Introduction
Prostate cancer (PCa) is the most common malignancy
among males in Europe and USA, with increasing incidence (1). Additionally, ~1/7 of Australian males
are diagnosed with PCa annually, at a mean age of 75 years; PCa is
considered to be the second most fatal malignancy among the
European and American male populations (2). In China, with the improvement of the
living standards, changes in diet composition and the progressive
aging of the population, the incidence of malignant tumors has
notably increased; PCa in particular is quickly becoming the
leading cause of mortality among middle-aged and elderly men
(3,4). At present, the main methods for
diagnosing PCa are digital rectal examination (DRE), measurement of
serum prostate-specific antigen (PSA) levels, magnetic resonance
imaging and transrectal ultrasound-guided prostatic biopsy
(5–8). In addition, DRE combined with PSA is
considered as the standard method for early screening of PCa
(6); however, the findings on DRE
may be subjective, and the value of DRE in the diagnosis of early
PCa without notable nodules is limited. On the contrary, PSA
detection is characterized by high sensitivity and relatively low
specificity, and the detection rate of PCa is only ~25%,
particularly when PSA is between 4–10 ng/ml (PSA diagnostic gray
zone) (9). Furthermore, PSA
detection often leads to inaccurate diagnoses and poor therapeutic
strategies for the treatment of PCa (10–12).
In addition, the exact molecular mechanism underlying the
development and progression of PCa remains unclear. Therefore,
elucidating this mechanism is crucial for the clinical diagnosis,
treatment and follow-up monitoring of patients with PCa.
MicroRNAs (miRNAs/miRs) are noncoding RNAs
comprising ~20–22 nucleotides in length and are highly conserved
among species, which act as key factors in tumor inhibition or
promotion via the regulation of oncogenes or tumor suppressors
(13). The association between
miRNAs and the development, diagnosis and treatment of PCa has
attracted notable attention (14–16).
For example, miR-206 was reported to exert antitumor effects on PCa
through regulating Annexin A2 and C-X-C motif chemokine 11
(17,18); miR-33a acts as a tumor suppressor
and was observed to be downregulated in PCa (19). Furthermore, miR-605 (20), miR-106a (21) and miR-483-5p (22) act as oncogenes in PCa. Conversely,
miR-154 (23), miR-331-3p
(24) and miR-625 (25) act as tumor suppressors in PCa.
miR-106b is a member of the miR-106b-25 family that is highly
expressed in laryngeal, gastric, breast and hepatic cancer, and has
important functions in regulating tumor cell migration, invasion
and proliferation (26–29). Furthermore, it was previously
reported that miR-106b induces apoptosis and inhibits invasion of
thyroid cancer cells by downregulating the expression of chromosome
1 open reading frame 24, suppresses the ability of Smad7 to enhance
epithelial-to-mesenchymal transition and promotes the metastasis of
esophageal cancer cells; inhibition of miR-106b induces apoptosis,
and suppresses proliferation and migration of renal cell carcinoma
cells (30–32). However, the role and the underlying
molecular mechanisms of miR-106b in PCa require further
investigation.
The aim of the present study was to investigate the
effects of miR-106b on PCa cell viability, migration and invasion
and the underlying mechanism of action, in order to determine
whether PCa may be a novel biomarker for the diagnosis and
treatment of PCa.
Materials and methods
Human tissue samples
The present study was approved by the Ethics
Committee of Ningbo First Hospital (Ningbo, China). A total of 40
patients with PCa from Ningbo First Hospital were investigated,
none of whom had received radiotherapy, chemotherapy or
immunotherapy prior to tumor resection. All the patients provided
written informed consent for their tissues to be used for research
purposes.
Cell culture and transfection
The human PCa cell line LNCaP was purchased from the
American Type Culture Collection (ATCC, Manassas, VA, USA), and
routinely cultured in RPMI-1640 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) and
1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.)
at 37°C in 5% CO2.
miR-106 inhibitors, miR-negative control (NC)
inhibitor, miR-106 mimics and miR-NC mimics were purchased from
Ambion (Thermo Fisher Scientific, Inc.). Inhibitors and mimics were
transfected at a concentration of 100 nM. LNCaP cells were seeded
into 6-well plates and transfected with 100 nM of miR-106
inhibitor, miR-NC inhibitor, miR-106 mimics and miR-NC mimics using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols for 48
h. The sequences of miR-106 inhibitor, miR-NC inhibitor, miR-106
mimics and miR-NC mimics were as follows: miR-106b mimics,
5′-TAAAGTGCTGACAGTGCAGAT-3′; miR-NC mimics,
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-106b inhibitor,
5′-AUCUGCUCAGCACUUUA-3′; miR-NC inhibitor,
5′-CAGUACUUUUGUGUAGUACAA-3′.
Target prediction and luciferase
assay
The prediction of the 3′-untranslated regions
(3′-UTRs) of la-related protein 4B (LAR4B) as a binding target of
miR-106b was checked by using TargetScan (version 7.1; www.targetscan.org/vert_71/). Subsequently, the
3′-UTR of LAR4B was mutated using a mutagenesis kit (Promega
Corporation, Madison, WI, USA) according to the manufacturer's
protocol. After transfection, cells were maintained at 37°C in 5%
CO2 for 48 h. Wild-type (WT) and mutant (Mut) sequences
of LAR4B were amplified and inserted into the pmirGLO vector
(Shanghai GenePharma Co., Ltd., Shanghai, China) to construct
luciferase reporter plasmids according to the manufacturer's
protocol (Promega Corporation). Cells were transfected with miR-106
mimics or miR-NC mimics and LAR4B 3′-UTR WT or LAR4B 3′-UTR Mut
plasmids. Cells were maintained at 37°C in 5% CO2 for 48
h after transfection. Luciferase activity was detected with a dual
luciferase reporter kit (Promega Corporation). Renilla
luciferase was used to normalize the luciferase activity.
Cell viability assay
Cells at a density of 5×103 cells/well
were seeded in 96-well plates and transfected. After incubation for
0, 12, 24 and 48 h, the transfected cells were treated with 0.5
mg/ml MTT solution and incubated in the dark at 37°C for 4 h.
Subsequently, the supernatants were removed and dimethyl sulfoxide
was added to dissolve the formazan crystals. Then, the optical
density at 490 nm was recorded using a microplate
spectrophotometer. The experiment was performed in triplicate.
Wound healing assay
Cells at density of 5×105 cells/well were
seeded into a 6-well plate. After cells attained 90% confluence,
the cell monolayer was scratched using a 10-µl pipette tip,
and the cells were cultured in a serum-free DMEM (Thermo Fisher
Scientific, Inc.) for cell recovery. Subsequently, the cells were
imaged at 48 h with an inverted light microscope (Olympus, Tokyo,
Japan; magnification, ×200) and samples were observed in five
randomly-selected fields of view.
Transwell invasion assay
Matrigel was diluted with serum-free medium (1:3)
and then added to the upper chambers (50 µl per well) and
allowed to form a gel for 30 min at 37°C with 5% CO2.
Then, transfected LNCaP cells (1×106 cells/well) were
seeded into the upper chamber with serum-free RPMI-1640 medium,
whereas RPMI-1640 supplemented with 10% FBS was added to the lower
chamber. After incubation for 48 h in 5% CO2 at 37°C,
the cells on the top of membranes were removed and the invading
cells were fixed with 70% ethanol at room temperature for 30 min
and stained with 0.5% crystal violet solution at room temperature
for 30 min, and counted using an with an inverted light microscope
(magnification, ×200) and samples were observed in five
randomly-selected fields of view.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA from PCa tissues and cell lines was
extracted with TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and miRNA was extracted using the miRcute miRNA
Isolation kit (Tiangen, Shanghai, China). TaqMan MicroRNA Reverse
Transcription kit and Taqman High-capacity cDNA kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) were used to reverse
transcribe miRNA and mRNA, respectively. The RT conditions were the
following: 5 min at 25°C, 30 min at 42°C and 5 min at 85°C. The
expression of miR-106b was determined by RT-qPCR using the TaqMan
miR kit (Applied Biosystems; Thermo Fisher Scientific, Inc.); the
mRNA expression of LAR4B, matrix metalloproteinase-2 (MMP2),
mothers against decapentaplegic homolog 2 (Smad2), cluster of
differentiation (CD)44 and Ki-67 was measured using a TaqMan
RT-qPCR kit (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The thermocycling conditions were the following: Initial
denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 60 sec. U6 and GAPDH were used as controls for
miRNA and mRNA, respectively. Data were acquired by using a HT-7900
TaqMan instrument (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The mRNA relative expression levels were calculated using
the 2−ΔΔCq method (33). The PCR primers were as follows:
miR-106b, 5′-TTTTCGCCCTTAGCGTGAAGA-3′ (forward) and
5′-GAGGCAGTCGAAGCTCTCG-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′
(forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); LARP4B,
5′-TGGTCCTATATCGCAAACCACT-3′ (forward) and
5′-GCACTACTCGCTTCCAAATGT-3′ (reverse); MMP2,
5′-GCTATGGACCTTGGGAGAA-3′ (forward) and 5′-TGGAAGCGGAATGGAAAC-3′
(reverse); Smad2, 5′-CATCAGCCAATGGCAAGTGAA-3′ (forward) and
5′-AGAACAGGGTCTGCATCCATCATA-3′ (reverse); CD44,
5′-ACAACTGGTGATGGAGACTCATCC-3′ (forward) and
5′-CAGAGTGGCTTATCATCTTGG-3′ (reverse); and Ki-67,
5′-GCAGGACTTCACTTGCTTCC-3′ (forward) and 5′-TCATTTGCGTTTGTTTCACG-3′
(reverse); GAPDH, 5′-ACAACTTTGGTATCGTGGAAGG-3′ (forward); and
5′-GCCATCACGCCACAGTTTC-3′ (reverse).
Western blot analysis
Total protein from PCa tissues and LNCaP cells was
extracted with radioimmunoprecipitation assay buffer (Beijing
Solarbio Science & Technology, Beijing, China), and the protein
concentration was measured using the BCA Protein Assay kit (Vazyme,
Piscataway, NJ, USA). Equal amounts of protein (10 µg) were
separated via 12% SDS-PAGE and transferred onto polyvinylidene
difluoride membranes. The membranes were then blocked with 5%
non-fat milk for 1 h, followed by incubation at 4°C overnight with
the following primary antibodies (1:1,000): LARP4B (cat. no.
ab197085; Abcam, Cambridge, MA, USA), MMP2 (cat. no. 40994; Cell
Signaling Technology, Inc.), Smad2 (cat. no. 8685; Cell Signaling
Technology, Inc.), CD44 (cat. no. 37259; Cell Signaling Technology,
Inc.), Ki-67 (cat. no. 4400; Cell Signaling Technology, Inc.) and
GAPDH (cat. no. 5174; Cell Signaling Technology, Inc.).
Subsequently, the membranes were washed with tris-buffered saline
with 0.1% Tween-20 three times (15 min each time) and incubated
with horseradish peroxidase-conjugated secondary antibodies
(1:2,000) for a further 2 h at room temperature including
anti-mouse (cat. no. 7076; Cell Signaling Technology, Inc.) and
anti-rabbit (cat. no. 5127; Cell Signaling Technology, Inc.).
Protein bands were examined via Quantity One software (version 4.5;
Bio-Rad Laboratories, Inc., Hercules, CA, USA) with an ECL kit
(Abcam). GAPDH was used as the loading control.
Statistical analysis
Each experiment was performed in triplicate. SPSS
19.0 software (IBM Corp., Armonk, NY, USA) was used to analyze the
experimental data. The data were presented as the mean ± standard
deviation. Statistical differences between two groups were analyzed
by a Student's t-test. Statistical differences among multiple
groups were analyzed by one-way analysis of variance followed by a
Bonferroni post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Clinicopathological features of the
patients
As presented in Table
I, the expression of miR-106b was significantly associated with
pT stage, histological grade and lymphatic metastasis, but had no
significant association with gender and age.
 | Table I.Expression of miR-106b in the tissues
of patients with PCa. |
Table I.
Expression of miR-106b in the tissues
of patients with PCa.
| Factors | Case | miR-106b
(mean) | P-value |
|---|
| Age, years |
|
| 0.631 |
|
≥60 | 22 | 2.164±0.112 |
|
|
<60 | 18 | 2.248±0.135 |
|
| Serum PSA,
ng/ml |
|
| 0.668 |
|
≥10 | 23 | 2.225±0.139 |
|
|
<10 | 17 | 2.148±0.087 |
|
| pT Stage |
|
| 0.007a |
|
≥T3 | 25 | 2.462±0.132 |
|
|
<T3 | 15 | 1.943±0.078 |
|
| Histological
grade |
|
| 0.028a |
|
Well-intermediate
differentiation | 10 | 1.861±0.104 |
|
| Poor
differentiation | 30 | 2.391±0.128 |
|
| Metastasis |
|
| 0.019a |
| No | 31 | 1.938±0.095 |
|
|
Yes | 9 | 2.399±0.116 |
|
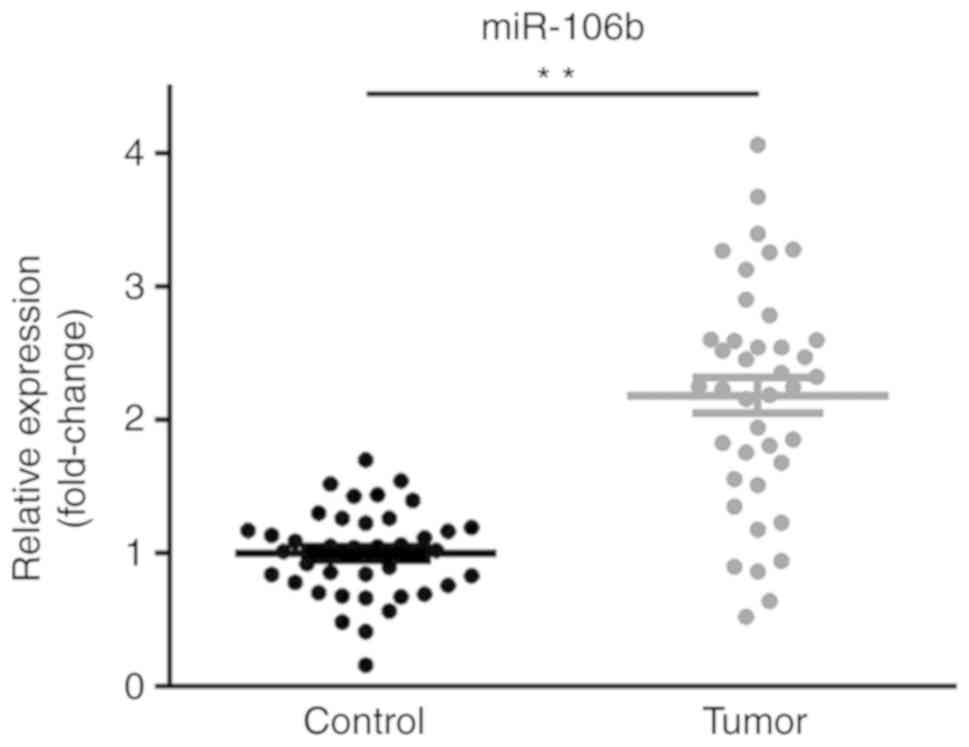
miR-106b is upregulated in patients
with PCa
To evaluate the expression and clinical value of
miR-106b in PCa, 40 pairs of PCa tissues and matched adjacent
normal tissues were collected and the expression of miR-106b was
examined by RT-qPCR. The results demonstrated that the expression
of miR-106b was significantly upregulated in PCa tissues relative
to the respective adjacent normal tissues (Fig. 1). Therefore, miR-106b may have the
potential as a novel biomarker for the diagnosis and prognosis of
PCa.
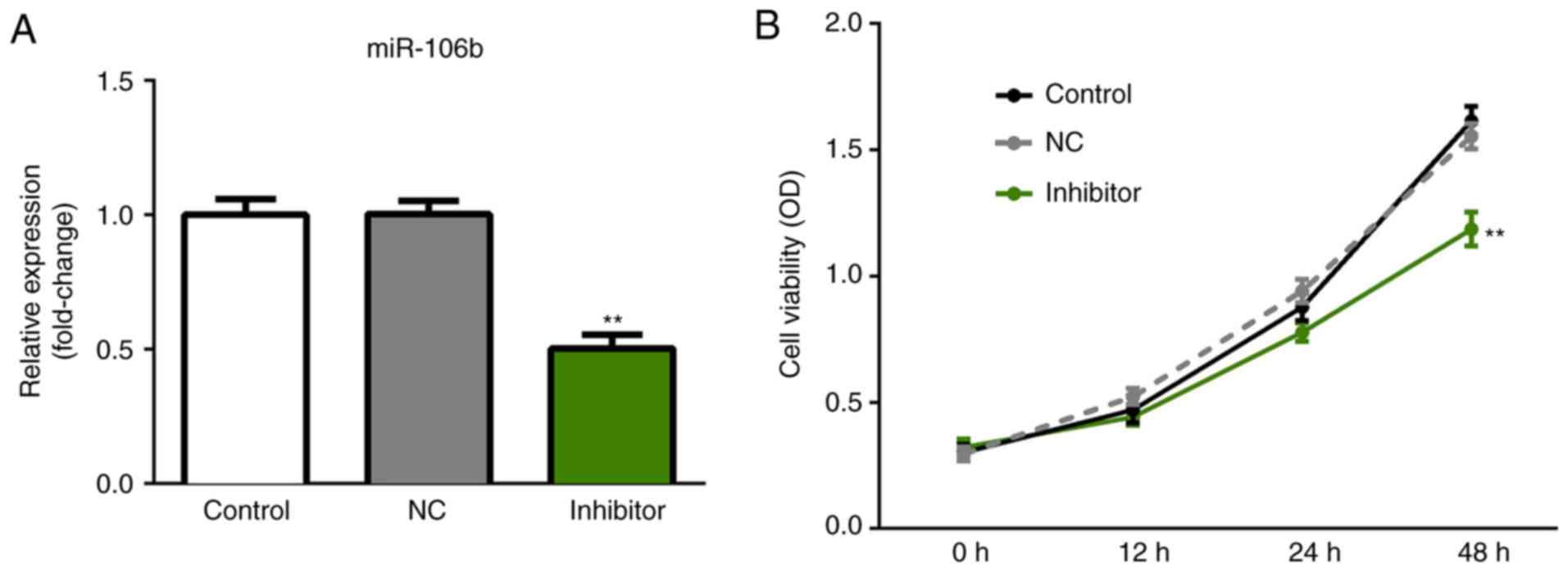
Downregulation of miR-106b inhibits in
PCa cell viability
To further investigate the role of miR-106b in the
progression and development of PCa, miR-NC inhibitor and miR-106b
inhibitor were respectively transfected into LNCaP cells, and
RT-qPCR was performed to evaluate the transfection efficiency after
48 h. The results demonstrated that the expression of miR-106b was
significantly decreased relative to the negative control group in
LNCaP cells (Fig. 2A).
Subsequently, an MTT assay was conducted to evaluate the effects of
miR-106b on the viability of LNCaP cells. As presented in Fig. 2B, inhibition of miR-106b exerted a
significant inhibitory effect on the viability of LNCaP cells
compared with the controls.
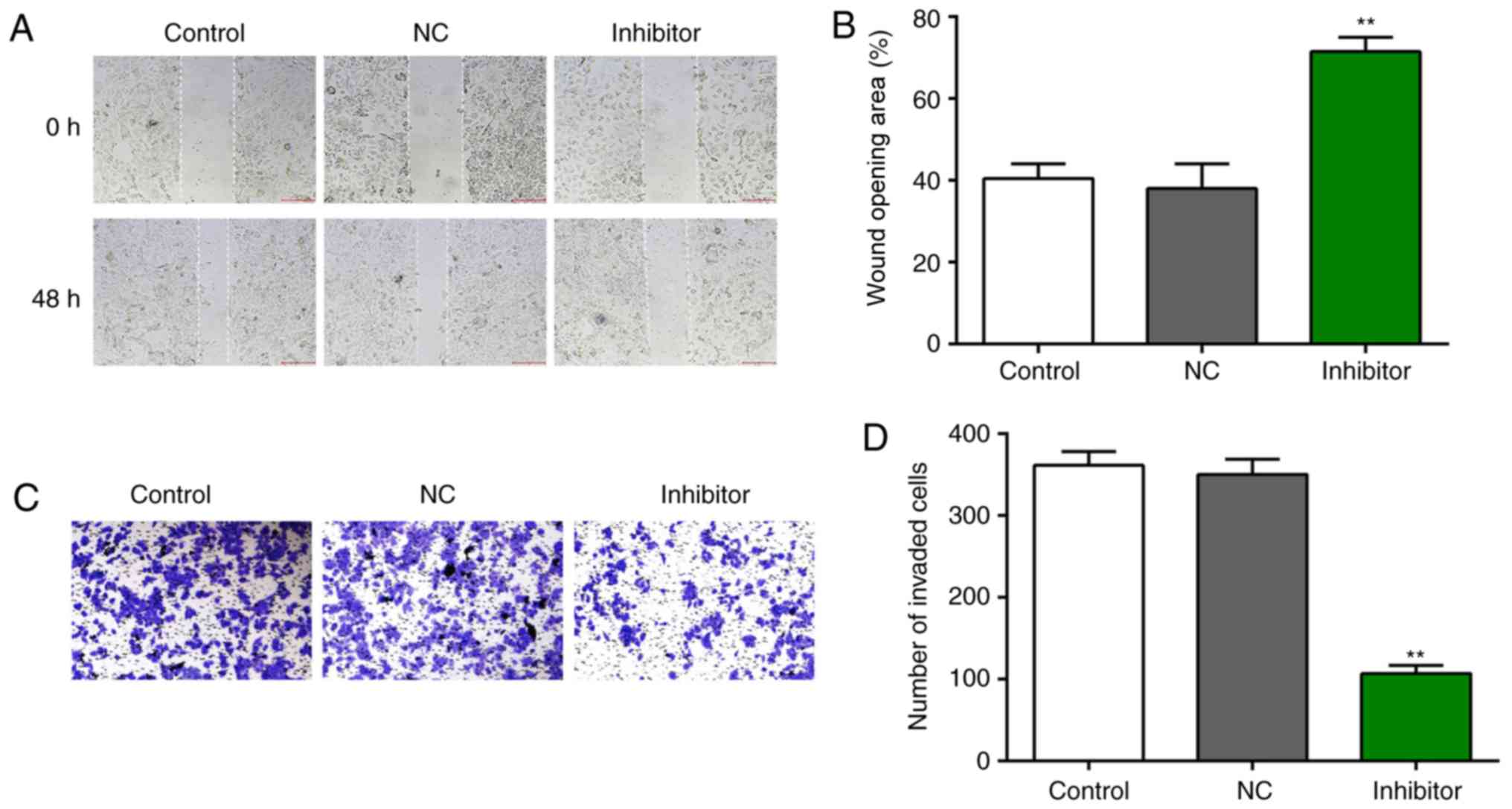
Inhibition of miR-106b suppresses the
migration and invasion of PCa cells
In addition, to investigate the effects of miR-106b
on the migration and invasion of PCa cells, wound healing and
Transwell invasion assays, were respectively performed with LNCaP
cells. The results revealed that knockdown of miR-106b
significantly suppressed LNCaP cell migration after 48 h (Fig. 3A and B) compared with the control.
Furthermore, the invasive ability of LNCaP cells was significantly
decreased following transfection with miR-106b inhibitors for 48 h
compared with the control (Fig. 3C and
D). Collectively, these results indicate that the
downregulation of miR-106b decreased the migration and invasive
abilities of LNCaP cells.
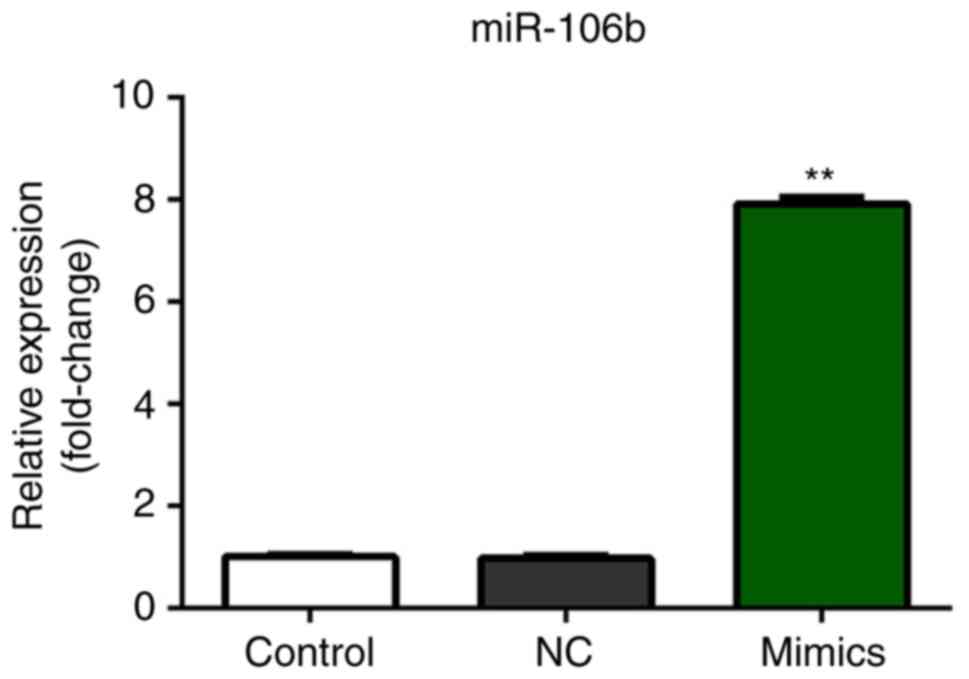
Expression of miR-106b
As presented in Fig.
4, the expression of miR-106b was significantly increased
following transfection with miR-106b mimics compared with the
control; no significant difference between the control and miR-NC
mimics groups was observed.
LARP4B is a direct target of
miR-106b
To further characterize the possible downstream
regulators of miR-106b affecting the development and progression of
PCa, TargetScan was used to predict the target genes regulated by
miR-106b. The results indicated that LARP4B is a potential target
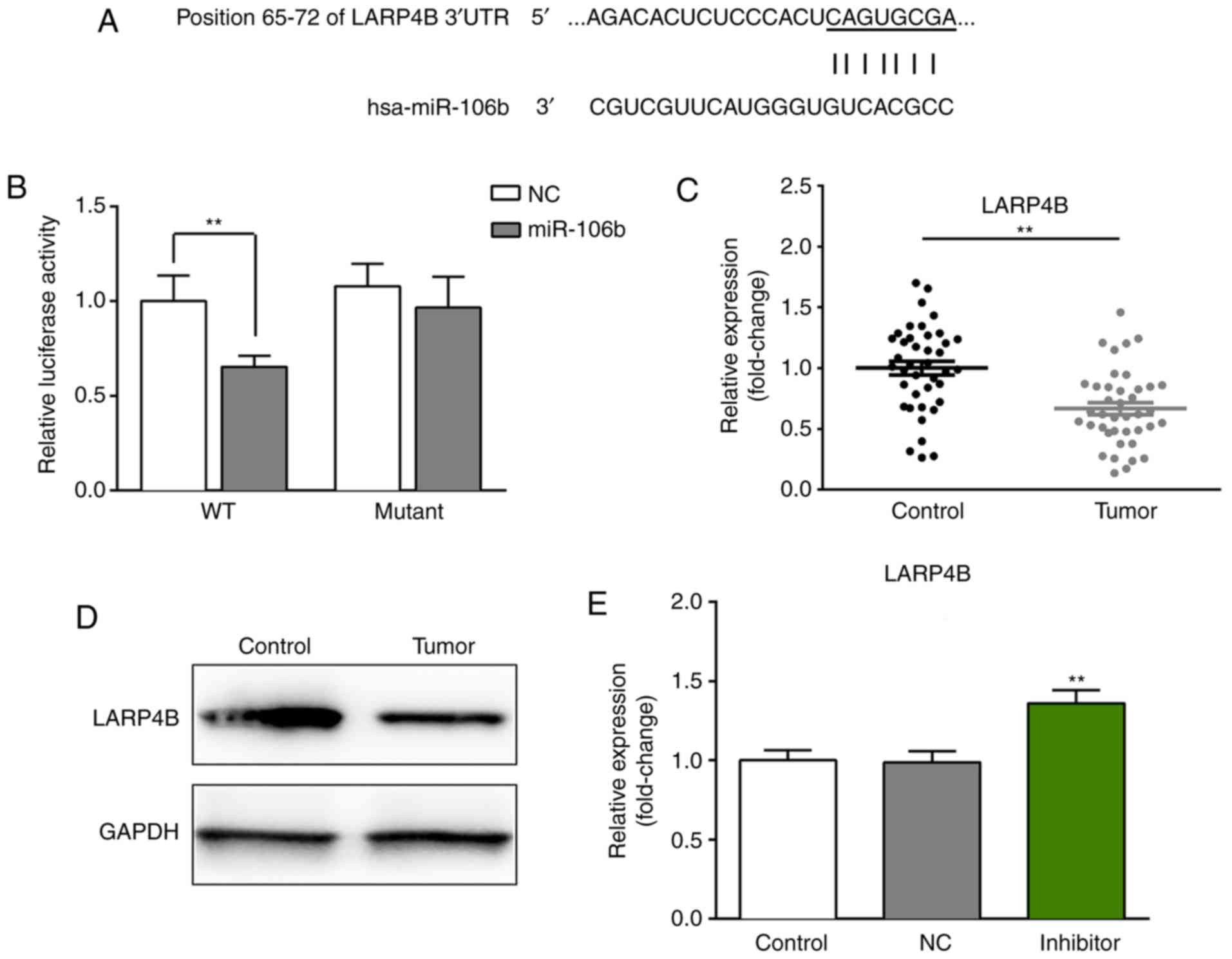
gene of miR-106b (Fig. 5A).
Furthermore, a luciferase reporter assay was used to further
confirm whether miR-106b directly targets LARP4B in LNCaP cells. As
presented in Fig. 5B, miR-106b
could directly bind to the 3′-UTR of LARP4B after co-transfection
of miR-106b mimic or miR-NC mimic, and the luciferase reporter
vector. Upregulation of miR-106b significantly suppressed the
luciferase activity of wild-type LARP4B 3′-UTR compared with the
miR-NC group, but not that of mutated LARP4B 3′-UTR.
In addition, to further confirm whether LARP4B is a
direct target gene of miR-106b, RT-qPCR and western blot analyses
were performed to evaluate the mRNA and protein levels of LARP4B in
PCa tissues and the matched adjacent normal tissues. As presented
in Fig. 5C and D, the expression
of LARP4B was decreased at the mRNA and protein level in PCa
tissues compared with in the matched adjacent normal tissues.
Furthermore, the expression of LARP4B in LNCaP cells
after transfection with miR-106b inhibitors was measured by
RT-qPCR, respectively. The results revealed that miR-106b
downregulation significantly increased the mRNA (Fig. 5E) and protein (Fig. 6E and F) expression of LARP4B
compared with the control.
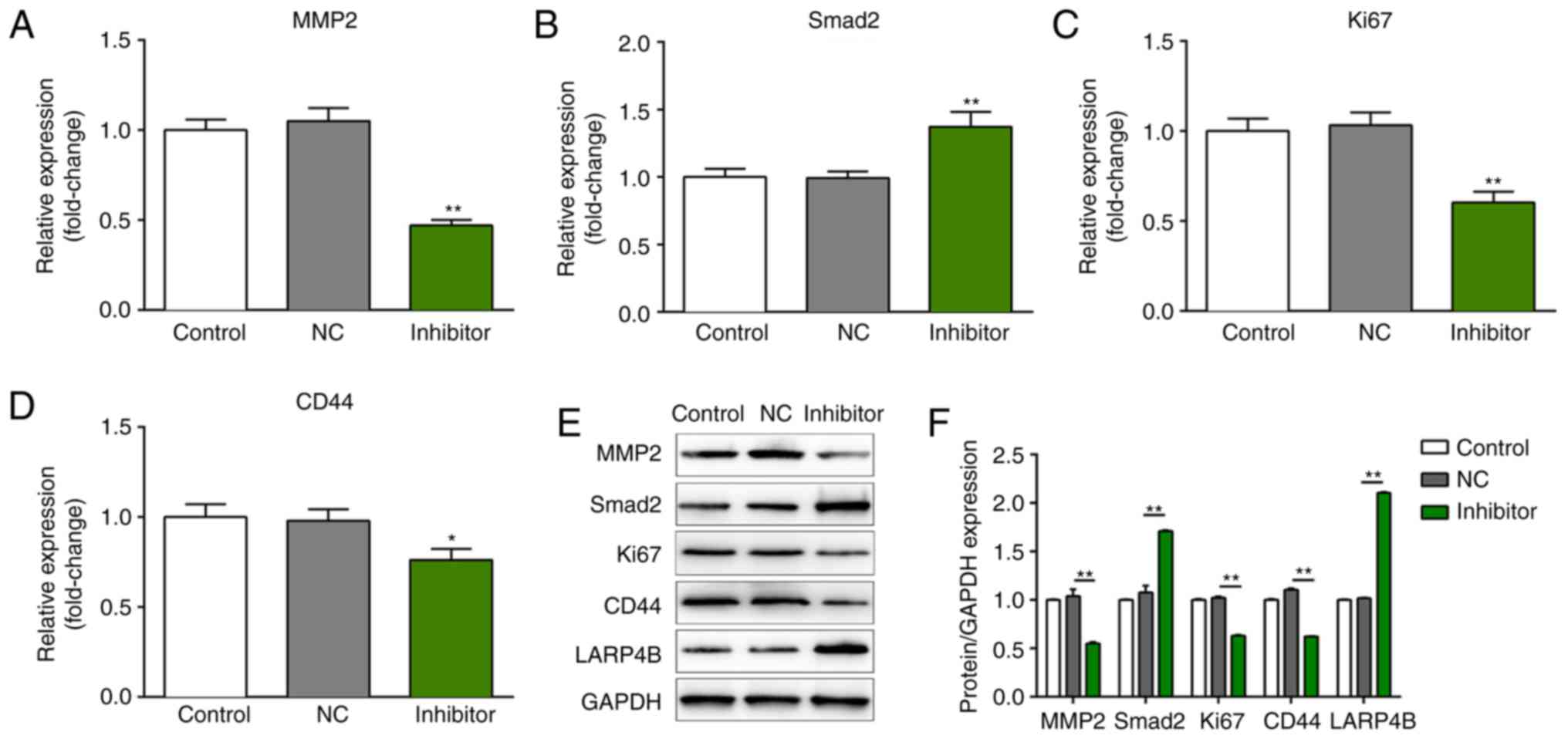 | Figure 6.Knockdown of miR-106b regulates the
expression of MMP2, Smad2, CD44 and Ki-67. (A-D) The mRNA and (E
and F) protein expression levels of MMP2, Smad2, Ki-67 and CD44 in
LNCaP cells after transfection with miR-106b, which were examined
by reverse transcription-quantitative polymerase chain reaction and
western blot analysis, repectively. The band intensity was
quantified by ImageJ software. The results are expressed as the
mean ± standard deviation of three independent experiments and each
was performed in triplicate. *P<0.05, **P<0.01 vs. control
group. CD, cluster of differentiation; MMP, matrix
metalloproteinase; Smad2, mothers against decapentaplegic homolog
2. |
Knockdown of miR-106b regulates the
expression of MMP2, Smad2, CD44 and Ki-67
To further confirm the role of miR-106b in the
progression and development of PCa, the expression of proteins
associated with the proliferation and metastasis of PCa, including
MMP2, Smad2, CD44 and Ki-67, was measured by RT-qPCR analysis and
western blotting. As presented in Fig.
6A-F, inhibition of miR-106b significantly suppressed the
expression of MMP2, CD44 and Ki-67, whereas it markedly increased
the expression of Smad2 compared with the control.
Discussion
miR-106b was reported to be aberrantly expressed in
numerous types of cancers, such as non-small cell lung cancer
(34), breast cancer (35) and renal cell cancer (36). Additionally, miR-106b was
differentially expressed in colorectal cancer (37), esophageal squamous cell carcinoma
(38) and hepatocellular cancer
(39). miR-106b was downregulated
in thyroid cancer tissues, while overexpressed miR-106b inhibited
the migration and invasion of thyroid cancer cells (30); however, miR-106b was upregulated in
gastric cancer, and downregulated miR-106b reduced the migration
and invasion of gastric cancer cells (27). Therefore, miR-106b may serve an
oncogenic or anti-tumor role in cancer. However, the possible roles
of miR-106b in prostate cancer require further investigation.
In the present study, miR-106b was overexpressed in
PCa tissues. The expression of miR-106b in PCa cells was
significantly decreased following treatment with miR-106b
inhibitor. We proposed that miR-106b may act as oncogene in PCa.
Previous studies showed that miR-106b was upregulated in PCa, and
was associated with the progression and prognosis of PCa,
suggesting that miR-106b could serve as novel clinical markers of
PCa (40,41). However, the underlying mechanisms
remain unclear.
LARPs are a class of RNA-binding proteins that are
located at different subcellular sites and interact with RNA,
serving an important role in cell transcription and translation
(42,43). In recent years, LARPs have been
reported to serve important roles in the proliferation,
differentiation, migration and angiogenesis of several malignant
tumors (44). LARP4B, a member of
the LARP family, acts as a tumor suppressor in glioma (45); however, its expression and
regulatory mechanism in PCa have not been elucidated. In the
present study, we determined that the expression of LARP4B was
downregulated in PCa tissues. Furthermore, LARP4B was reported as a
target gene of miR-106b. Additionally, downregulation of miR-106b
increased the expression of LARP4B. These results support LARP4B as
a critical downstream mediator of miR-106b, involved in the
progression and development of PCa.
We further investigated the potential roles of
miR-106b on the behavior of PCa cells. Downregulated miR-106b
suppressed the cell viability, migration and invasion of PCa cells.
In addition, knockdown of miR-106 downregulated the expression of
MMP2, Ki67 and CD44, but increased that of Smad2. Upregulated MMP2
promoted the migration and invasion of glioblastoma multiforme
cells (46). Mutations or
deletions in Smad2 can interrupt transforming growth factor (TGF)-β
signal transduction, and lead to reduced growth inhibition of
induced by TGF-β, resulting in tumor development (47–49).
CD44 is highly expressed in malignant tumor cells and its
expression is closely associated with patient prognosis (50–52).
miR-200b-3p regulated the proliferation and apoptosis of colorectal
cancer cells, and inhibited Ki-67 signaling (53). MMP2, Smad2, Ki-67 and CD44 serve an
important role in the proliferation, migration and invasion of
tumor cells (54–56). The role of miR-106b in regulating
the expression of MMP2, Smad2, Ki-67 and CD44 further suggested
that miR-106b could suppress the viability, migration, and invasion
of PCa cells.
In conclusion, the present study demonstrated that
miR-106b targets LAR4B to suppress cancer cell viability, migration
and invasion; thus; miR-106b may represent a novel target for the
treatment of patients with PCa. However, there was a limitation in
the present study, in which no evidence was provided to link LARP4B
exclusively to miR-106b and its function in cancer, which will be
further investigated in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WY drafted the manuscript. WY, JC and GW collected,
analyzed and interpreted the data. DZ conceived and designed the
present study.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Ningbo First Hospital (Ningbo, China). Patients
provided written informed cosnent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Colombet M, Soerjomataram I,
Dyba T, Randi G, Bettio M, Gavin A, Visser O and Bray F: Cancer
incidence and mortality patterns in Europe: Estimates for 40
countries and 25 major cancers in 2018. Eur J Cancer. 103:356–387.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ren SC, Chen R and Sun YH: Prostate cancer
research in China. Asian J Androl. 15:350–353. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Naji L, Randhawa H, Sohani Z, Dennis B,
Lautenbach D, Kavanagh O, Bawor M, Banfield L and Profetto J:
Digital rectal examination for prostate cancer screening in primary
care: A systematic review and meta-analysis. Ann Fam Med.
16:149–154. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liss MA, Chen H, Hemal S, Krane S, Kane
CJ, Xu J and Kader AK: Impact of family history on prostate cancer
mortality in white men undergoing prostate specific antigen based
screening. J Urol. 193:75–79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Durmus T, Baur A and Hamm B:
Multiparametric magnetic resonance imaging in the detection of
prostate cancer. Rofo. 186:238–246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boesen L, Nørgaard N, Løgager V, Balslev I
and Thomsen HS: A prospective comparison of selective
multiparametric magnetic resonance imaging fusion-targeted and
systematic transrectal ultrasound-guided biopsies for detecting
prostate cancer in men undergoing repeated biopsies. Urol Int.
99:384–391. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moghul M, Somani B, Lane T, Vasdev N,
Chaplin B, Peedell C, KandaSwamy GV and Rai BP: Detection rates of
recurrent prostate cancer: 68Gallium (Ga)-labelled
prostate-specific membrane antigen versus choline PET/CT scans. A
systematic review. Ther Adv Urol. 11:17562872188157932019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Teoh JY, Yuen SK, Tsu JH, Wong CK, Ho BSh,
Ng AT, Ma WK, Ho KL and Yiu MK: Prostate cancer detection upon
transrectal ultrasound-guided biopsy in relation to digital rectal
examination and prostate-specific antigen level: What to expect in
the Chinese population? Asian J Androl. 17:821–825. 2015.PubMed/NCBI
|
|
11
|
Javali TD, Dwivedi DK, Kumar R,
Jagannathan NR, Thulkar S and Dinda AK: Magnetic resonance
spectroscopy imaging-directed transrectal ultrasound biopsy
increases prostate cancer detection in men with prostate-specific
antigen between 4–10 ng/ml and normal digital rectal examination.
Int J Urol. 21:257–262. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kash DP, Lal M, Hashmi AH and Mubarak M:
Utility of digital rectal examination, serum prostate specific
antigen, and transrectal ultrasound in the detection of prostate
cancer: A developing country perspective. Asian Pac J Cancer Prev.
15:3087–3091. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McCann JV, Xiao L, Kim DJ, Khan OF,
Kowalski PS, Anderson DG, Pecot CV, Azam SH, Parker JS, Tsai YS, et
al: Endothelial miR-30c suppresses tumor growth via inhibition of
TGF-β-induced Serpine1. J Clin Invest. 130:1654–1670. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee J, Kwon MH, Kim JA and Rhee WJ:
Detection of exosome miRNAs using molecular beacons for diagnosing
prostate cancer. Artif Cells Nanomed Biotechnol. 46 (Suppl
3):S52–S63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Malla B, Aebersold DM and Dal Pra A:
Protocol for serum exosomal miRNAs analysis in prostate cancer
patients treated with radiotherapy. J Transl Med. 16:2232018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiaoli Z, Yawei W, Lianna L, Haifeng L and
Hui Z: Screening of target genes and regulatory function of miRNAs
as prognostic indicators for prostate cancer. Med Sci Monit.
21:3748–3759. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang N, Wang L, Liu J, Liu L, Huang J,
Chen X and Luo Z: MicroRNA-206 regulates the epithelial-mesenchymal
transition and inhibits the invasion and metastasis of prostate
cancer cells by targeting Annexin A2. Oncol Lett. 15:8295–8302.
2018.PubMed/NCBI
|
|
18
|
Wang Y, Xu H, Si L, Li Q, Zhu X, Yu T and
Gang X: MiR-206 inhibits proliferation and migration of prostate
cancer cells by targeting CXCL11. Prostate. 78:479–490. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Karatas OF, Wang J, Shao L, Ozen M, Zhang
Y, Creighton CJ and Ittmann M: miR-33a is a tumor suppressor
microRNA that is decreased in prostate cancer. Oncotarget.
8:60243–60256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou YJ, Yang HQ, Xia W, Cui L, Xu RF, Lu
H, Xue Z, Zhang B, Tian ZN, Cao YJ, et al: Down-regulation of
miR-605 promotes the proliferation and invasion of prostate cancer
cells by up-regulating EN2. Life Sci. 190:7–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo B, Kang N, Chen Y, Liu L and Zhang Y:
Oncogene miR-106a promotes proliferation and metastasis of prostate
cancer cells by directly targeting PTEN in vivo and in vitro.
Minerva Med. 109:24–30. 2018.PubMed/NCBI
|
|
22
|
Yang ZG, Ma XD, He ZH and Guo YX:
miR-483-5p promotes prostate cancer cell proliferation and invasion
by targeting RBM5. Int Braz J Urol. 43:1060–1067. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu C, Li J, Cheng G, Zhou H, Tao L, Cai
H, Li P, Cao Q, Ju X, Meng X, et al: miR-154 inhibits EMT by
targeting HMGA2 in prostate cancer cells. Mol Cell Biochem.
379:69–75. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Epis MR, Giles KM, Beveridge DJ,
Richardson KL, Candy PA, Stuart LM, Bentel J, Cohen RJ and Leedman
PJ: miR-331-3p and Aurora Kinase inhibitor II co-treatment
suppresses prostate cancer tumorigenesis and progression.
Oncotarget. 8:55116–55134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jackson BL, Grabowska A and Ratan HL:
MicroRNA in prostate cancer: Functional importance and potential as
circulating biomarkers. BMC Cancer. 14:9302014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Y, Wang K, Gao W, Zhang C, Huang F, Wen
S and Wang B: MicroRNA-106b regulates the tumor suppressor RUNX3 in
laryngeal carcinoma cells. FEBS Lett. 587:3166–3174. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang TS, Yang XH, Chen X, Wang XD, Hua J,
Zhou DL, Zhou B and Song ZS: MicroRNA-106b in cancer-associated
fibroblasts from gastric cancer promotes cell migration and
invasion by targeting PTEN. FEBS Lett. 588:2162–2169. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Smith AL, Iwanaga R, Drasin DJ, Micalizzi
DS, Vartuli RL, Tan AC and Ford HL: The miR-106b-25 cluster targets
Smad7, activates TGF-β signaling, and induces EMT and tumor
initiating cell characteristics downstream of Six1 in human breast
cancer. Oncogene. 31:5162–5171. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Tan W, Neo TW, Aung MO, Wasser S,
Lim SG and Tan TM: Role of the miR-106b-25 microRNA cluster in
hepatocellular carcinoma. Cancer Sci. 100:1234–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carvalheira G, Nozima BH and Cerutti JM:
microRNA-106b-mediated down-regulation of C1orf24 expression
induces apoptosis and suppresses invasion of thyroid cancer.
Oncotarget. 6:28357–28370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dai F, Liu T, Zheng S, Liu Q, Yang C, Zhou
J, Chen Y, Sheyhidin I and Lu X: MiR-106b promotes migration and
invasion through enhancing EMT via downregulation of Smad 7 in
Kazakh's esophageal squamous cell carcinoma. Tumour Biol.
37:14595–14604. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Chen D, Su Z, Li Y, Liu J, Jin L,
Shi M, Jiang Z, Qi Z, Gui Y, et al: MicroRNA-106b functions as an
oncogene in renal cell carcinoma by affecting cell proliferation,
migration and apoptosis. Mol Med Rep. 13:1420–1426. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wei K, Pan C, Yao G, Liu B, Ma T, Xia Y,
Jiang W, Chen L and Chen Y: miR-106b-5p promotes proliferation and
inhibits apoptosis by regulating BTG3 in non-small cell lung
cancer. Cell Physiol Biochem. 44:1545–1558. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li N, Miao Y, Shan Y, Liu B, Li Y, Zhao L
and Jia L: miR-106b and miR-93 regulate cell progression by
suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell
Death Dis. 8:e27962017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu J, Wei JH, Feng ZH, Chen ZH, Wang YQ,
Huang Y, Fang Y, Liang YP, Cen JJ, Pan YH, et al: miR-106b-5p
promotes renal cell carcinoma aggressiveness and stem-cell-like
phenotype by activating Wnt/β-catenin signalling. Oncotarget.
8:21461–21471. 2017.PubMed/NCBI
|
|
37
|
Ni S, Weng W, Xu M, Wang Q, Tan C, Sun H,
Wang L, Huang D, Du X and Sheng W: miR-106b-5p inhibits the
invasion and metastasis of colorectal cancer by targeting CTSA.
Onco Targets Ther. 11:3835–3845. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang J, Chen D, Liang S, Wang J, Liu C,
Nie C, Shan Z, Wang L, Fan Q and Wang F: miR-106b promotes cell
invasion and metastasis via PTEN mediated EMT in ESCC. Oncol Lett.
15:4619–4626. 2018.PubMed/NCBI
|
|
39
|
Yen CS, Su ZR, Lee YP, Liu IT and Yen CJ:
miR-106b promotes cancer progression in hepatitis B
virus-associated hepatocellular carcinoma. World J Gastroenterol.
22:5183–5192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liang H, Studach L, Hullinger RL, Xie J
and Andrisani OM: Down-regulation of RE-1 silencing transcription
factor (REST) in advanced prostate cancer by hypoxia-induced
miR-106b~25. Exp Cell Res. 320:188–199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hudson RS, Yi M, Esposito D, Glynn SA,
Starks AM, Yang Y, Schetter AJ, Watkins SK, Hurwitz AA, Dorsey TH,
et al: MicroRNA-106b-25 cluster expression is associated with early
disease recurrence and targets caspase-7 and focal adhesion in
human prostate cancer. Oncogene. 32:4139–4147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Merret R, Martino L, Bousquet-Antonelli C,
Fneich S, Descombin J, Billey E, Conte MR and Deragon JM: The
association of a La module with the PABP-interacting motif PAM2 is
a recurrent evolutionary process that led to the
neofunctionalization of La-related proteins. RNA. 19:36–50. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hussain RH, Zawawi M and Bayfield MA:
Conservation of RNA chaperone activity of the human La-related
proteins 4, 6 and 7. Nucleic Acids Res. 41:8715–8725. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Stavraka C and Blagden S: The La-related
proteins, a family with connections to cancer. Biomolecules.
5:2701–2722. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Koso H, Yi H, Sheridan P, Miyano S, Ino Y,
Todo T and Watanabe S: Identification of RNA-binding protein LARP4B
as a tumor suppressor in glioma. Cancer Res. 76:2254–2264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu J, Yang J, Yu L, Rao C, Wang Q, Sun C,
Shi C, Hua D, Zhou X, Luo W, et al: miR-361-5p inhibits glioma
migration and invasion by targeting SND1. Onco Targets Ther.
11:5239–5252. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bao Y, Chen Z, Guo Y, Feng Y, Li Z, Han W,
Wang J, Zhao W, Jiao Y, Li K, et al: Tumor suppressor microRNA-27a
in colorectal carcinogenesis and progression by targeting SGPP1 and
Smad2. PLoS One. 9:e1059912014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tu B, Peng ZX, Fan QM, Du L, Yan W and
Tang TT: Osteosarcoma cells promote the production of pro-tumor
cytokines in mesenchymal stem cells by inhibiting their osteogenic
differentiation through the TGF-β/Smad2/3 pathway. Exp Cell Res.
320:164–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fleming NI, Jorissen RN, Mouradov D,
Christie M, Sakthianandeswaren A, Palmieri M, Day F, Li S, Tsui C,
Lipton L, et al: SMAD2, SMAD3 and SMAD4 mutations in colorectal
cancer. Cancer Res. 73:725–735. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mattheolabakis G, Milane L, Singh A and
Amiji MM: Hyaluronic acid targeting of CD44 for cancer therapy:
From receptor biology to nanomedicine. J Drug Target. 23:605–618.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Prochazka L, Tesarik R and Turanek J:
Regulation of alternative splicing of CD44 in cancer. Cell Signal.
26:2234–2239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kinugasa Y, Matsui T and Takakura N: CD44
expressed on cancer-associated fibroblasts is a functional molecule
supporting the stemness and drug resistance of malignant cancer
cells in the tumor microenvironment. Stem Cells. 32:145–156. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen L, Wang X, Zhu Y, Zhu J and Lai Q:
miR-200b-3p inhibits proliferation and induces apoptosis in
colorectal cancer by targeting Wnt1. Mol Med Rep. 18:2571–2580.
2018.PubMed/NCBI
|
|
54
|
Yadav L, Puri N, Rastogi V, Satpute P,
Ahmad R and Kaur G: Matrix metalloproteinases and cancer-roles in
threat and therapy. Asian Pac J Cancer Prev. 15:1085–1091. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ok Atılgan A, Özdemir BH, Akçay EY, Ataol
Demirkan Ö, Tekindal MA and Özkardeş H: Role of tumor-associated
macrophages in the Hexim1 and TGFβ/SMAD pathway, and their
influence on progression of prostatic adenocarcinoma. Pathol Res
Pract. 212:83–92. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lee IH, Sohn M, Lim HJ, Yoon S, Oh H, Shin
S, Shin JH, Oh SH, Kim J, Lee DK, et al: Ahnak functions as a tumor
suppressor via modulation of TGFβ/Smad signaling pathway. Oncogene.
33:4675–4684. 2014. View Article : Google Scholar : PubMed/NCBI
|