Introduction
House dust mites (HDMs) are common indoor sources of
allergens (1,2). The primary species of HDM are
Dermatophagoides pteronyssinus, D. farinae, Euroglyphus
maynei and Blomia tropicalis (3,4),
with the former two being ubiquitous in home dust samples in
temperate and tropical regions (5,6). HDM
allergens constitute a major cause of allergic diseases (7,8),
with one-half of allergy sufferers exhibiting an allergic reaction
to HDM allergens (9,10).
Clinicians use HDM allergen proteins to diagnose and
treat HDM allergies (11,12). The HDM antigens used clinically are
obtained from a crude HDM extract (13,14).
As these crude extracts are a mixture of a small portion of
allergens and a number of unrelated impurities, their effects are
highly varied, including adverse side effects in certain patients
(15). Of the 39 HDM allergen
groups recognized in the World Health Organization and
International Union of Immunological Societies (WHO/IUIS) allergen
database, 33 include identified D. farinae allergens and
only 23 include D. pteronyssinus allergens (16). The identification of HDM allergens,
particularly the detection and naming of novel HDM allergens, has
direct significance for the diagnosis and treatment of HDM-induced
allergic diseases.
Previous studies have suggested that the major HDM
allergens belong to Group 1 (17,18),
Group 2 (19,20), Group 23 (21,22)
and Group 24 (23). A Group 23
allergen from D. pteronyssinus (Der p 23) was identified to
be a major allergen present in HDM feces in dust (24,25).
Der p 23 has been demonstrated to react with specific
immunoglobulin Es (sIgEs) from 74% of patients with D.
pteronyssinus allergies, which is smaller proportion compared
with the positive reaction rates of the two previously recognized
major HDM allergens, namely Der p 1 and Der p 2 (21). Der p 23 is a small, globular
protein stabilized by 2 disulfide bonds that is structurally
similar to other allergens, including Blot 12, in that it contains
carbohydrate-binding domains that bind chitin (26).
To the best of our knowledge, the Group 23 allergen
in D. farinae (Der f 23) had not been identified and
characterized prior to the present study. The aims of this study
were firstly to confirm the existence of Der f 23, and secondly to
characterize the sIgE binding activity of Der f 23, if such an
allergen was able to be isolated. In vitro IgE binding was
determined by IgE western blot analysis, dot blot assays and
ELISAs; in vivo reactivity was assayed with a skin prick
test (SPT). The identification of a novel Der f 23 allergen would
be clinically useful for the diagnosis and treatment of HDM-induced
allergic diseases.
Materials and methods
Materials
A D. farinae cDNA library preserved by the
School of Medicine, Shenzhen University (Shenzhen, China) was
employed. E. coli BL21 (DE3) plysS cells were purchased from
Merck KGaA, and the pMD 19-T vector was purchased from Takara
Biotechnology Co., Ltd. The pET-His and pET-His-DsbA vectors were
purchased from Wuhan Miaoling Bioscience & Technology Co., Ltd.
The DNA sequence encoding the Der f 23Δ P2 protein (P2) was
synthesized by Nanjing GenScript Biotech Corp. The Primer STAR HS
DNA polymerase was purchased from Takara Biotechnology Co., Ltd.
The lysozyme was purchased from Sangon Biotech Co., Ltd. The
nitrocellulose and polyvinylidene fluoride (PVDF) membranes were
purchased from Merck KGaA.
Serum samples from HDM-sensitive individuals,
referred to as HDM allergic sera, and non-allergic individuals were
provided by the First Affiliated Hospital of Guangzhou Medical
College. Sera from non-allergic individuals were used for control
group. A cohort of 129 subjects (65 males and 64 females; age
range, 18–55 years) were enrolled. Samples from 31 non-allergic
individuals were used as negative controls. The HDM-specific IgEs
within the sera samples were assayed using an ImmunoCAP system
(Thermo Fisher Scientific, Inc.). Ethical approval was obtained
from the First Affiliated Hospital of Guangzhou Medical College
(approval no. 2012-51). All procedures involving human participants
were in accordance with the ethical standards of the committee of
the First Affiliated Hospital of Guangzhou Medical College. All
participants voluntarily agreed to participate, and all provided
written informed consent.
Methods
cDNA cloning, protein expression and
purification
Der f 23 open reading frame (ORF) cDNA was amplified
by polymerase chain reaction (PCR) using the Primer STAR HS DNA
polymerase with the D. farinae cDNA library. The primers
used were: Forward, 5′-ATGAAATTCAACATAACTATCGC-3′; and reverse,
5′-TTATGTACATGTTAATTCTTTTTCA-3′. The PCR thermocycling conditions
were 94°C for 30 sec, 55°C for 30 sec and 72°C for 50 sec, for 30
cycles. The PCR product, confirmed by DNA sequencing (GenScript
Biotech Corporation), was subcloned into a pET-His vector, and then
transformed into Escherichia coli (E. coli) BL21
(DE3) plysS. E. coli were grown overnight in Luria-Bertani
medium (Thermo Fisher Scientific, Inc.) containing 100 mg/l
ampicillin at 37°C, and were induced by adding
isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration
of 0.5 mM. Following cultivation for additional 3 h at 37°C, E.
coli cells were harvested by centrifugation at 9,600 × g for 5
min at 4°C. Following mixing with the protein extraction buffer (20
mM PB, 150 mM NaCl and 1 mg/ml lysozyme), the harvested cells were
sonicated for 3 sec with 5-sec intervals in an ice bath for a total
of 20 min, and then centrifuged again at 9,600 g for 20 min at 4°C.
The recombinant protein in the soluble portion was purified with a
Ni-NTA column (cat. no. 17040303; GE Healthcare) and gel filtration
(HiLoad Superdex 16/600; cat. no. 28-9893-33; GE Healthcare).
IgE western blot analysis and IgE dot
blot assays
The protein concentration was determined by Bradford
assay (Sangon Biotech Co., Ltd). The protein samples were diluted
to 1 mg/ml and subjected to electro-transfer onto the PVDF membrane
through 12% SDS-PAGE (20 µg/lane) for the IgE western blot
analysis. Subsequently, the membranes were blocked overnight at 4°C
with 5% skim milk in TBS + 0.05% Tween-20 (TBST). Then, the
membranes were incubated with allergic sera (diluted in 1:10
vol/vol in PBS buffer) for 2 h at 37°C. Following washing, the
membranes were incubated with a horseradish peroxidase
(HRP)-conjugated anti-human IgE antibody for 2 h at 37°C (1:2,000;
cat. no. 9160-05; SouthernBiotech). The proteins were visualized
for 10 min using 3,3′-diaminobenzidine (1:10; cat. no. 34002;
Thermo Fisher Scientific, Inc.) at room temperature.
The IgE dot blot assay was performed as previously
described (23). The protein
samples were diluted to 1 mg/ml, and 2 µl samples were added to the
nitrocellulose membrane for the IgE dot blot assay. Subsequently,
the membranes were dried and blocked overnight at 4°C with 5% skim
milk in TBST. Then, the membranes were incubated with allergic sera
(diluted in 1:10 vol/vol in PBS buffer) for 2 h at 37°C. Following
washing, the membranes were incubated with the HRP-conjugated
anti-human IgE antibody for 2 h at 37°C (1:2,000; cat. no. 9160-05;
SouthernBiotech). The proteins were visualized for 10 min using
3,3′-diaminobenzidine (1:10; cat. no. 34002; Thermo Fisher
Scientific, Inc.).
IgE ELISA
The IgE ELISA assay developed in the present study
was performed as previously described (23). The isolated protein was added to
the ELISA plate at 100 ng/well and coated overnight at 4°C. The
coated plate was blocked with 5% skim milk in PBS + 0.1% Tween-20
(PBST) for 2 h at 37°C. Then, the plate was incubated with allergic
sera (diluted 1:10 vol/vol in 1% skim milk-PBST) for 2 h at 37°C.
IgEs were detected with the HRP-conjugated mouse anti-human IgE
antibody (1:2,000 in 1% skim milk-PBST; cat. no. 9160-05;
SouthernBiotech). Binding reactions were visualized by adding
tetramethyl benzidine substrate, and absorbance at 450 nm was
measured by a microplate reader (Bio-Rad Laboratories, Inc.).
SPTs
SPTs were performed with rDer f 23 protein
(10 mg/ml) and standardized D. farinae extract (ALK Abelló
A/S) in 10 patients with allergic rhinitis and/or asthma and 10
healthy controls (Table I)
(23). The response was observed
15 min after pricking. The result was considered positive when the
prick spot developed a wheal with a surrounding fleck; no visible
response was considered a negative result.
 | Table I.Skin reactivity to Der f
extracts and rDer f 23 protein (10 µg/ml). |
Table I.
Skin reactivity to Der f
extracts and rDer f 23 protein (10 µg/ml).
|
|
| Skin prick
testsa |
|---|
|
|
|
|
|---|
| Patient no. | Sex/age | Clinical
history | Der f
extracts | rDer f 23 |
|---|
| 1 | Female/39 | AR | 3+ | 2+ |
| 2 | Female/38 | AR+BA | 3+ | – |
| 3 | Male/25 | AR | 3+ | – |
| 4 | Female/40 | AR | 3+ | – |
| 5 | Male/52 | AR | 4+ | 2+ |
| 6 | Female/28 | BA | 3+ | – |
| 7 | Male/24 | AR | 4+ | 2+ |
| 8 | Female/23 | AR | 3+ | – |
| 9 | Male/38 | BA | 3+ | – |
| 10 | Male/44 | AR+BA | 3+ | – |
Sequence homology
The nucleotide sequences of Der f 23 (GenBank:
KU166910.1) and Der p 23 (GenBank: KP895831) were imported into
DNAMAN 8 software (version 8.0; Lynnon Biosoft) for alignment. The
sequences were saved in FASTA format for subsequent analysis.
Expression and purification of Der f
23ΔP2 and P2 proteins
Der f 23ΔP2 and P2 region gene sequences were
synthesized by Nanjing GenScript Biotech Corp., and each was
subcloned into the pET-His-DsbA vector. The recombinant proteins
were expressed and purified as aforementioned. Specifically, the
Der f 23ΔP2 protein was expressed in inclusion bodies and was
dissolved in 6 M urea with 25 mM β-mercaptoethanol. For the
renaturation process, the Der f 23∆P2 protein was dialyzed with PBS
at 4°C for 24 h using a dialysis membrane (cat. no. F132550; Sangon
Biotech Co., Ltd.). Additionally, soluble recombinant DsbA-P2
protein was obtained. The purified protein Der f 23 ΔP2 and DsbA-P2
were subjected to in vitro IgE binding assays.
Statistical analysis
Quantitative data are presented as means ± standard
error of the mean. Analyses were performed using GraphPad Prism 7
software (GraphPad Software, Inc.). Differences between the
allergic and control groups were determined by one-way ANOVA
followed by Dunnett's post-hoc test for multiple comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Amino acid sequence homology between
Der f 23 and Der p 23
The Der f23 ORF sequence was cloned using the D.
farinae cDNA library as a template, and specific Der f 23
sequence primers were designed based on the D. farinae
genome in the National Center for Biotechnology Information (NCBI)
database (Genome ID: 9138). DNA sequencing confirmed the sequence
of the ORF cDNA of Der f 23 to be 525 base pairs, and the sequence
was registered in the NCBI library (GenBank: KU166910.1). A
homology comparison conducted in DNAMAN 8 software indicated that
Der f 23 has 173 amino acids, whereas Der p 23 has 97 amino acids,
with the additional amino acids constituting 2 extra regions,
namely P1 (Phe26-Pro38) and P2 (Ser56-Thr117), as demonstrated in
Fig. 1.
Cloning, expression and purification
of Der f 23
To obtain the Der f 23 protein, a prokaryotic
pET-His vector was constructed and transformed into E. coli
for expression and purification. The Der f 23 ORF DNA sequence
obtained by PCR amplification with specific primers, as
demonstrated in Fig. 2A, was
cloned into a pET-His vector. The pET-His-Der f 23 vector was
identified by double digestion with BamH I and
HindIII (Fig. 2B).
Subsequently, the pET-His-Der f 23 vector was transformed into
E. coli BL21 (DE3) plysS cells for expression and
purification. SDS-PAGE analysis demonstrated that Der f 23 (~30
kDa) was successfully expressed in the supernatant following IPTG
induction and purification with Ni-NTA-resin (Fig. 2C and D).
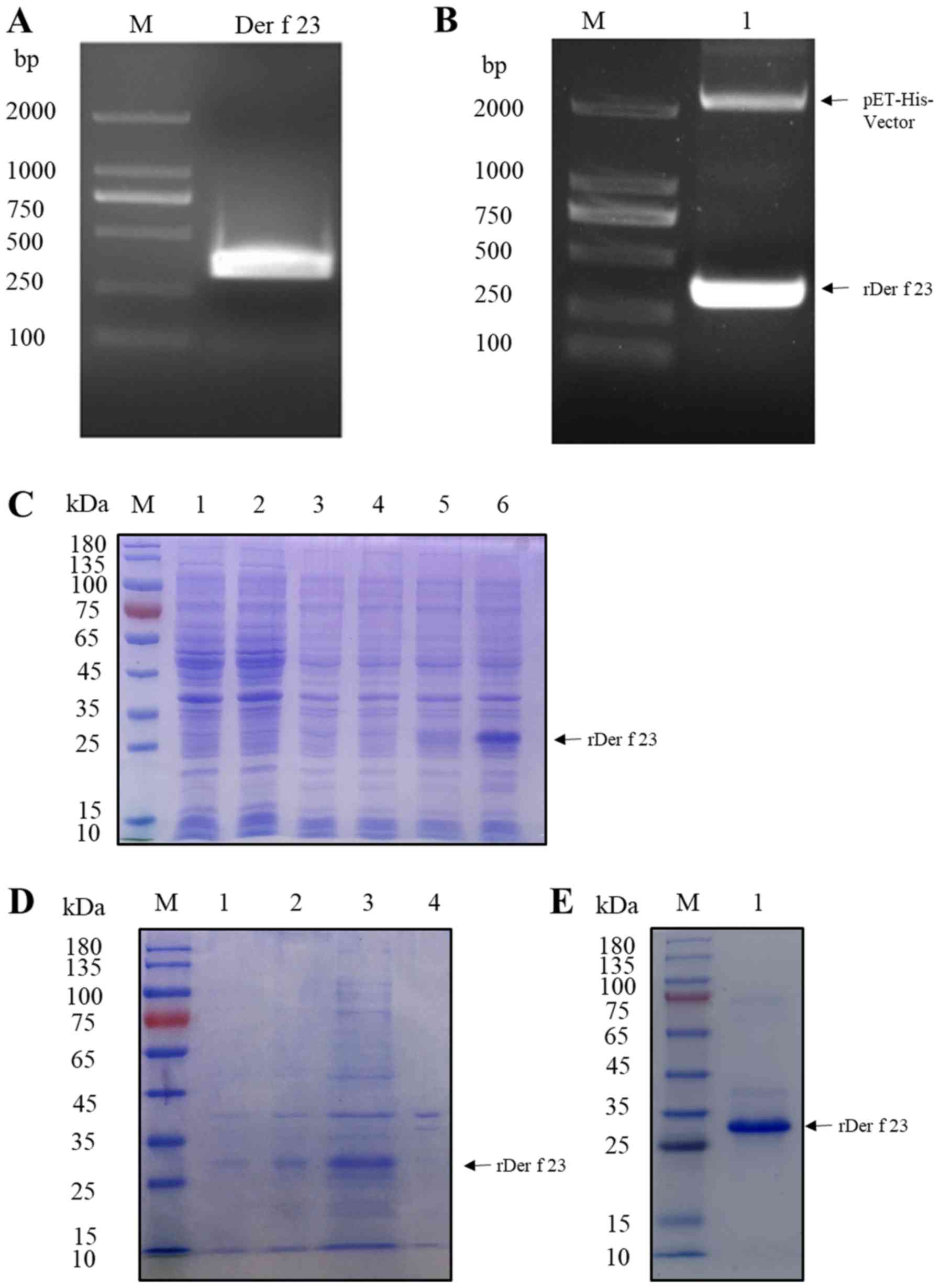 | Figure 2.Cloning, expression and purification
of rDer f 23. (A) Cloning of Der f 23 cDNA by polymerase chain
reaction. Lane M, DL2000 marker; lane 1, Der f 23 cDNA PCR product.
(B) Analysis of expression vector pET-His-Der f 23 by double enzyme
digestion. Lane M, DL2000 Marker; lane 1, pET-His-Der f 23 vector
digested by BamH I/HindIII. (C) Analysis of
purification of pET-His-Der f 23 by SDS-PAGE. Lane M, protein
marker; lane 1, cell lysate of E. coli without induction;
lane 2, cell lysate of E. coli following IPTG induction;
lane 3, E. coli transformed with pET-His-vector without
induction; lane 4m, cell lysate of pET-His-vector following IPTG
induction; lane 5, E. coli transformed with pET-Der f 23
prior to IPTG induction; lane 6, E. coli transformed with
pET-Der f 23 following IPTG induction. (D) Analysis of expression
of rDer f 23 in an E. coli system. Lane M, protein marker;
lane 1, E. coli transformed with pET-Der f 23 prior to IPTG
induction; lane 2, E. coli transformed with pET-Der f 23
following IPTG induction; lane 3, supernatant from E. coli
transformed with pET-His-Der f 23 plasmid following IPTG induction,
subsequent to ultrasonication; lane 4, sediment from E. coli
samples transformed with pET-His-Der f 23 plasmid following IPTG
induction, subsequent to ultrasonication. (E) SDS-PAGE of purified
rDer f 23. Lane M, protein marker; lane 1, purified rDer f 23.
E. coli, Escherichia coli; rDer f 23, recombinant
Dermatophagoides farinae 23 protein; IPTG,
isopropyl-β-D-thiogalactopyranoside. |
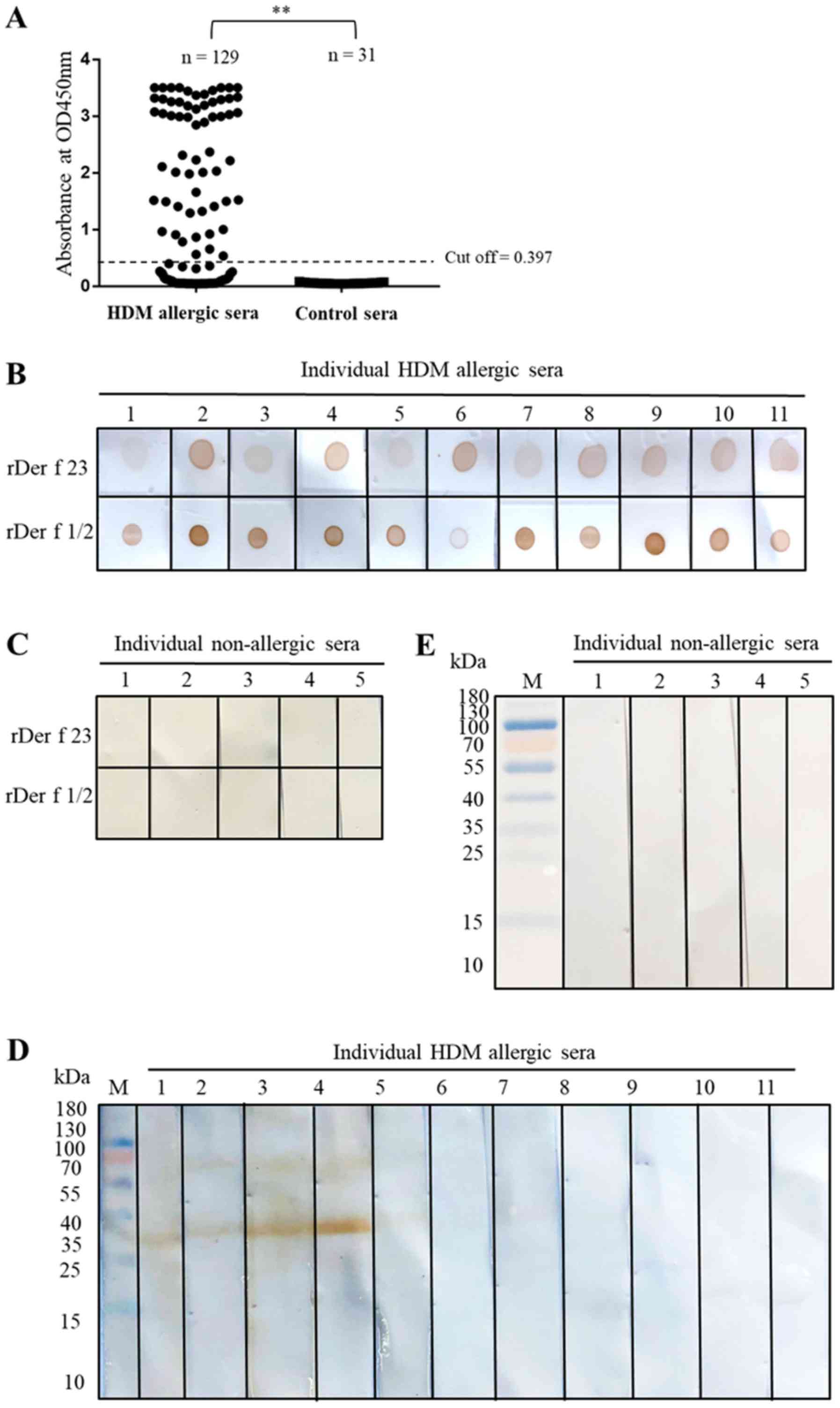
IgE binding of Der f 23
The IgE ELISA indicated that HDM allergic sera from
72/129 patients with HDM allergies (55.8%) exhibited sIgE binding
activity with Der f 23 (Fig. 3A).
None of the serum samples from 31 non-allergic individuals were
reactive (Fig. 3A). Data from the
IgE dot blot assays suggested that Der f 23 protein exhibited a
good sIgE binding capacity for HDM allergic sera, but not for sera
from non-allergic individuals. Der f 1/2 fusion protein was used as
positive control (Fig. 3B). In the
IgE western blot analysis, only 4/11 selected IgE ELISA reactive
sera exhibited sIgE reactivity to rDer f 23 (Fig. 3D), suggesting that sIgE binding
epitopes in Der f 23 may be conformation-dependent. As demonstrated
in Table I, 3/10 (30%) of patients
with HDM allergies exhibited a positive in vivo reaction to
Der f 23 in the SPTs. Based on these results, the WHO/IUIS Allergen
Nomenclature subcommittee published this allergenic HDM protein as
Der f 23 (16).
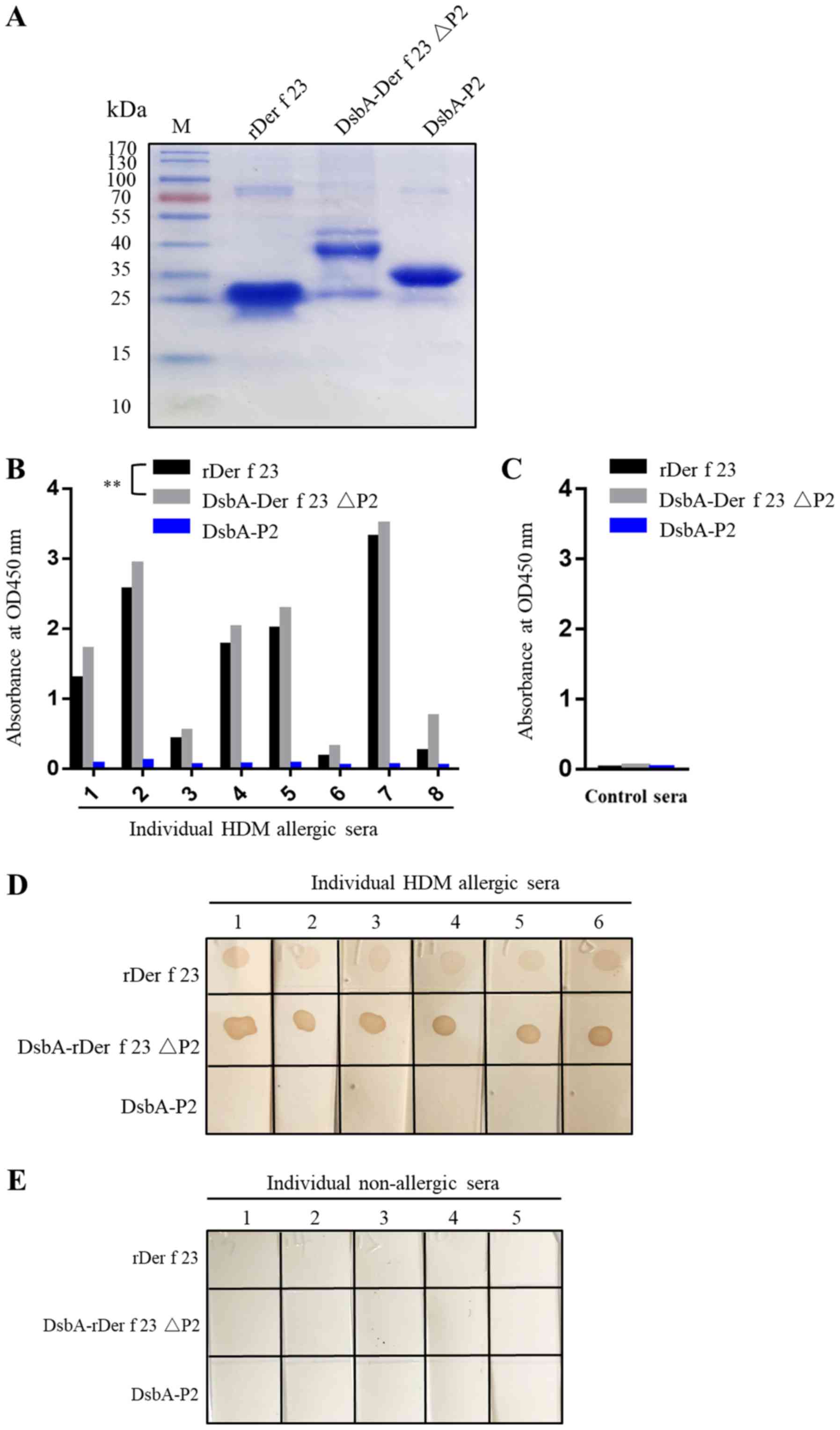
Involvement of P2 region
(Ser56-Thr117) in sIgE binding
The P2 region peptide and Der f 23 ΔP2 protein were
expressed in E. coli. DsbA-Der f 23ΔP2 was observed in the
form of inclusion bodies with a molecular weight of ~38 kDa and
DsbA-P2 was soluble with a molecular weight of ~35 kDa (Fig. 4A). The IgE ELISA demonstrated that
rDer f 23 and DsbA-Der f 23ΔP2 exhibited significant binding
activity, with all 8 HDM allergic sera tested, whereas P2 exhibited
no IgE binding reactivity with any of the sera (Fig. 4B). The DsbA protein served as a
negative control for sIgE binding activity (data not shown).
Notably, within the same positive sera, the optical density value
of DsbA-Der f 23ΔP2 was increased compared with that of rDer f 23
(P<0.05). Similarly, the dot blot assay results confirmed that
DsbA-Der f 23 ΔP2 exhibited significant sIgE binding reactivity,
while the P2 peptide did not. The color reaction was more marked
for DsbA-Der f 23 ΔP2 compared with the Der f 23. These data
indicated that the P2 region of Der f 23 may affect the Der f 23
protein structure and thereby attenuate the IgE binding ability of
Der f 23 in sera.
Discussion
The present study cloned, expressed and purified
rDer f 23 protein using an E. coli expression system. The
resultant recombinant protein exhibited sIgE binding activity in
vitro and in vivo, and was therefore included as Der f
23 in the WHO/IUIS allergen database (16). The present study identified that,
in comparison with Der p 23, Der f 23 contained an extra P2 region
that affected its IgE binding ability. In the IgE ELISAs, 72/129
positive sera (55.81%) from patients with HDM allergies exhibited
marked sIgE binding activity to rDer f 23 protein. These data
confirmed that Der f 23 is a major allergen, consistent with
previous results for Der p 23 (21).
The allergen components of the 2 HDM species D.
farinae and D. pteronyssinus are highly homologous,
encompassing 39 HDM allergen groups (16). Indeed, the Der p 23 allergen
identified in the present study had 71% amino acid sequence
homology with Der f 23. Additionally, it was identified that Der p
23 had 2 extra regions not present in Der f 23, namely a P1 region
(Phe26-Pro38) and a P2 region (Ser56-Thr117). These sequence
differences may be the underlying cause of a differential
allergenic effect between Der f 23 and Der p 23 proteins, and
therefore an allergic distinction between the 2 HDM species of
D. farinae and D. pteronyssinus. In light of recent
evidence demonstrating the existence of multiple Der f 23 isoforms
(27), we hypothesized that the
sequence difference between Der f 23 and Der p 23 may be due to the
insertion of an intron in the gene that encodes Der f 23.
The IgE binding ability of HDM allergens is based
primarily on B-cell epitopes of allergens, which may occur in
conformational or linear form. Conformational epitopes are more
favorable for IgE binding compared with linear epitopes (28). Conformational changes in IgEs
contribute to a decrease in their dissociation rates from
high-affinity IgE receptors (29).
B-cell epitopes are primarily conformational, including some that
are discontinuous, in which polypeptide chain folding draws distant
amino acid segments in the primary structure into close proximity
on the surface of the molecule (30,31).
These regions are able to form complementarity-determining regions
recognized by antibodies (32).
The results from the present study, that only 4/11 sera with a
positive IgE ELISA result also exhibited a positive IgE western
blot analysis result suggests that the IgE epitope of Der f 23 may
be predominantly a conformational epitope, consistent with the data
from Szalai et al (33),
which suggested that the B-cell epitopes of Der p 1 and Der p 2 are
conformational.
Despite their high degrees of homology, different
HDM allergen components within the same group have differing IgE
binding abilities determined by their amino acid sequences and
protein structures. For example, although Der p 1 and Der f 1 share
an extensive sequence identity, differences identified in their
crystal structures may explain the differences in human IgE
antibody responses to these allergens (34). The IgE ELISA experiment conducted
in the present study to examine the role of the P2 region of Der f
23, which is not present in Der p 23, in the allergenicity of Der f
23 indicated that the rDer f 23 and DsbA-Der f 23 ΔP2 proteins
exhibited positive sIgE binding activity with HDM allergic sera,
whereas DsbA-P2 did not. In addition, the optical density value at
450 nm for DsbA-Der f 23 ΔP2 reacting with sIgE from HDM allergic
sera was increased compared with that of rDer f 23. Furthermore,
the IgE dot blot experiment revealed a more marked reaction for
DsbA-Der f 23ΔP2 compared with rDer f 23 in HDM allergic sera from
6 individuals, consistent with the results of the western blot
analysis. We hypothesized that the P2 domain region affects the
spatial location of the IgE binding epitope, but this requires
additional studies to confirm. Taken together, these results
indicate that the addition of the P2 domain has the ability to
affect the allergenic capabilities of HDM allergens.
In conclusion, Der f 23 was identified and
characterized using sIgE binding assays in vitro, and Der f
23 immunogenicity was confirmed with in vivo SPTs. Based on
these data, Der f 23 was included in the WHO/IUIS allergen database
(21). The results of the present
study suggested that Der f 23 is an important allergen of D.
farinae that interacts with sIgEs by way of conformational
epitopes. An amino acid sequence difference between Der f 23 and
Der p 23, namely the P2 region that is present only in the former,
was demonstrated to affect the sIgE binding activity of Group 23
HDM allergens. These results contribute to the theoretical basis
for the diagnosis and treatment of HDM allergies.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by research
funding from the National Natural Science Foundation of China
(grant no. 81571570), and Guangdong Province (grant nos.
2018A050506083, 2014A030313563, 2016A030313039 and 2017A010105014)
and Shenzhen City (grant no. JCYJ20150626141652681 and 2016
Biochemistry Discipline Construction).
Availability of data and materials
All data generated or analyzed in the present study
are available from the corresponding author upon reasonable
request.
Authors' contributions
YH performed the experiments and wrote the draft
manuscript. CD, YS, JialC and ZZhan participated in the
experiments. JiajC and KJ designed the study and wrote the
manuscript. ZZhao made revisions to the manuscript and participated
in study design. All of the authors reviewed and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from the First
Affiliated Hospital of Guangzhou Medical College (approval no.
2012-51). All procedures involving human participants were in
accordance with the ethical standards of the committee of The First
Affiliated Hospital of Guangzhou Medical College. All participants
voluntarily agreed to participate, and all provided written
informed consent.
Patient consent for publication
All participants voluntarily agreed to participate,
and all provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HDM
|
house dust mite
|
|
Der f
|
Dermatophagoides farinae
|
|
Der p
|
Dermatophagoides
pteronyssinus
|
|
Der f 23
|
Group 23 allergen of
Dermatophagoides farinae
|
|
Der p 23
|
Group 23 allergen of Dermatophagoides
pteronyssinus
|
|
IPTG
|
isopropyl-β-D-thiogalactopyranoside
|
|
HRP
|
horseradish peroxidase
|
References
|
1
|
Iversen M, Korsgaard J, Hallas T and Dahl
RJ: Mite allergy and exposure to storage mites and house dust mites
in farmers. Clin Exp Allergy. 20:211–219. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Custovic A, Taggart SC and Woodcock A:
House dust mite and cat allergen in different indoor environments.
Clin Exp Allergy. 24:1164–1168. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krantz GW: A manual of acarology (Oregon
State University Book Stores). 194–195. 2010.
|
|
4
|
Kim HK, Yun YK and Ahn YJ: Fumigant
toxicity of cassia bark and cassia and cinnamon oil compounds to
Dermatophagoides farinae and Dermatophagoides pteronyssinus (Acari:
Pyroglyphidae). Exp Appl Acarol. 44:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arshad SH, Bateman B, Sadeghnejad A, Gant
C and Matthews SM: Prevention of allergic disease during childhood
by allergen avoidance: The Isle of Wight prevention study. J
Allergy Clin Immunol. 119:307–313. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Suh J, Vaccaro L and Bogart R: Methods for
controlling dust mites and the allergens produced by dust mites.
(US). 2002.
|
|
7
|
Lau S, Illi S, Sommerfeld C, Niggemann B,
Bergmann R, von Mutius E and Wahn U: Early exposure to house-dust
mite and cat allergens and development of childhood asthma: A
cohort study. Multicentre Allergy Study Group. Lancet.
356:1392–1397. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gelber LE, Seltzer LH, Bouzoukis JK,
Pollart SM, Chapman MD and Platts-Mills TA: Sensitization and
exposure to indoor allergens as risk factors for asthma among
patients presenting to hospital. Am Rev Respir Dis. 147:573–578.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim HS, Kang SH, Won S, Lee EK, Chun YH,
Yoon JS, Kim HH and Kim JT: Immunoglobulin E to allergen components
of house dust mite in Korean children with allergic disease. Asia
Pac Allergy. 5:156–162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Charpin D, Birnbaum J, Haddi E, Genard G,
Lanteaume A, Toumi M, Faraj F, Van der Brempt X and Vervloet D:
Altitude and allergy to house-dust mites. A paradigm of the
influence of environmental exposure on allergic sensitization. Am
Rev Respir Dis. 143:983–986. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pittner G, Vrtala S, Thomas WR, Weghofer
M, Kundi M, Horak F, Kraft D and Valenta R: Component-resolved
diagnosis of house-dust mite allergy with purified natural and
recombinant mite allergens. Clin Exp Allergy. 34:597–603. 2010.
View Article : Google Scholar
|
|
12
|
Custovic A, Taggart SC, Francis HC,
Chapman MD and Woodcock A: Exposure to house dust mite allergens
and the clinical activity of asthma. J Allergy Clin Immunol.
98:64–72. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boyce JA, Assa'a A, Burks AW, Jones SM,
Sampson HA, Wood RA, Plaut M, Cooper SF, Fenton MJ, Arshad SH, et
al: Guidelines for the diagnosis and management of food allergy in
the United States: Summary of the NIAID-sponsored expert panel
report. Nutrition. 27:253–267. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Novakova SM, Staevska MT, Novakova PI,
Yoncheva MD, Bratoycheva MS, Musurlieva NM, Tzekov VD and Nicolov
DG: Quality of life improvement after a three-year course of
sublingual immunotherapy in patients with house dust mite and grass
pollen induced allergic rhinitis: Results from real-life. Health
Qual Life Outcomes. 15:1892017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chapman MD, Smith AM, Vailes LD, Arruda
LK, Dhanaraj V and Pomés A: Recombinant allergens for diagnosis and
therapy of allergic diseases. J Allergy Clin Immunol. 106:409–418.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sub-Committee WIAN, . WHO/IUIS Allergen
Tree View. http://www.allergen.org/treeview.phpMay
23–2019
|
|
17
|
Chapman MD and Platts-Mills TA:
Purification and character-ization of the major allergen from
Dermatophagoides pteronyssinus-antigen P1. J Immunol. 125:587–592.
1980.PubMed/NCBI
|
|
18
|
Maeda S, Maeda S, Shibata S, Chimura N and
Fukata T: House dust mite major allergen Der f 1 enhances
proinflammatory cytokine and chemokine gene expression in a cell
line of canine epidermal keratinocytes. Vet Immunol Immunopathol.
131:298–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park GM, Lee SM, Lee IY, Ree HI, Kim KS,
Hong CS and Yong TS: Localization of a major allergen, Der p 2, in
the gut and faecal pellets of Dermatophagoides pteronyssinus. Clin
Exp Allergy. 30:1293–1297. 2010. View Article : Google Scholar
|
|
20
|
Ichikawa S, Hatanaka H, Yuuki T, Iwamoto
N, Kojima S, Nishiyama C, Ogura K, Okumura Y and Inagaki F:
Solution structure of Der f 2, the major mite allergen for atopic
diseases. J Biol Chem. 273:356–360. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weghofer M, Grote M, Resch Y, Casset A,
Kneidinger M, Kopec J, Thomas WR, Fernández-Caldas E, Kabesch M,
Ferrara R, et al: Identification of Der p 23, a peritrophin-like
protein, as a new major Dermatophagoides pteronyssinus allergen
associated with the peritrophic matrix of mite fecal pellets. J
Immunol. 190:3059–3067. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mueller GA, Randall TA, Glesner J,
Pedersen LC, Perera L, Edwards LL, DeRose EF, Chapman MD, London RE
and Pomés A: Serological, genomic, and structural analyses of the
major mite allergen Der p 23. Clin Exp Allergy. 46:365–376. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chan TF, Ji KM, Yim AK, Liu XY, Zhou JW,
Li RQ, Yang KY, Li J, Li M, Law PT, et al: The draft genome,
transcriptome, and microbiome of Dermatophagoides farinae reveal a
broad spectrum of dust mite allergens. J Allergy Clin Immunol.
135:539–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Banerjee S, Weber M, Blatt K, Swoboda I,
Focke-Tejkl M, Valent P, Valenta R and Vrtala S: Conversion of Der
p 23, a new major house dust mite allergen, into a hypoallergenic
vaccine. J Immunol. 192:4867–4875. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tovey ER, Chapman MD and Platts-Mills TA:
Mite faeces are a major source of house dust allergens. Nature.
289:592–593. 1981. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fanuel S, Tabesh S, Sadroddiny E and
Kardar GA: Analysis of predicted B and T-cell epitopes in Der p 23,
allergen from Dermatophagoides pteronyssinus. Bioinformation.
13:307–312. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Randall TA, Mullikin JC and Mueller GA:
The Draft Genome Assembly of Dermatophagoides pteronyssinus
supports identification of novel allergen isoforms in
Dermatophagoides species. Int Arch Allergy Immunol. 175:136–146.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gieras A, Cejka P, Blatt K, Focke-Tejkl M,
Linhart B, Flicker S, Stoecklinger A, Marth K, Drescher A,
Thalhamer J, et al: Mapping of conformational IgE epitopes with
peptide-specific monoclonal antibodies reveals simultaneous binding
of different IgE antibodies to a surface patch on the major birch
pollen allergen, Bet v 1. J Immunol. 186:5333–5344. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Holdom MD, Davies AM, Nettleship JE, Bagby
SC, Dhaliwal B, Girardi E, Hunt J, Gould HJ, Beavil AJ, McDonnell
JM, et al: Conformational changes in IgE contribute to its uniquely
slow dissociation rate from receptor FcεRI. Nat Struct Mol Biol.
18:571–576. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Laver WG, Air GM, Webster RG and
Smith-Gill SJ: Epitopes on protein antigens: Misconceptions and
realities. Cell. 61:553–556. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sela M: Antigenicity: Some molecular
aspects. Science. 166:1365–1374. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Crameri RJ: Correlating IgE reactivity
with three-dimensional structure. Biochem J. 376:e1–e2. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Szalai K, Fuhrmann J, Pavkov T, Scheidl M,
Wallmann J, Brämswig KH, Vrtala S, Scheiner O, Keller W, Saint-Remy
JM, Neumann D, et al: Mimotopes identify conformational B-cell
epitopes on the two major house dust mite allergens Der p 1 and Der
p 2. Mol Immunol. 45:1308–1317. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chruszcz M, Chapman MD, Vailes LD, Stura
EA, Saint-Remy JM, Minor W and Pomés A: Crystal structures of mite
allergens Der f 1 and Der p 1 reveal differences in surface-exposed
residues that may influence antibody binding. J Mol Biol.
386:520–530. 2009. View Article : Google Scholar : PubMed/NCBI
|