Introduction
According to data released by GLOBOCAN in 2012, the
incidence of colorectal cancer (CRC) was the third highest in
regards to all male malignant tumors and the second highest in
regards to female malignant tumors (1). Studies have shown that the
progression of CRC is a multi-stage, multi-factor process (1–3).
Effective inhibition of CRC metastasis is a major clinical issue.
It has been found that the invasion and migration of tumors are
closely related to epithelial-mesenchymal transition (EMT)
(4).
EMT refers to the phenomenon that epithelial cells
switch to mesenchymal cells under specific pathological conditions
(4). During the evolution of tumor
growth, EMT mainly includes changes in reconstruction of the
cytoskeleton, cell polarity, loss of adhesion between cells,
destruction of the tumor basement membrane and extracellular
matrix, resulting in high migration and invasion capacity,
anti-apoptosis and degradation of interstitial phenotypic
characteristics such as the extracellular matrix (5). Studies have shown that matrix
metalloproteinase (MMP) family genes, especially MMP-2 and
MMP-9, can alter the microenvironment of epithelial cells by
remodeling of the matrix membrane and extracellular matrix,
downregulating E-cadherin expression, upregulating vimentin
expression and promoting EMT progression (6). This process is usually triggered by a
variety of growth factors (such as epidermal growth factor) to
activate the cascade of signal transduction pathways such as Wnt,
resulting in enhanced anti-apoptotic and invasive capacities of
tumor-initiating cells, ultimately leading to tumor development
(7–9).
One of the most important signaling pathways for
regulating EMT is the β-catenin-dependent classical Wnt signaling
pathway (10). β-catenin is an
intercellular adhesion molecule, mainly located in the cell
membrane, which mediates intercellular adhesion and participates in
gene expression. However, the adhesion activity will disappear once
β-catenin is translocated to the cell nucleus or degraded (11). Overexpression of β-catenin is also
a major manifestation of activation of signaling pathways (12). β-catenin is a key regulator of the
Wnt/β-catenin signaling pathway. Wnt proteins bind to their
receptors, which trigger intracellular signal transduction, and
lead to β-catenin accumulation within the cell which translocates
into the nucleus and interacts with transcription factors (such as
TCF) to activate the transcription of the target genes such as
c-Myc and cyclin D1 (13,14).
Research has confirmed that the Wnt signaling pathway activates
genes related to the MMP family and leads to the development of EMT
(15).
Stanniocalcin 2 (STC2) is a secreted glycoprotein
that is expressed in a variety of tissues and organs and functions
in an autocrine or paracrine manner (16). Recent experiments have shown that
STC2 plays an important role in the development of cancer
including CRC (17,18). However, to the best of our
knowledge, whether or not STC2 regulates the Wnt signaling
pathway in CRC has not yet been studied. In the present study, CRC
cell line SW480 was used as the main research tool to investigate
whether or not the effect of STC2 on CRC proliferation,
migration and EMT is related to the Wnt signaling pathway.
Materials and methods
Reagents
Human CRC cell lines (SW480, SW620, HT29, LoVo and
HCT116) and human normal colorectal mucosal FHC cells were obtained
from the American Type Culture Collection (ATCC; Rockville, MD,
USA). SB216763 was purchased from Sigma-Aldrich (280744-09-4; HPLC
>98%; Merck KGaA, Darmstadt, Germany). CCK-8 assay kit was
obtained from Nanjing Jiancheng Bioengineering Institute (Nanjing,
China). Transfection reagent Lipofectamine® 3000 was
purchased from Invitrogen (Thermo Fisher Scientific, Inc., Waltham,
MA, USA).
Cell culture and transfection
Human CRC cell lines (SW480, SW620, HT29, LoVo and
HCT116) and human normal colorectal mucosal cells (FHC) were
cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific,
Inc.) containing 10% (v/v) fetal bovine serum (FBS; Invitrogen;
Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere
with 5% CO2. The non-specific control small interference
RNA (siRNA) and siSTC2 were obtained from Shanghai GenePharma Co.,
Ltd. (Shanghai, China). The siSTC2 was inserted into the pSUPER
vector (OligoEngine, Inc., Seattle, WA, USA), and the empty pSUPER
vector was used as a negative control. Lipofectamine®
(LFN) transfection (Thermo Fisher Scientific, Inc.) was performed
according to the manufacturer's instructions. In brief, the lipid
complex was prepared by combining 4 µl of the reagent LFN Plus with
2 µg of plasmid DNA suspended in 1 ml serum-free medium and
incubated at room temperature for 15 min. The solution was then
mixed with 40 µl of LFN in serum-free medium and further incubated
at room temperature for 15 min. Lipid compounds will first be
diluted in serum-free medium to produce a required concentration
for 5 ml volume, and then incubated with the cells in an incubator
with 5% CO2 at 37°C under 24 h for subsequent
experimentation.
Cell viability assay
Having transferred the SW480 cells at 0, 12, 24 and
48 h post-culture, 10 µl of CCK-8 solution was added into the wells
and the cells were incubated at 37°C for 2 h in an incubator with
5% CO2 in the dark. Subsequently, the optical density
(OD) of each well in the different cell groups at an absorbance of
450 nm was determined using a microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Cell viability was detected
using the CCK-8 assay kit according to the manufacturer's
protocol.
Wound healing assay
Having transferred the SW480 cells for 24 h, a
straight gap was created using a 200-µl sterile tip in the middle
of the well. The cells were washed with Dulbecco's modified Eagle's
medium (DMEM) for 2 times for smoothing the edges of the scratch
and removing floating cells. After being incubated in an incubator
(37°C, 5% CO2) for 0 and 24 h, the images of the
migrating cells were observed under a digital microscope (Keyence
Corp., Osaka, Japan), and the distance of cell migration was
measured by Image-Pro Plus Analysis software version 7 (Media
Cybernetics, Inc., Rockville, MD, USA). The experiment was repeated
3 times and the results are presented using mean values.
Transwell assay
A Transwell chamber with 8-µm pores (3413; Corning
Inc., Corning, NY, USA) was placed on a 24-well plate with a layer
of 50 µl Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) coated
onto the Transwell chamber. The SW480 cells were cultured in
serum-free medium for 12 h to eliminate the effects of the serum
and then resuspended in DMEM containing bovine serum albumin (BSA;
Sigma-Aldrich; Merck KGaA) with free FBS. A total of 100 µl of the
suspended cells was added to the Transwell chamber, and 400 µl of
DMEM containing 20% FBS was added to the basolateral chamber. The
cells were cultured for 24 h at 37°C in an incubator with 5%
CO2. The Transwell chamber was removed and the culture
solution in the Transwell was discarded and washed with
calcium-free phosphate-buffered saline (PBS) for 2 times.
Subsequently, the chamber was fixed in methanol solution for 30 min
and stained with 0.1% crystal violet for 20 min at room
temperature. Next, the chamber was washed several times with PBS,
and the upper chamber liquid was aspirated. The non-migratory cells
in the upper layer were gently wiped off using a cotton swab. The
microporous membrane was carefully removed using small tweezers,
dried with the bottom side up and then transferred to a glass slide
and sealed using a neutral gum. Images were observed and collected
by an inverted optical microscope (Keyence Corp.).
Western blot analysis
The SW480 cells were washed twice with PBS and added
to protein lysis buffer (RIPA; Cell Signaling Technology, Inc.,
Danvers, MA, USA) for 2 h on ice, centrifuged at 12,000 × g for 30
min at 4°C, and finally the supernatant was collected. The protein
concentration was tested using the BCA protein kit (Bio-Rad
Laboratories) and adjusted to a concentration of 5 µg/µl using 1X
loading and diethylpyrocarbonate (DEPC) water. A total of 6 µl (at
least 30 µg) of the samples were electrophoresed (80 V for 30 min
and then transferred to 120 V for 1.5 h) on 10% running gels, which
afterwards, was transferred to polyvinylidene fluoride (PVDF)
membranes (Bio-Rad Laboratories) on ice for 110 min at 110 V. The
membranes were blocked with 5% non-fat milk and eluted 3 times with
TBS for 5 min each time. The bands were then incubated overnight
with the corresponding primary antibody, washed with TBS 3 times
for 15 min, incubated with secondary antibodies [horseradish
peroxidase (HRP)-conjugated goat anti-mouse/rabbit IgG (dilution
1:2,000; cat. nos. sc-516102 and sc-2357; Santa Cruz Biotechnology,
Inc. Dallas, TX, USA)] for 2 h at room temperature, washed with TBS
3 times for 15 min each time, and furthermore washed once with
TBS/0.1% Tween-20 (TBST) for 15 min. Development was carried out
with a developer (EZ-ECL kit; Biological Industries Ltd., Kibbutz
Beit Haemek, Israel), and the gray-scale value of the strips were
analyzed and determined by ImageJ (version 5.0; Bio-Rad
Laboratories). The antibodies used in the present study were as
follows: Anti-GAPDH (mouse; dilution 1:1,000; cat. no. sc-47724;
Santa Cruz Biotechnology), anti-STC2 (rabbit; dilution 1:1,000;
cat. no. ab63057; Abcam, Cambridge, MA, USA), anti-E-cadherin
(mouse; 1:1,000; ab1416; Abcam), anti-vimentin (rabbit; dilution
1:1,000; cat. no. ab92547; Abcam), anti-MMP-2 (rabbit; dilution
1:1,000; cat. no. ab37150; Abcam), anti-MMP-9 (rabbit; dilution
1:1,000; cat. no. ab73734; Abcam).
RNA isolation and real-time PCR
The cell culture medium in each well was aspirated
as much as possible, and 1 ml of TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.) was added to the SW480 cells. The cells were
placed horizontally for a while and blown evenly. The cells
containing the lysate were transferred to a 1.5-ml EP tube and
allowed to stand at room temperature for 5 min. Chloroform (200 µl)
was added to each tube and inverted for 15 sec. After
emulsification, the cells were allowed to stand for 5 min. After
being centrifuged at 12,000 × g for 15 min at 4°C, the upper
aqueous phase was pipetted into a new 1.5 ml of EP and an equal
volume of isopropanol (~400 µl) was added to each tube and allowed
to stay at room temperature for 10 min. After being centrifuged at
12,000 × g for 15 min at 4°C, the supernatant was discarded and 1
ml of pre-cooled 75% ice ethanol was added. After being centrifuged
at 7,500 × g for 10 min at 4°C, the supernatant was discarded. An
appropriate amount of DEPC (20 µl) was added to dissolve the RNA.
The purity and concentration of RNA were tested at 260/280 nm using
the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies;
Thermo Fisher Scientific, Inc.). According to the program provided
by the manufacturer (Thermo Fisher Scientific, Inc.), a reverse
transcription cDNA kit was used to reverse transcribe 1 µg total
RNA for synthesis of cDNA (at 42°C for 60 min, at 70°C for 5 min,
at 4°C for preservation). SYBR-Green PCR Master Mix (Roche
Diagnostics, Basel, Switzerland) was used to perform quantitative
real-time polymerase chain reaction (qPCR) experiment using Opticon
Real-Time PCR Detection System (ABI 7500; Life Technologies; Thermo
Fisher Scientific, Inc.). The PCR cycle was as follows:
Pretreatment at 95°C for 10 min; followed by 40 cycles at 94°C for
15 sec, at 60°C for 1 min, finally at 60°C for 1 min and at 4°C for
preservation. The relative mRNA quantity was determined using
comparative cycle threshold (ΔΔCq) method (19). GAPDH expression was used for
normalization. The primer sequences used for RT-qPCR analysis are
listed in Table I.
 | Table I.Primers for qPCR. |
Table I.
Primers for qPCR.
| Genes | Forward | Reverse |
|---|
| STC2 |
5′-ATGCTACCTCAAGCACGACC-3′ |
5′-TCTGCTCACACTGAACC TGC-3′ |
| E-cadherin |
5′-AAGGCACAGCCTGTCGAAGCA-3′ |
5′-ACGTTGTCCCGGGTGTCATCCT-3′ |
| Vimentin |
5′-TGCCCTTAAAGGAACCAATGAG-3′ |
5′-AGGCGGCCAATAGTGTCTTG-3′ |
| MMP-2 |
5′-AGTTTCCATTCCGCTTCCAG-3′ |
5′-CGGTCGTAGTCCTCAGTGGT-3′ |
| MMP-9 |
5′-GTCCACCCTTGTGCTCTTCC-3′ |
5′-GACTCTCCACGCATCTCTGC-3′ |
| β-catenin |
5′-ATGCGGCTGCTGTTCTATTC-3′ |
5′-ACCAATGTCCAGTCCGAGAT-3′ |
| GAPDH |
5′-ACTTTGGTATCGTGGAAGGACTCAT-3′ |
5′-GTTTTTCTAGACGGCAGGTCAGG-3′ |
Statistical analyses
Data are presented as the means ± standard deviation
(SD). Differences between the multiple groups were assessed by
one-way analysis of variance (ANOVA) followed by Dunnett's post hoc
test. Differences were analyzed by GraphPad Prism 6 (GraphPad
Software, Inc., La Jolla, CA, USA) and all comparisons with a
P<0.05 were considered statistically significant.
Results
STC2 is highly expressed in CRC cell
lines
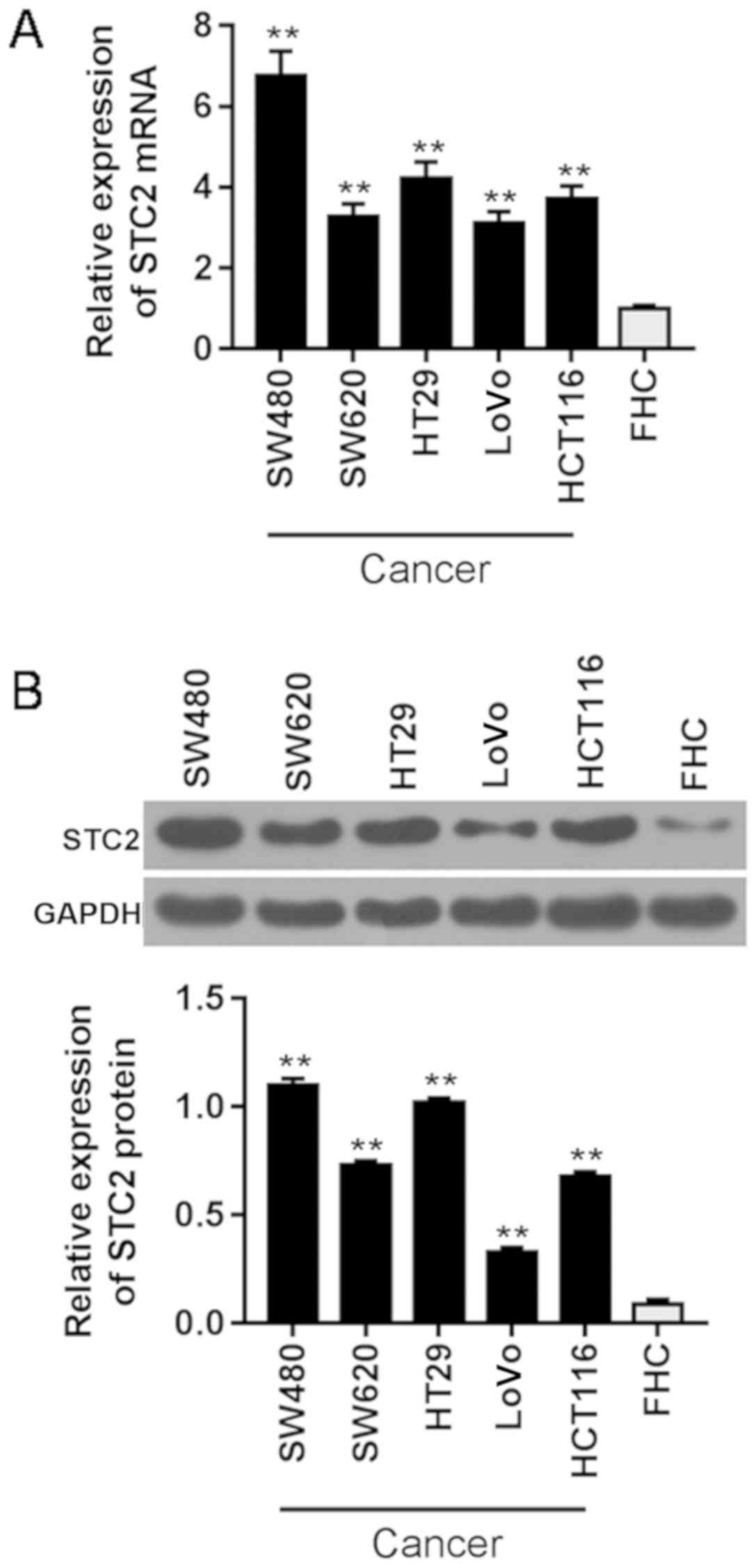
To select a suitable CRC cell line, qPCR (Fig. 1A) and western blot analysis
(Fig. 1B) were performed to
analyze the expression level of STC2 in the cell lines. We
discovered that STC2 was highly expressed in CRC cell lines (SW480,
SW620, HT29, LoVo and HCT116), particularly in SW480 cells. Thus,
SW480 cells were selected for subsequent experiments.
Viability, invasion and migration
potentials of the CRC cells are suppressed by siSTC2
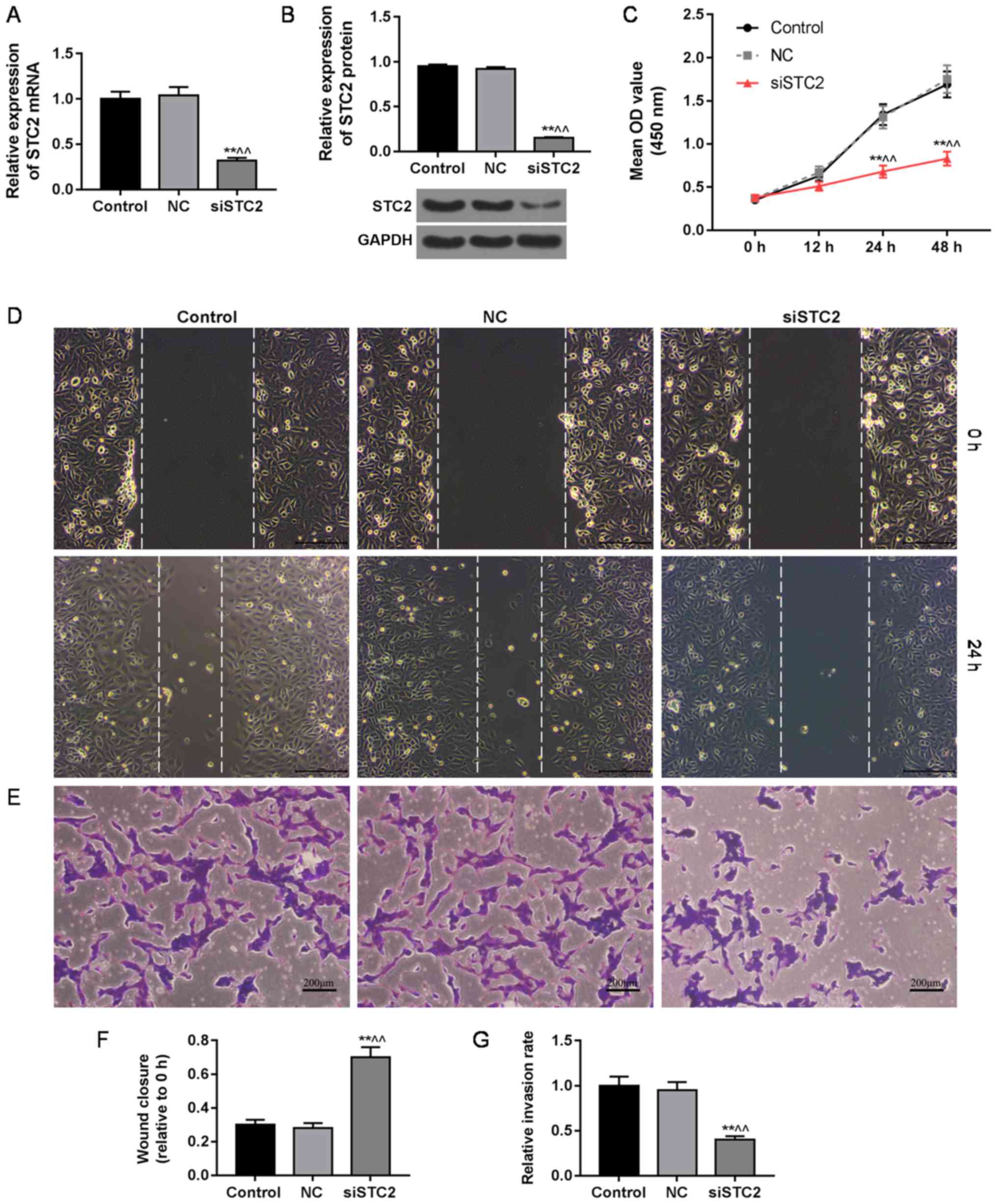
In the present study, the efficiency of siSTC2
transfection was confirmed by qPCR and western blot analysis. We
found that STC2 was silenced by plasmid transfection (Fig. 2A and B). Subsequently, we examined
the viability, invasion and migration of the cells after plasmid
transfection, and found that the cell viability was lower in the
siSTC2 group than that in the control or NC group after 24 and 48 h
of plasmid transfection (Fig. 2C),
and that the abilities of migration (Fig. 2D) and invasion (Fig. 2E) were decreased in the siSTC2
group, compared to the control or NC group. The wound closure gap
in the siSTC2 group was significantly wider than that in the
control and NC group at 24 h indicating reduced migration ability
(Fig. 2F). As shown in Fig. 2G, the rate of invasion in the
siSTC2 group was significantly lower than that in the control and
NC group.
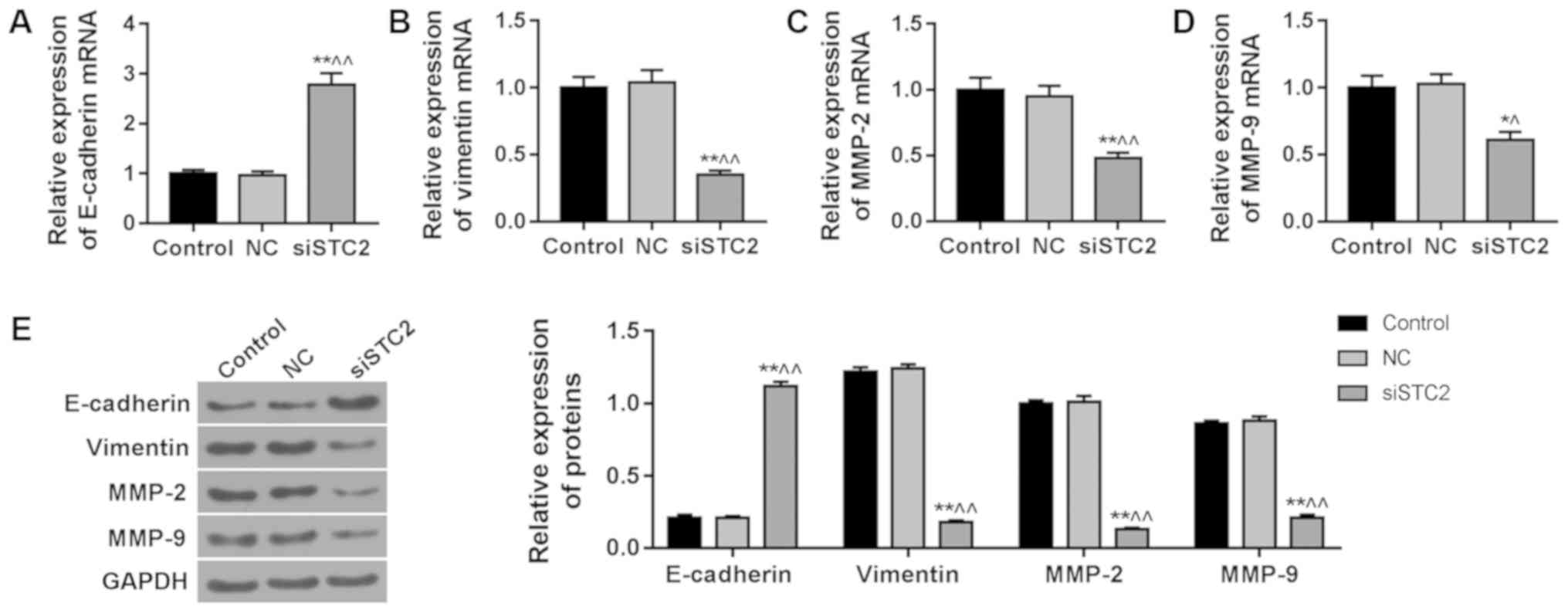
siSTC2 downregulates the expression of
vimentin, MMP-2 and MMP-9 and upregulates the expression of
E-cadherin in CRC cells
It has been reported that EMT is an important marker
of CRC development (20). The
results showed a significant increase in the expression of
E-cadherin (Fig. 3A) and a
reduction in the expressions of vimentin (Fig. 3B), MMP-2 (Fig. 3C) and MMP-9 (Fig. 3D) in the SW480 cells following
silencing of STC2. The protein level of E-cadherin (Fig. 3E) was upregulated and vimentin
(Fig. 3E), MMP-2 (Fig. 3E) and MMP-9 (Fig. 3E) were downregulated in the siSTC2
group, compared to control group.
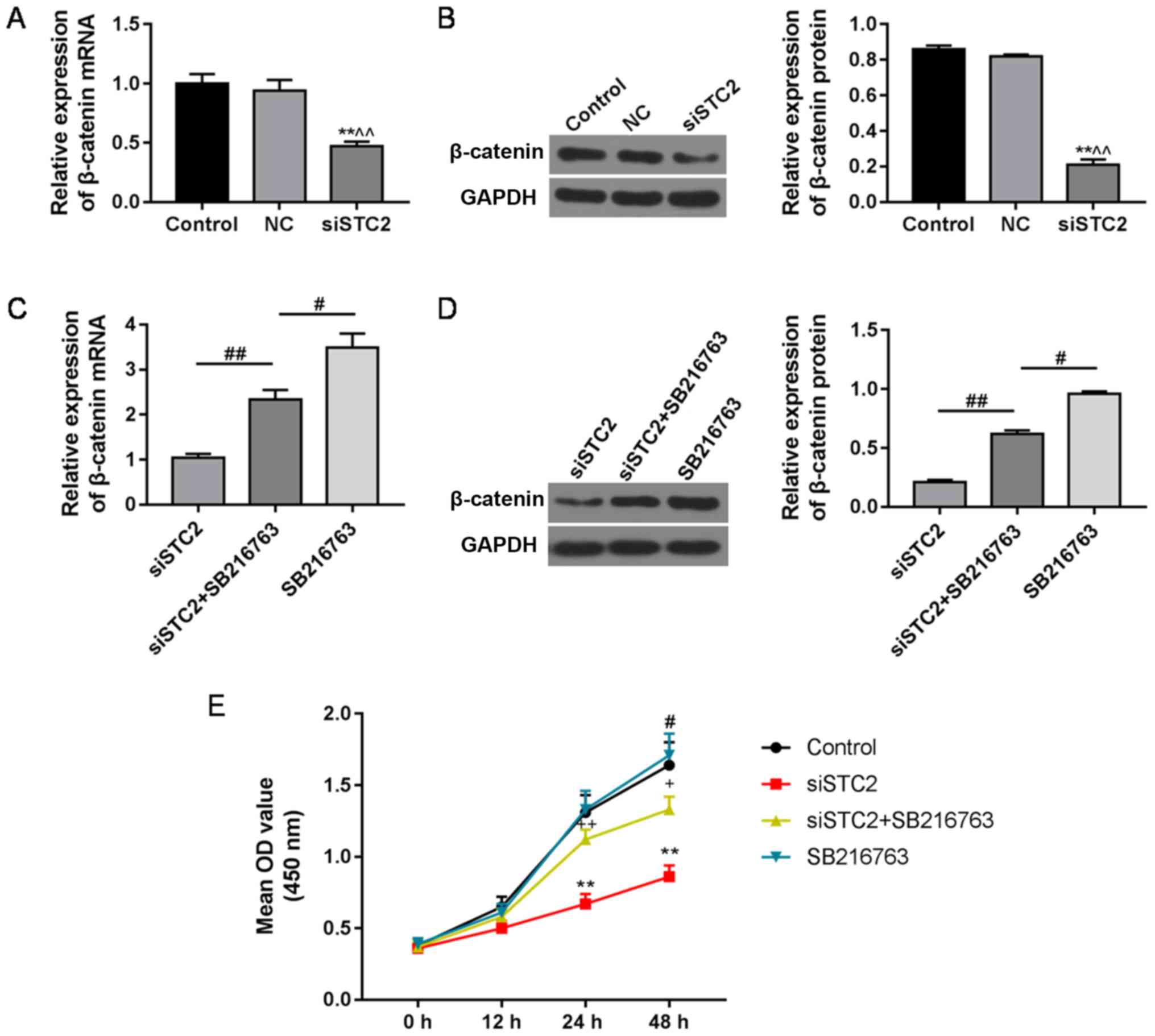
β-catenin is suppressed by siCST2, and
SB216763 treatment reverses this effect
In the present study, the expression of β-catenin
was significantly decreased by siSTC2 at the mRNA (Fig. 4A) and protein level (Fig. 4B). SB216763 (10 µM), a Wnt
activator, reversed the inhibitory effect of siSTC2 on β-catenin at
the mRNA (Fig. 4C) and protein
levels (Fig. 4D), and it also
reversed the inhibitory effect of siSTC2 on cell proliferation
(Fig. 4E).
Discussion
One of the most common malignancies in the clinic is
colon cancer, the incidence of which is the third highest worldwide
(21). It is highly important to
identify new colorectal cancer (CRC) treatment targets. It has been
reported that stanniocalcin 2 (STC2) is involved in the progression
of various types of cancer, such as gastric and breast cancer
(22,23). The differential expression levels
of STC2 have certain guiding significance for the
prediction, metastasis and prognosis of various malignant tumors
(24). Studies have shown that
STC2 is involved in the development of CRC (25,26).
However, its mechanism of action is not well understood. Therefore,
the present study aimed to further explore the mechanism of action
of STC2 in CRC.
Studies have found that the expression level of
STC2 in CRC patients is higher than that in normal tissues,
and that the expression level of STC2 is related to tumor
size and pathological grade (25,27,28).
In the present study, it was demonstrated that STC2 was
obviously increased in CRC cell lines. The results indicated that
STC2 may play a vital role in the occurrence and development
of CRC. Studies have shown that abnormal proliferation, invasion
and migration of tumor cells are common conditions in malignant
tumors (29–32). Therefore, the ability of tumor
invasion and migration is a detrimental biological feature of
malignant tumors, and it is also the most prominent clinical
manifestation of malignant tumors (33). It has been found that
overexpression of STC2 promotes CRC tumorigenesis by
activating the AKT/ERK signaling pathway (26). In the present study, we found that
silencing of STC2 reduced the cell viability and the migration and
invasion abilities, when compared to the control or NC group. These
results suggested that STC2 silencing may inhibit the progression
of CRC. One study has shown that epithelial-mesenchymal transition
(EMT) plays a vital role in tumor invasion and migration (34). Therefore, we explored the effect of
STC2 on EMT.
Previous studies on EMT have focused on cancer
metastasis profiles (35,36). In fact, the progression of
adenocarcinoma from normal intestinal mucosa to adenoma (adenoma
mucosa) and eventually to CRC is closely correlated with the EMT
process, and the expression changes in a series of genes, such as
E-cadherin and vimentin (37,38).
MMPs are a group of Zn2+-dependent endopeptidase that
can degrade the extracellular matrix (ECM), and promote tumor
invasion and migration, proliferation, differentiation and
apoptosis of tumor cells (39).
Degradation of the extracellular matrix mainly involves the matrix
metalloproteinase family, particularly MMP-2 and
MMP-9 (40–42). It has been reported that
MMP-2 and MMP-9 are vital for the metastasis of CRC
(43). We hypothesized that
knockdown of STC2 would inhibit EMT. Therefore, we investigated the
biomarkers of human colon mucosal epithelial cells. Chen et
al found that STC2 overexpression could promote the occurrence
of EMT in CRC in vitro and in vivo (26). Similarly, we found that silencing
of STC2 upregulated E-cadherin expression and downregulated
vimentin and MMP-2/-9 expression. Our findings confirmed that the
EMT process mediated by STC2 was associated with the
occurrence and development of colon cancer.
Studies have shown that the main signaling pathways
involved in the process of EMT are the Wnt/β-catenin signaling
pathway (44,45). As the Wnt/β-catenin signaling
pathway is widely involved in cell proliferation, differentiation
and metabolism (46), we explored
the STC2-mediated Wnt/β-catenin molecular signaling pathway and
found that the expression of β-catenin was decreased in the
STC2-silenced SW480 cells. β-catenin is a central molecule of the
Wnt signaling pathway. In the presence of Wnt signal stimulation,
Fratl mediates glycogen synthetase kinase 3β (GSK-3β) dissociation
from Axin, and prevents the phosphorylation of β-catenin. β-catenin
is not degraded and accumulates in the cytoplasm. Inhibition of the
Wnt/β-catenin signaling pathway in colon cancer has been found to
alleviate the degree of deterioration of colon cancer (47–49).
In the present study, with the activation of the Wnt/β-catenin
signal pathway, the Wnt activator SB2617763 was found to reverse
the inhibitory effect of siSTC2 on Wnt/β-catenin and cell
proliferation. SB216763 has been shown to specifically inhibit
GSK-3α and GSK-3β, thereby increasing the level of
dephosphorylation (active) of β-catenin (50,51).
Briefly, these findings suggest that Wnt/β-catenin may be involved
in the regulation of CRC following silencing of STC2.
In conclusion, it was demonstrated that STC2
was highly expressed in CRC cell lines, particularly SW480 cells.
STC2 silencing inhibited cell viability, invasion and migration,
compared to the control cells. Meanwhile, E-cadherin was increased,
while vimentin, MMP-2 and MMP-9 were decreased in the
CRC cells by siSTC2. Unsurprisingly, β-catenin was suppressed by
siSTC2, however, SB216763 reversed the level of β-catenin. Taken
together, these data showed that siSTC2 treatment conferred a
strong protective effect on CRC. This study suggests that STC2
silencing suppresses the migration of CRC cells and the occurrence
of EMT. In addition, the effect of STC2 on CRC may be
related to the Wnt/β-catenin signaling pathway. These results
provide novel insight into the identification of targeted
therapeutic strategies for CRC. However, the present study also has
some limitations. Due to the limited experimental funds, we only
explored the effect of STC2 on the proliferation, migration
and invasion of the CRC SW480 cell line. Whether or not STC2 has
the same effect on other CRC cell lines remains to be studied, and
we will further conduct gain-of-function experiments in regards to
STC2 in future studies. In addition, it is necessary to
investigate the role of the Wnt/β-catenin pathway in the
STC2-mediated effects, so as to better understand the mechanism of
STC2 in CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Zhejiang
Provincial Traditional Chinese Medicine Science Research Foundation
(grant no. 2016ZB037).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
QL substantially contributed to the conception and
design of the study. XZ and ZF acquired the data, analyzed and
interpreted the data. QL and ZP drafted the manuscript or
critically revised it for important intellectual content. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
institutional and/or national research committee and with the 1964
Helsinki declaration and its later amendments or comparable ethical
standards. No human or animals were involved in this research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Noreen F, Röösli M, Gaj P, Pietrzak J,
Weis S, Urfer P, Regula J, Schär P and Truninger K: Modulation of
age- and cancer-associated DNA methylation change in the healthy
colon by aspirin and lifestyle. J Natl Cancer Inst. 106(pii):
dju1612014.PubMed/NCBI
|
|
3
|
Pelser C, Arem H, Pfeiffer RM, Elena JW,
Alfano CM, Hollenbeck AR and Park Y: Prediagnostic lifestyle
factors and survival after colon and rectal cancer diagnosis in the
National Institutes of Health (NIH)-AARP diet and health study.
Cancer. 120:1540–1547. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang W, Liu Z, Zhou G, Tian A and Sun N:
Magnetic gold nanoparticle-mediated small interference RNA
silencing Bag-1 gene for colon cancer therapy. Oncol Rep.
35:978–984. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hay ED: The mesenchymal cell, its role in
the embryo, and the remarkable signaling mechanisms that create it.
Dev Dyn. 233:706–720. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao JH, Luo Y, Jiang YG, He DL and Wu CT:
Knockdown of β-catenin through shRNA cause a reversal of EMT and
metastatic phenotypes induced by HIF-1α. Cancer Invest. 29:377–382.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Shi J, Chai K, Ying X and Zhou BP:
The role of Snail in EMT and tumorigenesis. Curr Cancer Drug
Targets. 13:963–972. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reinacher-Schick A, Baldus SE, Romdhana B,
Landsberg S, Zapatka M, Mönig SP, Hölscher AH, Dienes HP, Schmiegel
W and Schwarte-Waldhoff I: Loss of Smad4 correlates with loss of
the invasion suppressor E-cadherin in advanced colorectal
carcinomas. J Pathol. 202:412–420. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu CY, Tsai YP, Wu MZ, Teng SC and Wu KJ:
Epigenetic reprogramming and post-transcriptional regulation during
the epithelial-mesenchymal transition. Trends Genet. 28:454–463.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Z, Yao L, Yang J, Wang Z and Du G:
PI3K/Akt and HIF-1 signaling pathway in hypoxiaischemia (Review).
Mol Med Rep. 18:3547–3554. 2018.PubMed/NCBI
|
|
11
|
Miyoshi K and Hennighausen L:
Beta-catenin: A transforming actor on many stages. Breast Cancer
Res. 5:63–68. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maruyama K, Ochiai A, Akimoto S, Nakamura
S, Baba S, Moriya Y and Hirohashi S: Cytoplasmic beta-catenin
accumulation as a predictor of hematogenous metastasis in human
colorectal cancer. Oncology. 59:302–309. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou W, Li Y, Gou S, Xiong J, Wu H, Wang
C, Yan H and Liu T: MiR-744 increases tumorigenicity of pancreatic
cancer by activating Wnt/β-catenin pathway. Oncotarget.
6:37557–37569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Margariti A, Zampetaki A, Xiao Q, Zhou B,
Karamariti E, Martin D, Yin X, Mayr M, Li H, Zhang Z, et al:
Histone deacetylase 7 controls endothelial cell growth through
modulation of beta-catenin. Circ Res. 106:1202–1211. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jia S, Qu T, Wang X, Feng M, Yang Y, Feng
X, Ma R, Li W, Hu Y, Feng Y, et al: KIAA1199 promotes migration and
invasion by Wnt/β-catenin pathway and MMPs mediated EMT progression
and serves as a poor prognosis marker in gastric cancer. PLoS One.
12:e01750582017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen WN and Zhu GJ: Progress in the
research of stanniocalcin. Sheng Li Ke Xue Jin Zhan. 39:225–228.
2008.(In Chinese). PubMed/NCBI
|
|
17
|
Chang AC, Jellinek DA and Reddel RR:
Mammalian stanniocalcins and cancer. Endocr Relat Cancer.
10:359–373. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yeung BH, Law AY and Wong CK: Evolution
and roles of stanniocalcin. Mol Cell Endocrinol. 349:272–280. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karagiannis GS, Berk A, Dimitromanolakis A
and Diamandis EP: Enrichment map profiling of the cancer invasion
front suggests regulation of colorectal cancer progression by the
bone morphogenetic protein antagonist, gremlin-1. Mol Oncol.
7:826–839. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arigami T, Uenosono Y, Ishigami S,
Yanagita S, Hagihara T, Haraguchi N, Matsushita D, Hirahara T,
Okumura H, Uchikado Y, et al: Clinical significance of
stanniocalcin 2 expression as a predictor of tumor progression in
gastric cancer. Oncol Rep. 30:2838–2844. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Coulson-Gilmer C, Humphries MP, Sundara
Rajan S, Droop A, Jackson S, Condon A, Cserni G, Jordan LB, Jones
LJ, Kanthan R, et al: Stanniocalcin 2 expression is associated with
a favourable outcome in male breast cancer. J Pathol Clin Res.
4:241–249. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Parris TZ, Kovacs A, Aziz L, Hajizadeh S,
Nemes S, Semaan M, Forssell-Aronsson E, Karlsson P and Helou K:
Additive effect of the AZGP1, PIP, S100A8 and UBE2C molecular
biomarkers improves outcome prediction in breast carcinoma. Int J
Cancer. 134:1617–1629. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hashemzadeh S, Arabzadeh AA, Estiar MA,
Sakhinia M, Mesbahi N, Emrahi L, Ghojazadeh M and Sakhinia E:
Clinical utility of measuring expression levels of Stanniocalcin 2
in patients with colorectal cancer. Med Oncol. 31:2372014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen B, Zeng X, He Y, Wang X, Liang Z, Liu
J, Zhang P, Zhu H, Xu N and Liang S: STC2 promotes the
epithelial-mesenchymal transition of colorectal cancer cells
through AKT-ERK signaling pathways. Oncotarget. 7:71400–71416.
2016.PubMed/NCBI
|
|
27
|
Ieta K, Tanaka F, Yokobori T, Kita Y,
Haraguchi N, Mimori K, Kato H, Asao T, Inoue H, Kuwano H and Mori
M: Clinicopathological significance of stanniocalcin 2 gene
expression in colorectal cancer. Int J Cancer. 125:926–931. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miyazaki S, Kikuchi H, Iino I, Uehara T,
Setoguchi T, Fujita T, Hiramatsu Y, Ohta M, Kamiya K, Kitagawa K,
et al: Anti-VEGF antibody therapy induces tumor hypoxia and
stanniocalcin 2 expression and potentiates growth of human colon
cancer xenografts. Int J Cancer. 135:295–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Leve F, Peres-Moreira RJ, Binato R,
Abdelhay E and Morgado-Diaz JA: LPA Induces colon cancer cell
proliferation through a cooperation between the ROCK and STAT-3
pathways. PLoS One. 10:e01390942015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshida GJ: Metabolic reprogramming: The
emerging concept and associated therapeutic strategies. J Exp Clin
Cancer Res. 34:1112015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang J and Sun X: MicroRNA-375 inhibits
the proliferation, migration and invasion of kidney cancer cells by
triggering apoptosis and modulation of PDK1 expression. Environ
Toxicol Pharmacol. 62:227–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chu GC and Chung LW: RANK-mediated
signaling network and cancer metastasis. Cancer Metastasis Rev.
33:497–509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Probst OC, Karayel E, Schida N, Nimmerfall
E, Hehenberger E, Puxbaum V and Mach L: The mannose
6-phosphate-binding sites of M6P/IGF2R determine its capacity to
suppress matrix invasion by squamous cell carcinoma cells. Biochem
J. 451:91–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kitamura T and Taketo MM: Keeping out the
bad guys: Gateway to cellular target therapy. Cancer Res.
67:10099–10102. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Škovierová H, Okajčeková T, Strnádel J,
Vidomanová E and Halašová E: Molecular regulation of
epithelial-to-mesenchymal transition in tumorigenesis (Review). Int
J Mol Med. 41:1187–1200. 2018.PubMed/NCBI
|
|
36
|
Gurzu S, Silveanu C, Fetyko A, Butiurca V,
Kovacs Z and Jung I: Systematic review of the old and new concepts
in the epithelial-mesenchymal transition of colorectal cancer.
World J Gastroenterol. 22:6764–6775. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vincan E, Brabletz T, Faux MC and Ramsay
RG: A human three-dimensional cell line model allows the study of
dynamic and reversible epithelial-mesenchymal and
mesenchymal-epithelial transition that underpins colorectal
carcinogenesis. Cells Tissues Organs. 185:20–28. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen X, Halberg RB, Burch RP and Dove WF:
Intestinal adenomagenesis involves core molecular signatures of the
epithelial-mesenchymal transition. J Mol Histol. 39:283–294. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Han X, Fang X, Lou X, Hua D, Ding W, Foltz
G, Hood L, Yuan Y and Lin B: Silencing SOX2 induced
mesenchymal-epithelial transition and its expression predicts liver
and lymph node metastasis of CRC patients. PLoS One. 7:e413352012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
He Q, Li H, Meng F, Sun X, Feng X, Chen J,
Li L and Liu J: Methionine sulfoxide reductase B1 regulates
hepatocellular carcinoma cell proliferation and invasion via the
mitogen-activated protein kinase pathway and epithelial-mesenchymal
transition. Oxid Med Cell Longev. 2018:52879712018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ryu ES, Kim MJ, Shin HS, Jang YH, Choi HS,
Jo I, Johnson RJ and Kang DH: Uric acid-induced phenotypic
transition of renal tubular cells as a novel mechanism of chronic
kidney disease. Am J Physiol Renal Physiol. 304:F471–F480. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lemieux E, Bergeron S, Durand V, Asselin
C, Saucier C and Rivard N: Constitutively active MEK1 is sufficient
to induce epithelial-to-mesenchymal transition in intestinal
epithelial cells and to promote tumor invasion and metastasis. Int
J Cancer. 125:1575–1586. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Park KS, Kim SJ, Kim KH and Kim JC:
Clinical characteristics of TIMP2, MMP2, and MMP9 gene
polymorphisms in colorectal cancer. J Gastroenterol Hepatol.
26:391–397. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fu W, Tao T, Qi M, Wang L, Hu J, Li X,
Xing N, Du R and Han B: MicroRNA-132/212 upregulation inhibits
TGF-β-mediated epithelial-mesenchymal transition of prostate cancer
cells by targeting SOX4. Prostate. 76:1560–1570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang JX, Zhai JF, Yang XT and Wang J:
MicroRNA-132 inhibits migration, invasion and
epithelial-mesenchymal transition by regulating TGFβ1/Smad2 in
human non-small cell lung cancer. Eur Rev Med Pharmacol Sci.
20:3793–3801. 2016.PubMed/NCBI
|
|
46
|
Teichroeb JH, Kim J and Betts DH: The role
of telomeres and telomerase reverse transcriptase isoforms in
pluripotency induction and maintenance. RNA Biol. 13:707–719. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim HJ, Moon SJ, Kim SH, Heo K and Kim JH:
DBC1 regulates Wnt/β-catenin-mediated expression of MACC1, a key
regulator of cancer progression, in colon cancer. Cell Death Dis.
9:8312018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li X, Hu W, Zhou J, Huang Y, Peng J, Yuan
Y, Yu J and Zheng S: CLCA1 suppresses colorectal cancer
aggressiveness via inhibition of the Wnt/beta-catenin signaling
pathway. Cell Commun Signal. 15:382017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Han X, Zheng J, Wang Y and Gao Z:
miRNA-29a inhibits colon cancer growth by regulation of the
PTEN/Akt/GSK3β and Wnt/β-catenin signaling pathways. Oncol Lett.
16:2638–2644. 2018.PubMed/NCBI
|
|
50
|
Zhang Y, Seid K, Obermayr F, Just L and
Neckel PH: Activation of Wnt signaling increases numbers of enteric
neurons derived from neonatal mouse and human progenitor cells.
Gastroenterology. 153:154–165.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mao D, Qiao L, Lu H and Feng Y: B-cell
translocation gene 3 overexpression inhibits proliferation and
invasion of colorectal cancer SW480 cells via Wnt/β-catenin
signaling pathway. Neoplasma. 63:705–716. 2016. View Article : Google Scholar : PubMed/NCBI
|