Introduction
In total, ~80% of primary liver cancers are
hepatocellular carcinomas (HCCs) (1). The onset of HCC is concealed, and
radiotherapy, chemotherapy and surgical treatment do not reliably
improve the prognosis of patients, leading to a high mortality rate
(1,2). Malignant proliferation and escape
from apoptosis are the most important pathological features of HCC
(1,3). Therefore, studying the molecular
mechanisms regulating the malignant proliferation and apoptosis of
HCC are required for developing effective therapeutic
interventions.
The sirtuin family are class III histone
deacetylases of nicotinamide adenine dinucleotide (NAD+) (4). Previous studies have demonstrated
that sirtuin family members regulate gene expression by
deacetylating various non-histone proteins, contribute to various
important physiological activities, including cell differentiation,
apoptosis, aging and energy metabolism, and serve important roles
in the formation and development of various tumors (5–8).
Sirtuin (SIRT)6 is localized in nuclear heterochromatin, and
exhibits adenosine diphosphate-ribosyltransferase and deacetylase
activities (9). At present, the
effects of SIRT6 on the proliferation and apoptosis of HCC remain
unclear, and the functions of SIRT6 in promotion or inhibition
during progression of tumor development are controversial (10); however, SIRT6 has exhibited the
potential as a candidate target in the treatment of tumors
(11,12). Therefore, the study of SIRT6 in HCC
is urgent and valuable.
Mitogen-activated protein kinase (MAPK) is an
important transmitter of signal transduction from the cell membrane
to the inner nucleus (13).
Extracellular-signal regulated protein kinase (ERK) and c-Jun
N-terminal kinase (JNK) are two subfamilies of MAPK; abnormal
activation of these kinases is associated with the development of
liver cancer (13).
Phosphorylation of ERK can activate a variety of target molecules,
including c-Jun, c-Fos and G1/S-specific cyclin-D1, promoting the
development of liver cancer (14).
Additionally, the JNK signaling pathway can promote the expression
of downstream pathway proteins c-Jun, P53 and P21, induce the
proliferation and angiogenesis of liver cancer, and contribute
towards the regulation of the cell cycle and apoptosis of liver
cancer cells (14). Therefore, the
activation of the ERK1/2 signaling pathway may be a mechanism
underlying the development of HCC.
The present study aimed to evaluate the expression
of SIRT6 in various HCC cell lines, and investigate the role of
SIRT6 in cell proliferation and apoptosis, further determining the
involvement of the ERK1/2 signal pathway. The present study
provided novel insight into the role of SIRT6 in HCC, and the
potential underlying mechanisms.
Materials and methods
Cell culture, plasmid, grouping and
transfection
HL-7702, MIHA, Hep3B, Huh-7, MHCC-97H, MHCC-97L,
MHCC-LM6, MHCC-LM3, YY-8103, SK-hep-1 cell lines were all obtained
from the Type Culture Collection of the Chinese Academy of
Sciences. The HL-7702 cell line is a normal liver cell line, and
was used as a control group. HL-7702 and MIHA cells were cultured
in RPMI-1640 medium (Thermo Fisher Scientific, Inc.); Hep3B, Huh-7,
MHCC-97H, MHCC-97L, MHCC-LM6, MHCC-LM3, YY-8103, SK-hep-1 cell
lines were cultured in DMEM (Thermo Fisher Scientific, Inc.).
Medium was supplemented with 10% fetal bovine serum (HyClone; GE
Healthcare Life Sciences), and cells were cultured at 37°C with 5%
CO2. Cells were subcultured every 2–3 days. The SIRT6
overexpression plasmid, the negative control (NC) plasmid (empty
pcDNA3.1 vector), the plasmid containing small interfering RNA
against SIRT6 (siSIRT6) and the plasmid containing NC-siRNA were
synthesized by Shanghai GenePharma Co., Ltd. The sequences of
oligonucleotides were as follow: siSIRT6,
5′-TCATGACCCGGCTCATGAA-3′; NC-siRNA, 5′-TCACCCATCGGTACGTGAA-3′.
Huh-7 cell line was divided into 3 groups: Control (blank); NC or
siNC (cells transfected with the NC plasmid or NC-siRNA plasmid);
and SIRT6 overexpression or siSIRT6 groups (cells transfected with
the SIRT6 overexpression or siSIRT6 plasmid). To further explore
whether SIRT6 affects the proliferation and apoptosis of HCC cells
by regulating the ERK1/2 pathway, the cells were treated with 10 µM
U0126 at 37°C for 24 h, and U0126 treatment and transfection
occurred simultaneously. Huh-7 cells were also separated into
control (blank), NC (cells transfected with the NC plasmid), SIRT6
(cells transfected with the SIRT6 overexpression plasmid), U0126
(cells treated with 10 µM U0126), SIRT6 + U0126 (cells transfected
with the SIRT6 overexpression plasmid and treated with 10 µM U0126)
groups for the measurement of cell proliferation using ERK1/2
inhibitor U0126. Transfection was conducted using Lipofectamine™
2000 transfection reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols. Following
transfection for 48 h, cells were used to subsequent
experiments.
Cell proliferation assay
Cell proliferation was assayed using a Cell Counting
Kit-8 (Sigma-Aldrich; Merck KGaA) at 0, 24 and 48 h following
transfection. Procedures were performed according to the
manufacturer's protocols. Cells (3×103 cells/well) were
then incubated in a 96-well plate for 1 h at 37°C, and the optical
density (OD) was measured at 450 nm using a microplate reader
(Multiskan™; Thermo Fisher Scientific, Inc.).
Measurement of apoptosis via flow
cytometry
Following transfection for 48 h, Huh-7 cells were
washed with PBS and dissociated with 0.25% trypsin for 2 min at
37°C. Cells (1–5×105) were centrifuged at 800 × g for 5
min and collected. Using an Annexin V-FITC Apoptosis
Staining/Detection kit (Abcam), 195 µl Annexin V binding solution,
6 µl Annexin V and 4 µl propidium iodide were added to cells, and
then mixed gently by pipetting. Cells were incubated at room
temperature for 20 min in the dark, and immediately analyzed using
a flow cytometer (BD FACSCanto II; BD Biosciences) and FACSDiva
software version 6.1.2 (BD Biosciences) to detect the apoptosis
rate. Flow cytometry demonstrated that the advanced apoptotic cells
were in the upper right quadrant, and the early apoptotic cells
were in the lower right quadrant. The apoptotic rate was the sum of
the early and advanced apoptotic rates.
Plate clone formation assay
Transfected cells were collected and inoculated into
a 100-mm dish at a concentration of 200 cells/dish following
transfection for 48 h. G418 (700 µg/ml; Abcam) was added to the
medium and mixed to detect positive cell clones for 3 weeks until
visible cell clones emerged. Fresh medium was replaced every 3
days. The supernatant was then discarded, and the cells were gently
washed with PBS twice. Cells were fixed with methanol for 15 min at
room temperature, and stained with crystal violet for 10–30 min at
room temperature. Cells were then washed slowly with running water
and dried. Each cell clone on the dishes was counted and
photographed under a light microscope (magnification, ×40).
Assessment of mRNA levels via reverse
transcription- quantitative (RT-q)PCR
Total RNA was extracted using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) and 1 µg of RNA of
each sample was transcribed into cDNA using an iScript™ cDNA
Synthesis kit (Bio-Rad Laboratories, Inc.). The reverse
transcription reaction was performed at 42°C for 15 min, followed
by reverse transcriptase inactivation at 85°C for 15 sec. A Fast
Start Universal SYBR-Green Master kit (Roche Diagnostics) was used
to perform qPCR. The primers used for the reaction are presented in
Table I. The reaction system was
the following: 2X SYBR-Green Master Mix (12.5 µl); cDNA template (2
µl); forward primer (10 µM; 1 µl); reverse primer (10 µM; 1 µl);
and ddH2O (8.5 µl). qPCR was conducted as follows: 95°C
for 10 min, then 40 cycles of 95°C for 15 sec, 60°C for 1 min and
72°C for 3 min. qPCR was conducted in a CFX96 Touch™ system (cat.
no. 6093; Bio-Rad Laboratories, Inc.). The 2−ΔΔCq method
(15) was used to calculate the
relative mRNA expression in samples; GAPDH was used as an internal
reference.
 | Table I.Primers used for quantitative
PCR. |
Table I.
Primers used for quantitative
PCR.
| Genes | Primers |
|---|
| SIRT6 | F:
5′-GCAGTCTTCCAGTGTGGTGT-3′ |
|
| R:
5′-CCATGGTCCAGACTCCGT-3′ |
| Bcl-2 | F:
5′-TTGAGGAAGTGAACATTTCGGTG-3′ |
|
| R:
5′-AGGTTCTGCGGACTTAGGTC-3′ |
| Bax | F:
5′-GCGAGTGTCTCAAGCGCATC-3′ |
|
| R:
5′-CCAGTTGAAGTTGCCGTCAGAA-3′ |
| GAPDH | F:
5′-ATGGTGAAGGTCGGTGTGAA-3′ |
|
| R:
5′-TGGAAGATGGTGATGGGCTT-3′ |
Extraction of total protein and
western blotting
Cells were collected and washed with PBS, and RIPA
buffer (Beijing Solarbio Science & Technology Co., Ltd.) was
added to lyse the cells according to the manufacturer's protocols.
Cells were then centrifuged at 4°C and 16,000 × g for 15 min to
remove the cell debris and supernatant, and total protein was
collected. The concentration of total protein was determined using
a Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific, Inc.).
Standard protein was diluted to 1, 0.5, 0.25, 0.125 and 0.0625 g/ml
respectively, then 2 µl samples and standard proteins were added to
a 96-well plate. BCA reagent was subsequently added, and plates
were incubated at 37°C for 30 min. The OD at 562 nm was detected
using a microplate reader. A standard curve was drawn, and the
concentration of the total protein was then calculated.
Total protein (30 µg) from each sample was denatured
at 95°C for 10 min, and proteins were separated via 10% SDS-PAGE at
100 V for 2 h. Proteins were transferred onto PVDF membranes using
a wet transfer electrophoresis tank (Bio-Rad Laboratories, Inc.) at
90 V for 2 h, and then blocked in 5% non-fat milk for 1 h at room
temperature. Membranes were then incubated overnight at 4°C with
the following primary antibodies: Anti-SIRT6 (1:2,000; cat. no.
ab191385; Abcam); anti-cleaved-caspase-3 (Asp175; 1:2,000; cat. no.
9664; Cell Signaling Technology, Inc.); anti-Bcl-2 (1:2,000; cat.
no. ab32124; Abcam); anti-Bax (1:2,000; cat. no. ab32503; Abcam);
anti-GAPDH (1:2,000; cat. no. ab181602; Abcam); anti-ERK1/2
(1:2,000; cat. no. ab17942; Abcam); and anti-phosphorylated
(p)-ERK1 (T202) + ERK2 (T185; 1:2,000; cat. no. ab201015; Abcam).
Membranes were washed three times with PBS-0.05% Tween 20 (PBST;
Beijing Solarbio Science & Technology Co., Ltd.) for 5 min, and
the horseradish peroxidase-conjugated secondary antibody (1:2,000;
cat. no. ab6721; Abcam) was added. Membranes were then incubated
for 1 h at RT, and bands were visualized using Pierce™ ECL Plus
western blotting substrate (Thermo Fisher Scientific, Inc.)
following three washes in PBST. The densitometry was performed
using the Bio-Rad ChemiDoc system with Image Lab software version
6.0 (Bio-Rad Laboratories, Inc.).
Statistical analysis
Data were analyzed using GraphPad Prism 7.0 software
(GraphPad Software, Inc.). All data were presented as the mean ±
standard deviation. One-way ANOVA followed by Tukey's post hoc test
were used to compare between groups. P<0.05 was considered to
indicate statistical significance.
Results
SIRT6 is overexpressed in HCC cell
lines
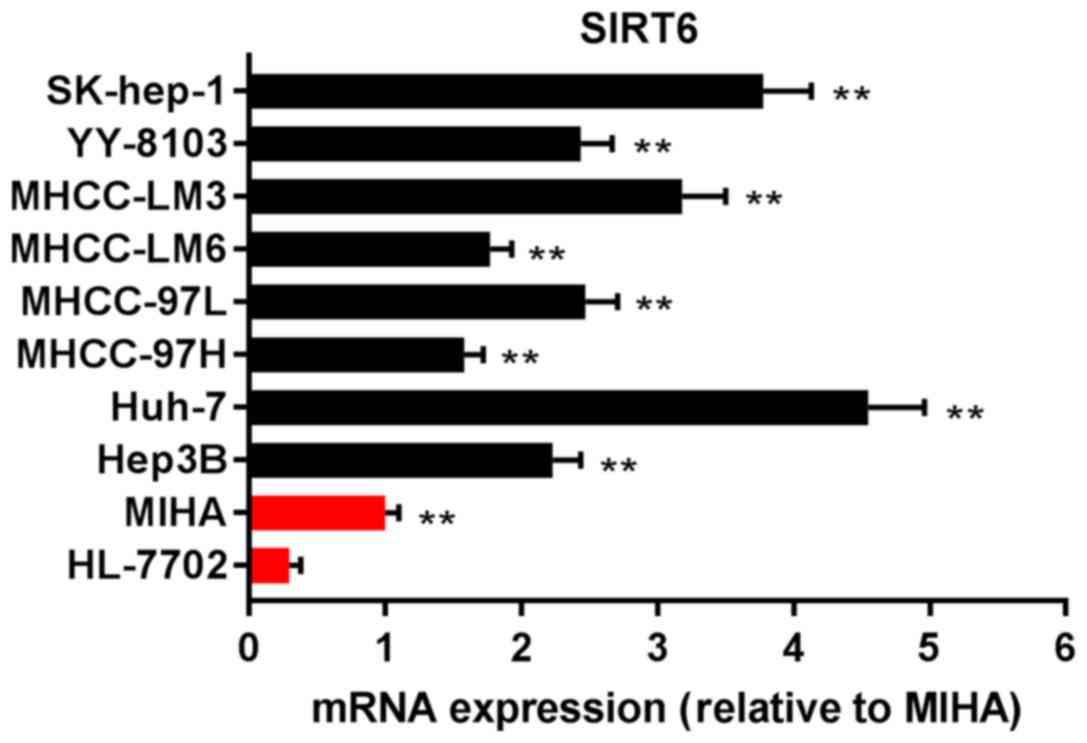
To investigate the expression of SIRT6 in HCC cell
lines, MIHA, HL7702, Hep3B, Huh-7, MHCC-97H, MHCC-97L, MHCC-LM6,
MHCC-LM3, YY-8103 and SK-hep-1 cell lines were purchased, and the
mRNA expression of SIRT6 was measured via RT-qPCR analysis. As
presented in Fig. 1, the
expression of SIRT6 mRNA in Hep3B, Huh-7, MHCC-97H, MHCC-97L,
MHCC-LM6, MHCC-LM3, YY-8103 and SK-hep-1 cells was significantly
upregulated compared with HL-7702 cells, normal human liver cells
set as the control (P<0.01). The results indicated that the
expression of SIRT6 was elevated in HCC.
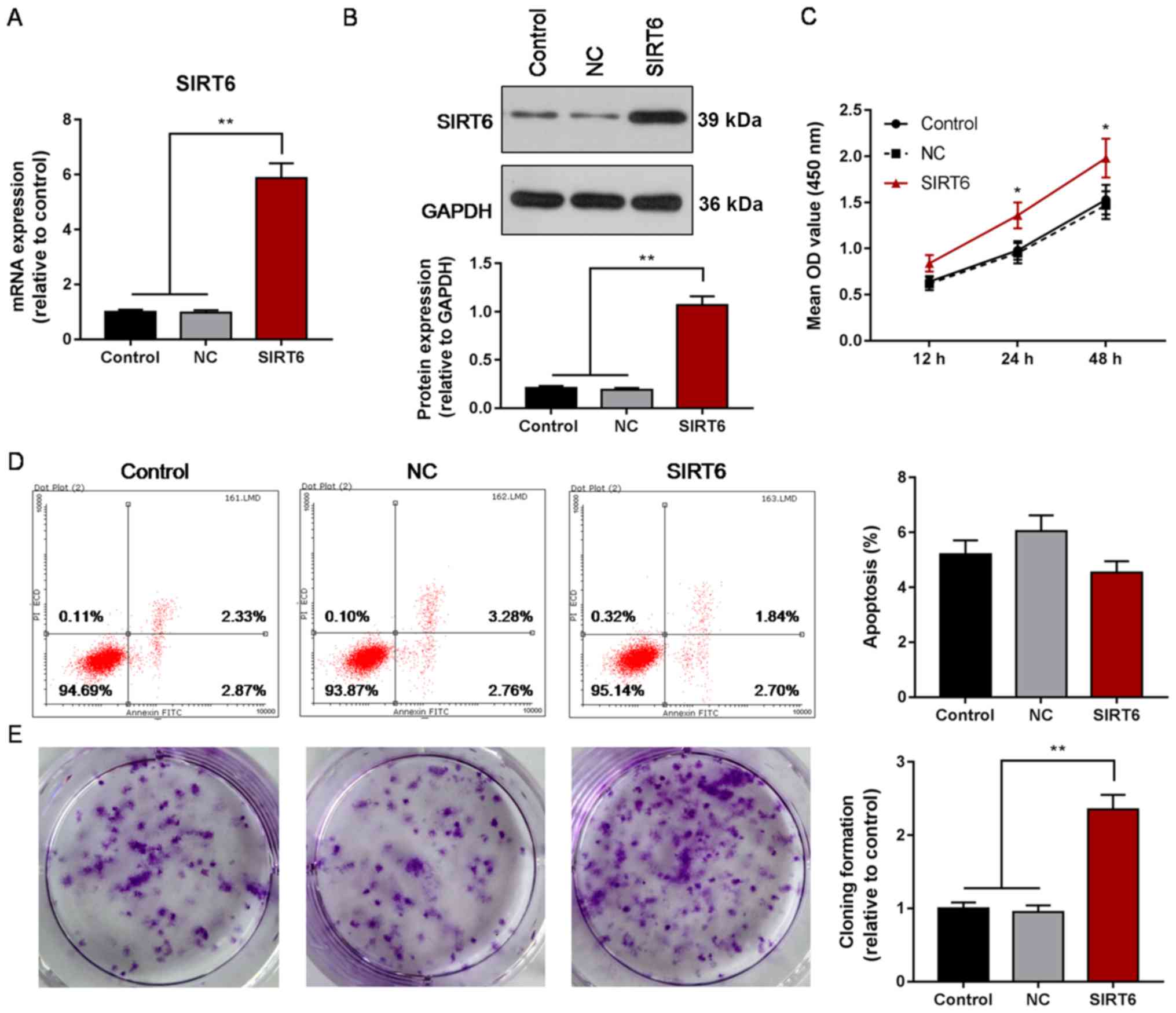
Overexpression of SIRT6 increases the
proliferation of Huh-7 cells
To investigate the effects of upregulated expression
of SIRT6 on the proliferation and apoptosis of HCC cells, the
proliferation, cloning efficiency and apoptosis of Huh-7 cells was
evaluated following overexpression of SIRT6. Overexpression of
SIRT6 mRNA and protein was demonstrated in Huh-7 cells (P<0.01;
Fig. 2A and B), and the
proliferation of transfected Huh-7 cells was significantly
increased compared with the control or NC groups at 24 and 48 h
following transfection (P<0.05; Fig. 2C). Cloning efficiency was increased
in the SIRT6 overexpression group compared with the control or NC
groups (P<0.01; Fig. 2E);
however, the apoptosis rate was not significantly affected by SIRT6
overexpression (Fig. 2D). The
results suggested that overexpression of SIRT6 may promote the
proliferation of HCC; however, its effects on apoptosis require
further investigation.
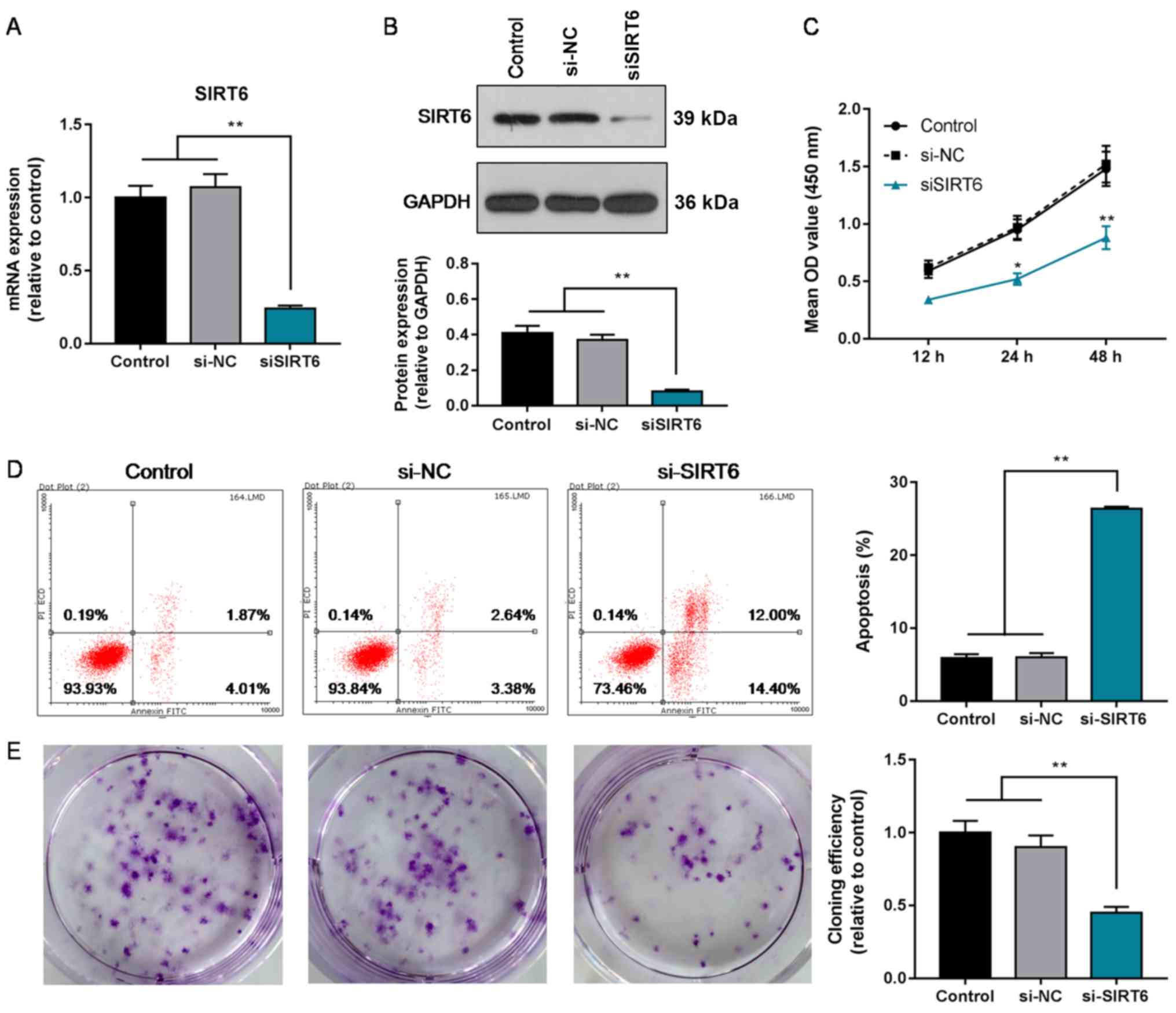
Knockdown of SIRT6 decreases the
proliferation, and increases the apoptosis rate of Huh-7 cells
To further determine the role of SIRT6 in HCC, the
effects of silencing SIRT6 on the proliferation and apoptosis of
Huh-7 cells was investigated. It was revealed that SIRT6 expression
was suppressed in Huh-7 cells following transfection with siSIRT6
(P<0.01; Fig. 3A and B).
Additionally, the proliferation of cells in the siSIRT6 group was
significantly decreased compared with the control and si-NC groups
at 24 and 48 h (P<0.05; Fig.
3C). Furthermore, the cloning efficiency of siSIRT6-transfected
cells was significantly reduced compared with the control and si-NC
groups as well (P<0.01; Fig.
3E). The apoptosis rate was also significantly increased in
siSIRT6 the group compared with the controls (P<0.01; Fig. 3D). Collectively, the results
indicated that the downregulation of SIRT6 decreased the
proliferation and increased the apoptosis of HCC cells, opposing
effects to the results observed following the overexpression of
SIRT6.
Intrinsic apoptosis pathway
suppression and activation following SIRT6 overexpression or
knockdown, respectively, in Huh-7 cells
To evaluate the activity of the intrinsic apoptosis
pathway, the mRNA expression of Bcl-2 and Bax was determined via
RT-qPCR analysis, and protein levels of Bcl-2, Bax and
cleaved-caspase-3 were measured via western blotting. It was
revealed that the expression of Bcl-2 in the SIRT6 overexpression
group was significantly upregulated compared with the control or NC
groups, and the expression of Bax and cleaved-caspase-3 was
significantly downregulated (P<0.05; Fig. 4A, C and E). Opposing results were
observed following siSIRT6-mediated knockdown of SIRT6 (P<0.01;
Fig. 4B, D and F). The results
indicated that overexpression of SIRT6 downregulated the intrinsic
apoptosis pathway in HCC cells, whereas silencing SIRT6 induced
opposing effects.
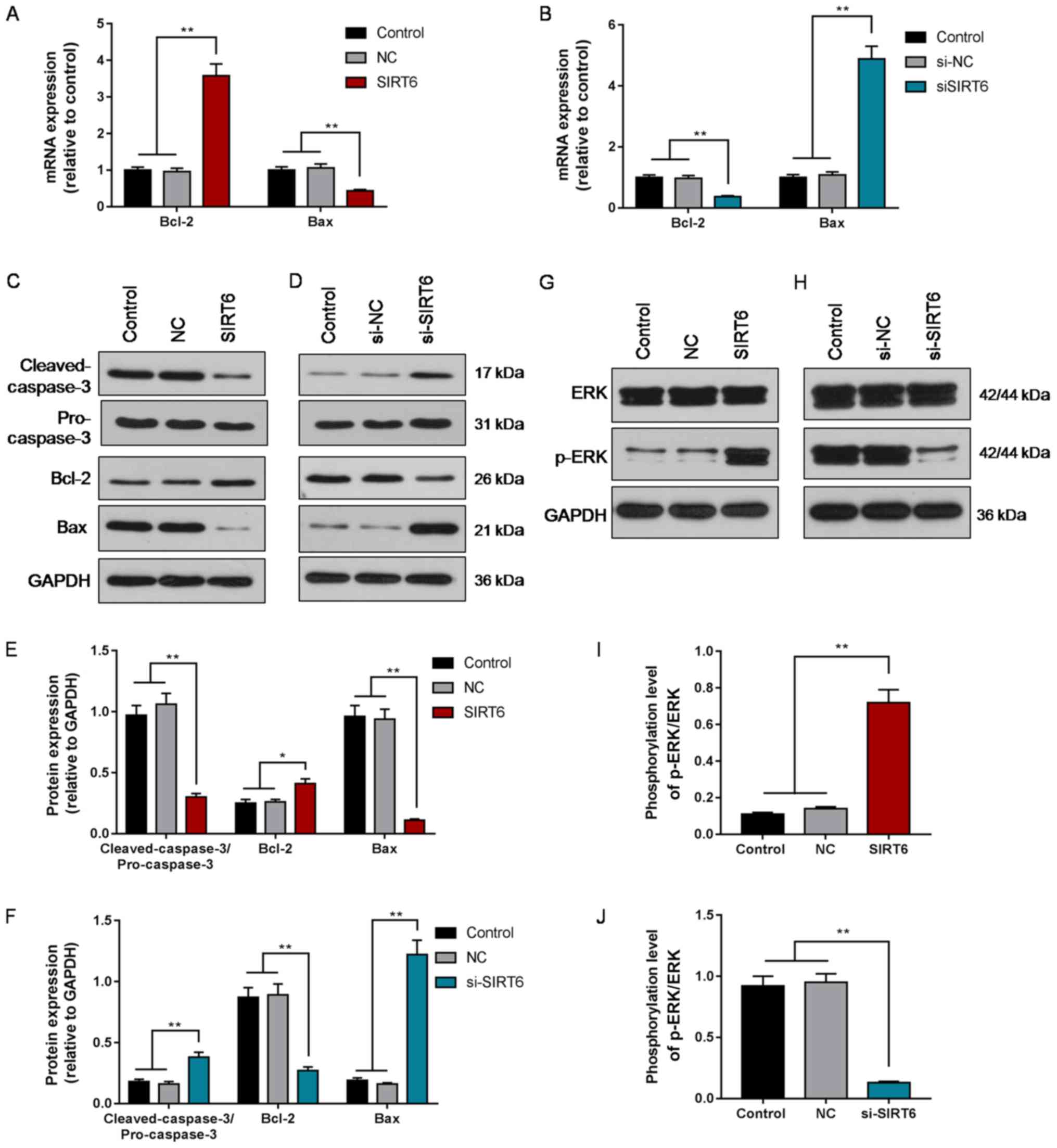 | Figure 4.Expression of Bcl-2, Bax,
cleaved-caspase-3, ERK1/2, and p-ERK1/2 following overexpression or
silencing of SIRT6 in hepatocellular carcinoma cells. (A) mRNA
expression of Bcl-2 and Bax in Huh-7 cells following SIRT6
overexpression. (B) mRNA expression of Bcl-2 and Bax in Huh-7 cells
following SIRT6 silencing. (C) Protein levels of cleaved-caspase-3,
Bcl-2, Bax in Huh-7 cells following SIRT6 overexpression. (D)
Protein levels of cleaved-caspase-3, Bcl-2, Bax in Huh-7 cells
following SIRT6 silencing. (E) Quantification of (C). (F)
Quantification of (D). (G) Protein levels of ERK1/2 and p-ERK1/2 in
Huh-7 cells following SIRT6 overexpression. (H) Protein levels of
ERK1/2 and p-ERK1/2 in Huh-7 cells following SIRT6 silencing. (I)
Quantification of (G). (J) Quantification of (H). Data are
presented as the mean ± standard deviation. *P<0.05,
**P<0.01. SIRT6, sirtuin 6; NC, negative control; siSIRT6, small
interfering RNA against SIRT6; p-, phosphorylated. |
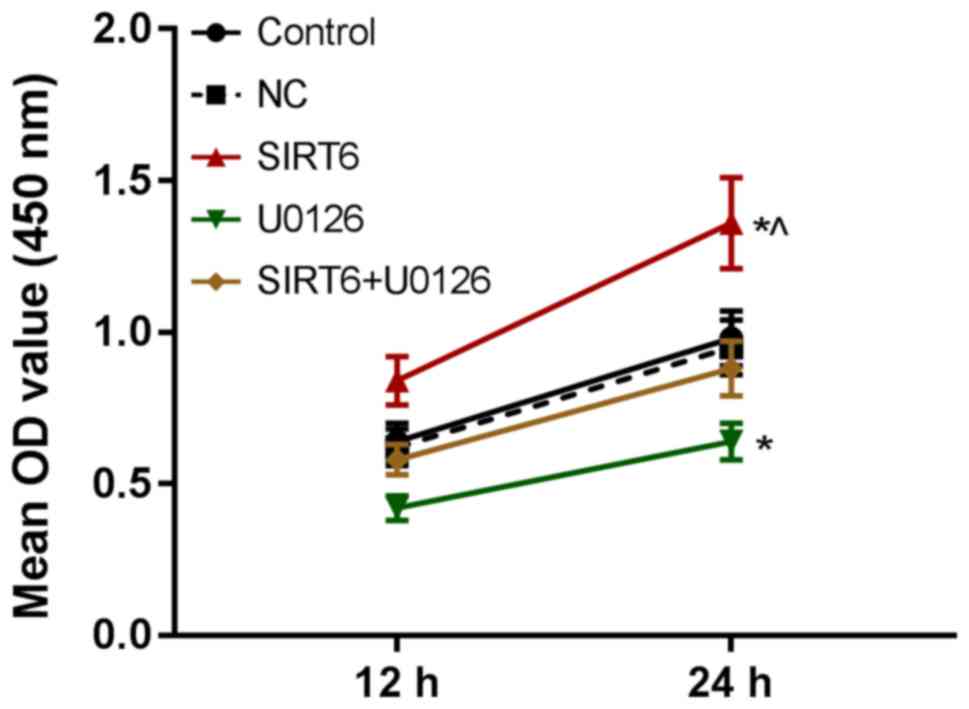
Activation of the ERK1/2 signal
pathway may be involved in the SIRT6-mediated regulation of the
proliferation of Huh-7 cells
To investigate the mechanisms underlying the effects
of SIRT6 on proliferation and apoptosis, the expression of ERK1/2
and p-ERK1/2 were determined following overexpression or knockdown
of SIRT6. Additionally, the role of the ERK1/2 signal pathway in
the effects of SIRT6 was explored by measuring the effects of the
ERK1/2 inhibitor U0126 on the proliferation of SIRT6-overexpressing
Huh-7 cells. It was demonstrated that the phosphorylation of ERK1/2
was significantly increased in the SIRT6 overexpression group
(P<0.01; Fig. 4G and I), and
decreased in the siSIRT6 group compared with the control and NC
groups (P<0.01; Fig. 4H and J).
Additionally, it was revealed that the proliferation of Huh-7 cells
was significantly decreased in the SIRT6 + U0126 group compared
with the SIRT6 group (P<0.05), and was significantly decreased
in the U0126 group compared with the control (P<0.05; Fig. 5). Collectively, the results
suggested that the ERK1/2 signal pathway was activated by
overexpression of SIRT6 and downregulated by knockdown of SIRT6,
and that the ERK1/2 signal pathway may be involved in the
SIRT6-mediated regulation of HCC cell proliferation.
Discussion
In the present study, the expression of SIRT6 was
measured in various HCC and normal liver cell lines, and the
effects of altering SIRT6 expression on the proliferation and
apoptosis of Huh-7 cell lines was evaluated. Furthermore, the
involvement of the ERK1/2 signal pathway in the SIRT6-regulated
proliferation of Huh-7 cells was investigated, providing novel
insight into potential target mechanisms for the treatment of
HCC.
The results of the present study demonstrated that
SIRT6 was overexpressed in various HCC cell lines. Ran et al
(16) also reported that SIRT6 was
highly expressed in Huh-7, HepG2, PLC/PRF/5, SMMC-7721, Hep3B and
SK-Hep-1 cell lines. Collectively, these studies have demonstrated
the overexpression of SIRT6 in a number of HCC cell lines,
indicating the value of studying SIRT6 in HCC.
The present study revealed that Huh-7 cell
proliferation and cloning efficiency were increased after SIRT6 was
overexpressed, and that they were decreased after the SIRT6 was
silenced. The apoptosis rate of cells in the SIRT6 overexpression
group was not significantly different to those in the control
group; however, it was significantly increased following siSIRT6
transfection. Therefore, the results indicated that the expression
of SIRT6 was positively associated with the proliferation, and
negatively associated with the apoptosis of HCC cells. Feng et
al (17) reported that SIRT6
promoted the tumorigenicity of HCC cells. Song et al
(18) revealed that following
knockout of SIRT6 by CRISPR/Cas-9, HCC cells exhibited decreased
viability and invasiveness. These reports are consistent with the
observations of the present study; however, Wang et al
(19) demonstrated that the
overexpression of SIRT6 attenuated HepG2 and HCCLM3 cell
proliferation. Similarly, Zhang and Qin (20) reported that knockdown of SIRT6
promoted the growth of HepG2 cells, whereas the overexpression of
SIRT6 inhibited HepG2 cell growth. It is proposed that this may be
due to SIRT6 performing distinct functions in different HCC cell
lines and tumor cells, HepG2 is a hepatoblastoma cell line
(21); however, this requires
further investigation (22,23).
The intrinsic apoptosis pathway is regulated by the
Bcl-2 protein family, and involves mitochondrial outer membrane
permeabilization (MOMP) via Bax (24). MOMP results in the release of
proapoptotic intermembrane space proteins which ultimately promote
apoptosome formation, resulting in caspase-9 engagement, and
caspase-3 and −7 activation, leading to the apoptosis of cells
(24,25). Thus, the expression levels of
Bcl-2, Bax and cleaved-caspase-3 are indicators of the activation
of intrinsic apoptosis pathway (26,27).
Tumor cells suppress apoptosis via various mechanisms, including
promoting the expression of Bcl-2 or downregulating proapoptotic
proteins such as Bax (28,29). Sui et al (30) reported that overexpression of Rab31
in Huh-7 cells promoted growth by increasing the Bcl-2/Bax ratio.
The present study demonstrated that the expression of Bcl-2 was
upregulated, whereas the levels of Bax and cleaved-caspase-3 were
downregulated following overexpression of SIRT6, with opposing
effects observed following silencing of SIRT6. The results
indicated that activation of the intrinsic apoptosis pathway may be
negatively associated with the expression of SIRT6 (31).
ERK is a notable serine/threonine protein kinase in
the MAPK family (32). Abnormal
activation of ERK results in the upregulation of a series of target
genes such as matrix metalloproteinase-2 (MMP-2) and vascular
endothelial growth factor, promoting the proliferation and invasion
of various tumor cells (33,34).
The results indicated that the ERK1/2 signal pathway was activated
following overexpression of SIRT6 and deactivated by the
downregulation of SIRT6. After using the ERK1/2 signal pathway
inhibitor U0126, the SIRT6 overexpression-induced increase in Huh-7
cell proliferation was attenuated. Huang et al (35) revealed that paxillin promoted Bcl-2
activation via paxillin-mediated ERK activation, which was
associated with tumor formation efficacy in mice in a study of
colorectal cancer. Bai et al (12) also reported that the overexpression
of SIRT6 promoted the migration and invasion of non-small cell lung
cancer cells via the ERK1/2/MMP-9 pathway. These reports indicated
that overexpression of SIRT6 can activate the ERK1/2 pathway and
therefore suppresses the intrinsic apoptosis pathway, promoting the
development of HCC. Conversely, Wang et al (19) observed that the overexpression of
SIRT6 reduced the expression of p-ERK in HepG2 and HCCLM3 cells.
Zhang and Qin (20) revealed that
overexpression of SIRT6 inhibited ERK1/2, and that inhibiting the
pathway with U0126 attenuated the tumor-suppressive effects of
SIRT6 overexpression. Thus, further investigation is required to
determine the precise roles of SIRT6 and the ERK1/2 signaling
pathway in HCC.
SIRT6 is an NAD(+)-dependent deacetylase. A previous
study identified SIRT6 protein as a potential drug target (36). Progress has been made in the study
of the SIRT6 structure and its inhibitors, and certain small
molecules with improved inhibition of the biological functions of
STIR6 have been discovered (37–39).
The present findings suggested that SIRT6 is a potential target for
the treatment of liver cancer; however, this study also possessed
certain shortcomings. Huh-7 cells were selected for further
experiments as they exhibited the highest SIRT6 expression out of
the 8 HCC cell lines; however, these proliferation and apoptosis
results can be more fully demonstrated if multiple HCC cell lines
were used. Animal experiments should be conducted in future studies
to validate the present findings in vivo. In addition, ERK
is involved in the transduction of numerous signaling pathways,
including the MAPK,PI3K/AKT and STAT pathways. It was demonstrated
that SIRT6 contributed to tumor development by regulating ERK
phosphorylation; the downstream mechanisms should be explored in
subsequent studies.
In conclusion, SIRT6 regulated the proliferation and
apoptosis of HCC cells via the regulation of the ERK1/2 pathway,
affecting the activation of the intrinsic apoptosis pathway. The
present study revealed that SIRT6 may be a potential target in the
gene therapy of HCC; however, the role of SIRT6 in HCC requires
further validation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
CZ, YY and KT made substantial contributions to the
conception and design of the study. QH and KT was involved in data
acquisition, data analysis and interpretation. CZ and YY drafted
the manuscript and critically revised it. All authors gave final
approval of the version to be published, and agreed to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kennedy AS and Sangro B: Nonsurgical
treatment for localized hepatocellular carcinoma. Curr Oncol Rep.
16:3732014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Blander G and Guarente L: The Sir2 family
of protein deacetylases. Annu Rev Biochem. 73:417–435. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vaquero A, Scher M, Lee D,
Erdjument-Bromage H, Tempst P and Reinberg D: Human SirT1 interacts
with histone H1 and promotes formation of facultative
heterochromatin. Mol Cell. 16:93–105. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang G, Hao F and Hu X: Downregulation of
microRNA-155 stimulates sevoflurane-mediated cardioprotection
against myocardial ischemia/reperfusion injury by binding to SIRT1
in mice. J Cell Biochem; 2019, View Article : Google Scholar
|
|
7
|
Zhang HX, Li YN, Wang XL, Ye CL, Zhu XY,
Li HP, Yang T and Liu YJ: Probucol ameliorates EMT and lung
fibrosis through restoration of SIRT3 expression. Pulm Pharmacol
Ther. 1018032019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jeong SG and Cho GW: The tubulin
deacetylase sirtuin-2 regulates neuronal differentiation through
the ERK/CREB signaling pathway. Biochem Biophys Res Commun.
482:182–187. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ming M, Han W, Zhao B, Sundaresan NR, Deng
CX, Gupta MP and He YY: SIRT6 promotes COX-2 expression and acts as
an oncogene in skin cancer. Cancer Res. 74:5925–5933. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vitiello M, Zullo A, Servillo L, Mancini
FP, Borriello A, Giovane A, Della Ragione F, D'Onofrio N and
Balestrieri ML: Multiple pathways of SIRT6 at the crossroads in the
control of longevity, cancer, and cardiovascular diseases. Ageing
Res Rev. 35:301–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Y, Xie QR, Wang B, Shao J, Zhang T,
Liu T, Huang G and Xia W: Inhibition of SIRT6 in prostate cancer
reduces cell viability and increases sensitivity to
chemotherapeutics. Protein Cell. 4:702–710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bai L, Lin G, Sun L, Liu Y, Huang X, Cao
C, Guo Y and Xie C: Upregulation of SIRT6 predicts poor prognosis
and promotes metastasis of non-small cell lung cancer via the
ERK1/2/MMP9 pathway. Oncotarget. 7:40377–40386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Zhao GD, Shi Z, Qi LL, Zhou LY and
Fu ZX: The Ras/Raf/MEK/ERK signaling pathway and its role in the
occurrence and development of HCC. Oncol Lett. 12:3045–3050. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ni Z, Wang B, Dai X, Ding W, Yang T, Li X,
Lewin S, Xu L, Lian J and He F: HCC cells with high levels of Bcl-2
are resistant to ABT-737 via activation of the ROS-JNK-autophagy
pathway. Free Radic Biol Med. 70:194–203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ran LK, Chen Y, Zhang ZZ, Tao NN, Ren JH,
Zhou L, Tang H, Chen X, Chen K, Li WY, et al: SIRT6 overexpression
potentiates apoptosis evasion in hepatocellular carcinoma via
BCL2-associated X protein-dependent apoptotic pathway. Clin Cancer
Res. 22:3372–3382. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Feng XX, Luo J, Liu M, Yan W, Zhou ZZ, Xia
YJ, Tu W, Li PY, Feng ZH and Tian DA: Sirtuin 6 promotes
transforming growth factor-β1/H2O2/HOCl-mediated enhancement of
hepatocellular carcinoma cell tumorigenicity by suppressing
cellular senescence. Cancer Sci. 106:559–566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song S, Yang Y, Liu M, Liu B, Yang X, Yu
M, Qi H, Ren M, Wang Z, Zou J, et al: MiR-125b attenuates human
hepatocellular carcinoma malignancy through targeting SIRT6. Am J
Cancer Res. 8:993–1007. 2018.PubMed/NCBI
|
|
19
|
Wang Y, Pan T, Wang H, Li L, Li J, Zhang D
and Yang H: Overexpression of SIRT6 attenuates the tumorigenicity
of hepatocellular carcinoma cells. Oncotarget. 8:76223–76230.
2017.PubMed/NCBI
|
|
20
|
Zhang ZG and Qin CY: Sirt6 suppresses
hepatocellular carcinoma cell growth via inhibiting the
extracellular signal-regulated kinase signaling pathway. Mol Med
Rep. 9:882–888. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar
|
|
22
|
Etchegaray JP, Zhong L and Mostoslavsky R:
The histone deacetylase SIRT6: At the crossroads between
epigenetics, metabolism and disease. Curr Top Med Chem.
13:2991–3000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lerrer B, Gertler AA and Cohen HY: The
complex role of SIRT6 in carcinogenesis. Carcinogenesis.
37:108–118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kalkavan H and Green DR: MOMP, cell
suicide as a BCL-2 family business. Cell Death Differ. 25:46–55.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Green DR and Llambi F: Cell death
signaling. Cold Spring Harb Perspect Biol. 7:2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khodapasand E, Jafarzadeh N, Farrokhi F,
Kamalidehghan B and Houshmand M: Is Bax/Bcl-2 ratio considered as a
prognostic marker with age and tumor location in colorectal cancer?
Iran Biomed J. 19:69–75. 2015.PubMed/NCBI
|
|
27
|
Pastor-Idoate S, Rodríguez-Hernández I,
Rojas J, Fernández I, Garcia-Gutierrez MT, Ruiz-Moreno JM,
Rocha-Sousa A, Ramkissoon YD, Harsum S, MacLaren RE, et al: BAX and
BCL-2 polymorphisms, as predictors of proliferative
vitreoretinopathy development in patients suffering retinal
detachment: The Retina 4 project. Acta Ophthalmol. 93:e541–e549.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hassan M, Watari H, AbuAlmaaty A, Ohba Y
and Sakuragi N: Apoptosis and molecular targeting therapy in
cancer. Biomed Res Int. 2014:1508452014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goldar S, Khaniani MS, Derakhshan SM and
Baradaran B: Molecular mechanisms of apoptosis and roles in cancer
development and treatment. Asian Pac J Cancer Prev. 16:2129–2144.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sui Y, Zheng X and Zhao D: Rab31 promoted
hepatocellular carcinoma (HCC) progression via inhibition of cell
apoptosis induced by PI3K/AKT/Bcl-2/BAX pathway. Tumour Biol.
36:8661–8670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Morigi M, Perico L and Benigni A: Sirtuins
in renal health and disease. J Am Soc Nephrol. 29:1799–1809. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xing X, Gu X, Ma T and Ye H: Biglycan
up-regulated vascular endothelial growth factor (VEGF) expression
and promoted angiogenesis in colon cancer. Tumour Biol.
36:1773–1780. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miyake M, Goodison S, Lawton A,
Gomes-Giacoia E and Rosser CJ: Angiogenin promotes tumoral growth
and angiogenesis by regulating matrix metallopeptidase-2 expression
via the ERK1/2 pathway. Oncogene. 34:890–901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun Y, Liu WZ, Liu T, Feng X, Yang N and
Zhou HF: Signaling pathway of MAPK/ERK in cell proliferation,
differentiation, migration, senescence and apoptosis. J Recept
Signal Transduct Res. 35:600–604. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang CC, Wu DW, Lin PL and Lee H:
Paxillin promotes colorectal tumor invasion and poor patient
outcomes via ERK-mediated stabilization of Bcl-2 protein by
phosphorylation at Serine 87. Oncotarget. 6:8698–8708.
2015.PubMed/NCBI
|
|
36
|
Kim JH, Lee JM, Kim JH and Kim KR:
Fluvastatin activates sirtuin 6 to regulate sterol regulatory
element-binding proteins and AMP-activated protein kinase in HepG2
cells. Biochem Biophys Res Commun. 503:1415–1421. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Parenti MD, Grozio A, Bauer I, Galeno L,
Damonte P, Millo E, Sociali G, Franceschi C, Ballestrero A,
Bruzzone S, et al: Discovery of novel and selective SIRT6
inhibitors. J Med Chem. 57:4796–4804. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sociali G, Galeno L, Parenti MD, Grozio A,
Bauer I, Passalacqua M, Boero S, Donadini A, Millo E, Bellotti M,
et al: Quinazolinedione SIRT6 inhibitors sensitize cancer cells to
chemotherapeutics. Eur J Med Chem. 102:530–539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Damonte P, Sociali G, Parenti MD, Soncini
D, Bauer I, Boero S, Grozio A, Holtey MV, Piacente F, Becherini P,
et al: SIRT6 inhibitors with salicylate-like structure show
immunosuppressive and chemosensitizing effects. Bioorg Med Chem.
25:5849–5858. 2017. View Article : Google Scholar : PubMed/NCBI
|