of
AMP-activated protein kinase α1 in 17α-ethinylestradiol-induced
cholestasis in rats. Arch Toxicol. 91:481–494. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li X, Liu R, Zhang L and Jiang Z: The
emerging role of AMP-activated protein kinase in cholestatic liver
diseases. Pharmacol Res. 125:105–113. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Moustafa T, Fickert P, Magnes C, Guelly C,
Thueringer A, Frank S, Kratky D, Sattler W, Reicher H, Sinner F, et
al: Alterations in lipid metabolism mediate inflammation, fibrosis,
and proliferation in a mouse model of chronic cholestatic liver
injury. Gastroenterology. 142:140–151.e12. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Leuenberger N, Pradervand S and Wahli W:
Sumoylated PPARalpha mediates sex-specific gene repression and
protects the liver from estrogen-induced toxicity in mice. J Clin
Invest. 119:3138–3148. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li T and Chiang JY: Nuclear receptors in
bile acid metabolism. Drug Metab Rev. 45:145–155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen P, Li D, Chen Y, Sun J, Fu K, Guan L,
Zhang H, Jiang Y, Li X, Zeng X, et al: p53-mediated regulation of
bile acid disposition attenuates cholic acid-induced cholestasis in
mice. Br J Pharmacol. 174:4345–4361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang H, Li TW, Ko KS, Xia M and Lu SC:
Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression
and apoptosis during cholestasis in mouse and human hepatocytes.
Hepatology. 49:860–870. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wilkins BJ, Lorent K, Matthews RP and Pack
M: p53-mediated biliary defects caused by knockdown of cirh1a, the
zebrafish homolog of the gene responsible for North American Indian
Childhood Cirrhosis. PLoS One. 8:e776702013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
So J, Khaliq M, Evason K, Ninov N, Martin
BL, Stainier D and Shin D: Wnt/β-catenin signaling controls
intrahepatic biliary network formation in zebrafish by regulating
notch activity. Hepatology. 67:2352–2366. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Okabe H, Yang J, Sylakowski K, Yovchev M,
Miyagawa Y, Nagarajan S, Chikina M, Thompson M, Oertel M, Baba H,
et al: Wnt signaling regulates hepatobiliary repair following
cholestatic liver injury in mice. Hepatology. 64:1652–1666. 2016.
View Article : Google Scholar : PubMed/NCBI
|