Introduction
Head and neck cancer, including oral squamous cell
carcinoma (OSCC), is the sixth leading cancer worldwide (1). More than 400,000 people succumb to
OSCC every year, and the 5-year survival rate for patients with
OSCC is relatively poor (2,3).
Recently, medical technology for OSCC treatment has been greatly
improved, and treatments of OSCC mainly include surgery, radiation
or a combination of methods (4).
However, more than half of OSCC patients are diagnosed in advanced
stages. Due to the high recurrence rate and distant metastases in
patients with advanced OSCC, it is worthwhile to improve current
medical treatment approaches, and the exploration of the molecular
mechanism in the development process of OSCC is greatly
required.
MicroRNAs (miRNAs/miRs) are a class of small
non-coding RNA molecules that function in RNA silencing and
post-transcriptional regulation of gene expression (5). Due to their important role in the
pathogenesis of cancer, miRNAs are regarded as biomarkers and
therapeutic targets in various types of cancer. Researchers
revealed that the aberrant expression of miR-18a-5p was associated
with the occurrence and development of multiple malignant cancers,
such as lung (6), esophageal
(7) and gastric cancer (8). However, the role and mechanism of
miR-18a-5p in OSCC are still unclear. It is now generally accepted
that the transforming growth factor-β (TGF-β) signaling pathway
also plays an important role in the development of OSCC (9). However, the underlying mechanism
concerning the involvement of TGF-β in the progression of OSCC is
not yet fully understood. Smad2 as a potent tumor-suppressive gene,
is also an important component of TGF-β signal transduction
(10).
The purpose of the present study was to investigate
the role of miR-18a-5p in OSCC cells in vitro, and to
explore the molecular mechanism. We hope to provide more
theoretical basis and treatment strategies for the diagnosis and
treatment of OSCC.
Materials and methods
Cell culture and cell
transfection
The OSCC cell line (SCC9) and the primary normal
human oral keratinocyte (HOK) cells were originally acquired from
the American Type Culture Collection (ATCC, Manassas, VA, USA).
SCC9 cells were grown in RPMI-1640 medium containing 10% fetal
bovine serum (FBS; both from Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and 1% penicillin-streptomycin solution
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and incubated in
an incubator at 37°C with 5% CO2. HOK cells were grown
in RPMI-1640 medium containing 15% FBS, and incubated at 37°C with
5% CO2 (11).
SCC9 cells (3×l04 cells/well) were
transiently transfected with the negative control (NC) (forward:
5′-UUCUCCGAACGUGUCACGUTT-3′; and reverse:
5′-ACGUGACACGUUCGGAGAATT-3′); the miR-18a-5p mimic (forward:
5′-UAAGGUGCAUCUAGUGCAGAUAG-3′; and reverse:
5′-AUCUGCACUAGAUGCACCUUAUU-3′) or the miR-18a-5p inhibitor
(5′-CUAUCUGACUAGAUGCACCUUA-3′) respectively using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Transfection efficiency was detected
48 h after the transfection experiment.
MTT assay
MTT assays were performed to assess cell viability.
Logarithmic phase cells were seeded in a 96-well plate with
1×104 cells/well and incubated at 37°C in a 5%
CO2 incubator for 12, 24 or 48 h respectively, after
which 20 µl of MTT solution (5 mg/ml in distilled water) was added
to each well, and cells were incubated for a further 2 h at 37°C
with 5% CO2. The absorbance was assessed at a wavelength
of 450 nm using a microplate reader.
Cell apoptosis assay
After specific treatment, SCC9 cells were collected
and washed with cold PBS at least three times. SCC9 cell apoptosis
was assessed by cell apoptosis assay. In brief, SCC9 cells
(1×106) from various groups (control, NC, mimic and
inhibitor) were firstly re-suspended in binding buffer, and then
labeled with Annexin V-FITC and propidium iodide (PI) (BD
Biosciences, San Diego, CA, USA) in line with the manufacturer's
instructions. Flow cytometry (BD FACS Aria; BD Biosciences,
Franklin Lakes, NJ, USA) was applied to analyze cell apoptosis. The
experiment was repeated at least three times.
Transwell assay
In vitro invasion assays were performed using
Transwell plates (BD Biosciences) with 8-µm pores. The SCC9 cells
(1×104 cells) in RPMI-1640 medium were added to the
upper chamber of the Transwell plates. Then RPMI-1640 medium
containing 20% FBS as a chemo-attractant was added to the lower
chamber. After 48 h of incubation, cells on the upper surface were
removed using cotton wool and the cells on the bottom surface of
the membrane were fixed with methanol and stained with 0.5% crystal
violet. Images were captured at an ×200 magnification and the cells
were counted using a light microscope (Olympus Corporation, Tokyo,
Japan).
Wound-healing assay
For the wound-healing assay, confluent monolayers of
SCC9 cells cultured in 24-well plates were mechanically wounded
using a 10-µl pipette tip. The wells were washed to remove cellular
debris and the cells were allowed to migrate for 48 h.
Representative images were captured at an ×200 magnification under
an inverted microscope (Olympus Corporation). The experiments were
repeated at least three times. This assay was performed 48 h after
transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract the total RNA from the
cells. GAPDH or U6 was used as an internal control for mRNA or
miRNA expression. cDNAs were generated by using the PrimeScript™ RT
reagent kit (Takara Bio, Inc.) in line with the manufacturer's
instructions. SYBR Premix Ex Taq (Takara Bio, Inc.) was carried out
to analyze the synthesized cDNAs according to the manufacturer's
instructions. Primer sequences used for qPCR were obtained as
required and listed as following: miR-18a-5p forward,
5′-ACGTAAGGTGCATCTAGTGCAGATA-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′; α-smooth muscle actin (α-SMA) forward,
5′-GTGTTGCCCCTGAAGAGCAT-3′ and reverse,
5′-GCTGGGACATTGAAAGTCTCA-3′; E-cadherin forward,
5′-CGAGAGCTACACGTTCACGG-3′ and reverse,
5′-GGGTGTCGAGGGAAAAATAGG-3′; vimentin forward,
5′-GACGCCATCAACACCGAGTT-3′ and reverse,
5′-CTTTGTCGTTGGTTAGCTGGT-3′; collagen I forward,
5′-GGCTTCCCTGGTCTTCCTGG-3′ and reverse, 5′-CCAGGGGGTCCAGCCAAT-3′;
TGF-β forward, 5′-GTCCCTGAAGTCAGCTGCATA-3′ and reverse,
5′-TGGGACAGTCCAGTTCTTCAT-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; GAPDH forward,
5′-GGAGTCCACTGGCGTCTTCA-3′ and reverse,
5′-GTCATGAGTCCTTCCACGATACC-3′. The 2ΔΔCq method
(12) was performed for the
calculation of the relative expression of the genes.
Western blot analysis
After specific treatment, total cellular proteins
from SCC9 cells were extracted using RIPA Buffer (Auragene,
Changsha, China). A BCA protein quantitative kit (Thermo Fisher
Scientific, Inc.) was used to assess the concentration of protein
samples. An equal amount of protein samples (30 µg/lane) were
resolved by 12% SDS-PAGE and then transferred onto PVDF membranes.
The membranes were blocked with 5% non-fat milk at room temperature
for 1 h, followed by incubation with primary antibodies: Smad2
(cat. no. 5339; 1:1,000; Cell Signaling Technology, Inc., Danvers,
MA, USA), Smad4 (cat. no. 46,535; 1:1,000; Cell Signaling
Technology, Inc.), Smad7 (cat. no. ab90086; dilution: 1: 1,000;
Abcam, Cambridge, MA, USA), collagen I (cat. no. ab34710; 1:1,000;
Abcam), TGF-β (cat. no. 3709; 1:1,000; Cell Signaling Technology,
Inc.), α-SMA (cat. no. 68,463; 1:1,000; Cell Signaling Technology,
Inc.), vimentin (cat. no. 12826; 1:1,000; Cell Signaling
Technology, Inc.), E-cadherin (cat. no. 3195; 1:1,000; Cell
Signaling Technology, Inc.), and β-actin (cat. no. 12620; 1:1,000;
Cell Signaling Technology, Inc.) at 4°C overnight. Subsequently,
the membranes were incubated with anti-rabbit immunoglobulin G
horseradish peroxidase (HRP)-conjugated secondary antibodies (cat
no. 7074; 1:2,000; Cell Signaling Technology, Inc.) at room
temperature for 2 h. Notably, secondary antibodies were not used
when the primary antibodies were HRP-conjugated. To visualize the
protein blots, an ECL kit (Applygen Technologies, Inc., Beijing,
China) was used according the manufacturer's protocol.
Densitometric semi-quantification was performed with ImageJ 1.38X
software (National Institutes of Health, Bethesda, MD, USA).
Dual luciferase reporter assay
To predict the targets of miR-18a-5p, TargetScan
bioinformatics software (www.targetscan.org/vert_71) was used, and results
revealed that Smad2 was a potential target of miR-18a-5p. To
confirm the direct binding sites, the wild-type (WT-Smad2) and
mutant (MUT-Smad2) 3′-untranslated region (3′-UTR) of Smad2 were
cloned into a pmiR-RB-Report™ dual luciferase reporter gene plasmid
vector (Guangzhou RiboBio Co., Ltd., Guangzhou, China). To
point-mutate the miR-18a-5p binding domain on the 3′UTR of Smad2, a
QuikChange Site-Directed Mutagenesis Kit (Stratagene; Agilent
Technologies, Inc., Santa Clara, CA, USA) was used as per the
manufacturer's instructions. SCC9 cells were co-transfected with
100 ng WT-Smad2 or 100 ng MUT-Smad2 and 50 nM miR-18a-5p (miR
mimic) or 50 nM negative control (NC) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocols.
Luciferase activity was assessed, 48 h later, using the
Dual-luciferase assay system (Promega Corporation, Madison, WI,
USA) according to the manufacturer's protocol. Luciferase activity
was normalized to Renilla luciferase activity.
Statistical analysis
Experiments were performed at least three times. All
data were displayed as the mean ± SD. SPSS 18.0 statistical
software (SPSS, Chicago, IL, USA) was performed for statistical
analyses. Comparisons between groups were performed by using
Student's t-test or one-way analysis of variance followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Upregulation of miR-18a-5p is revealed
in OSCC cells
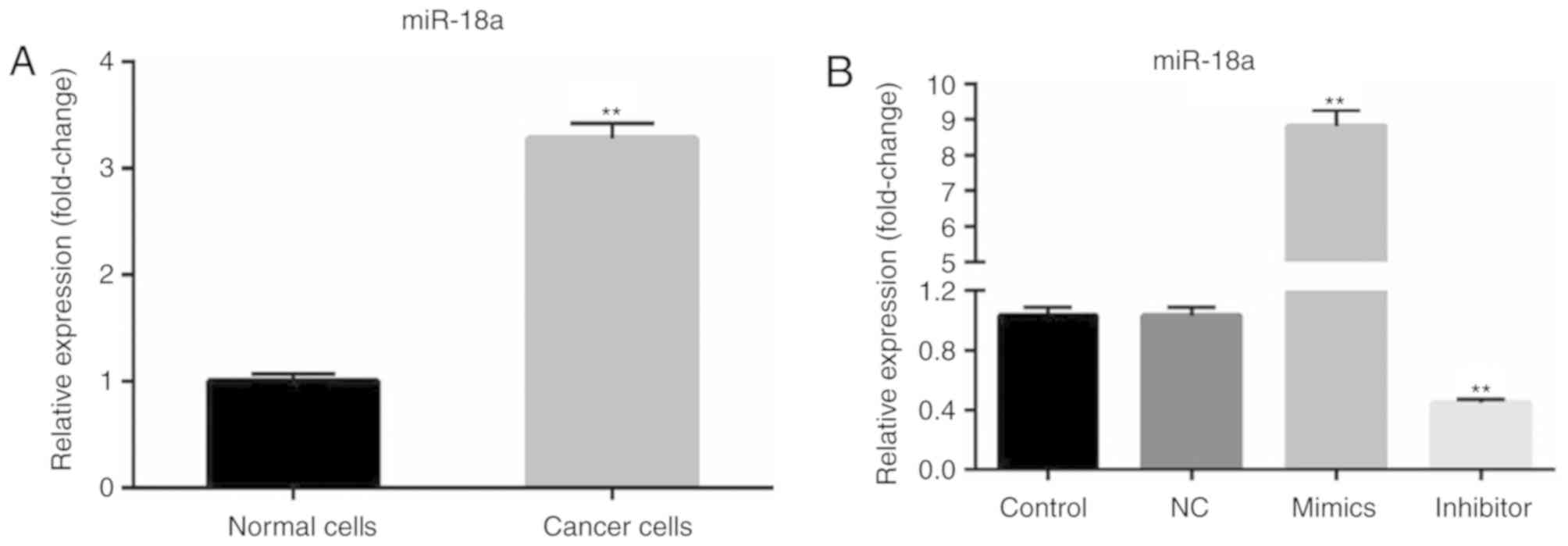
Using the RT-qPCR method, the expression level of
miR-18a-5p in OSCC cells (SCC9 cell line) was determined. RT-qPCR
analysis revealed that compared with primary normal human oral
keratinocyte (HOK) cells, miR-18a-5p was upregulated in SCC9 cells
(Fig. 1A). Then, SCC9 cells were
transfected with the NC, miR-18a-5p mimics or miR-18a-5p inhibitor.
It was revealed that miR-18a-5p was significantly upregulated in
the miR-18a-5p mimic-transfected SCC9 cells and downregulated in
miR-18a-5p inhibitor-transfected SCC9 cells (Fig. 1B).
Effect of miR-18a-5p on the cell
viability of SCC9 cells in vitro
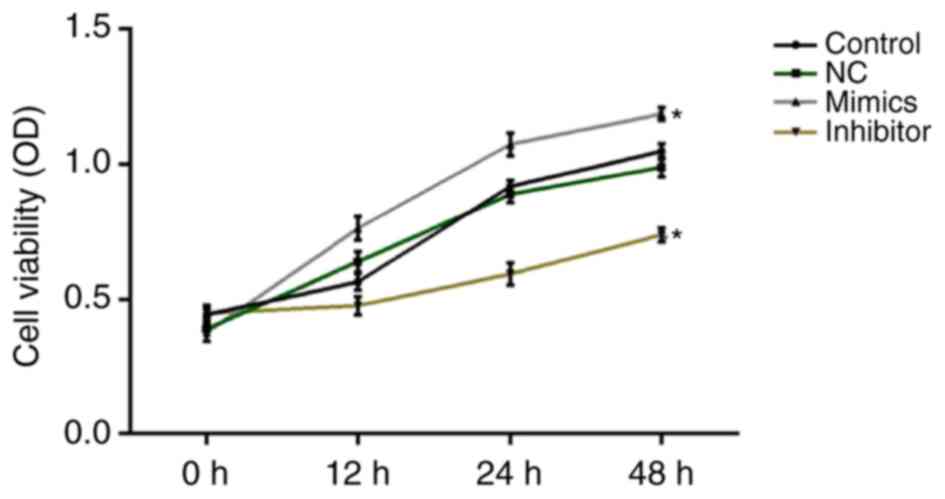
At 0, 12, 24 and 48 h after cell transfection, cell
viability was assessed using MTT assays. The results revealed that
miR-18a-5p mimics significantly increased SCC9 cell viability,
while miR-18a-5p inhibitor significantly decreased SCC9 cell
viability at all the time-points (Fig.
2).
Effect of miR-18a-5p on apoptosis of
SCC9 cells in vitro
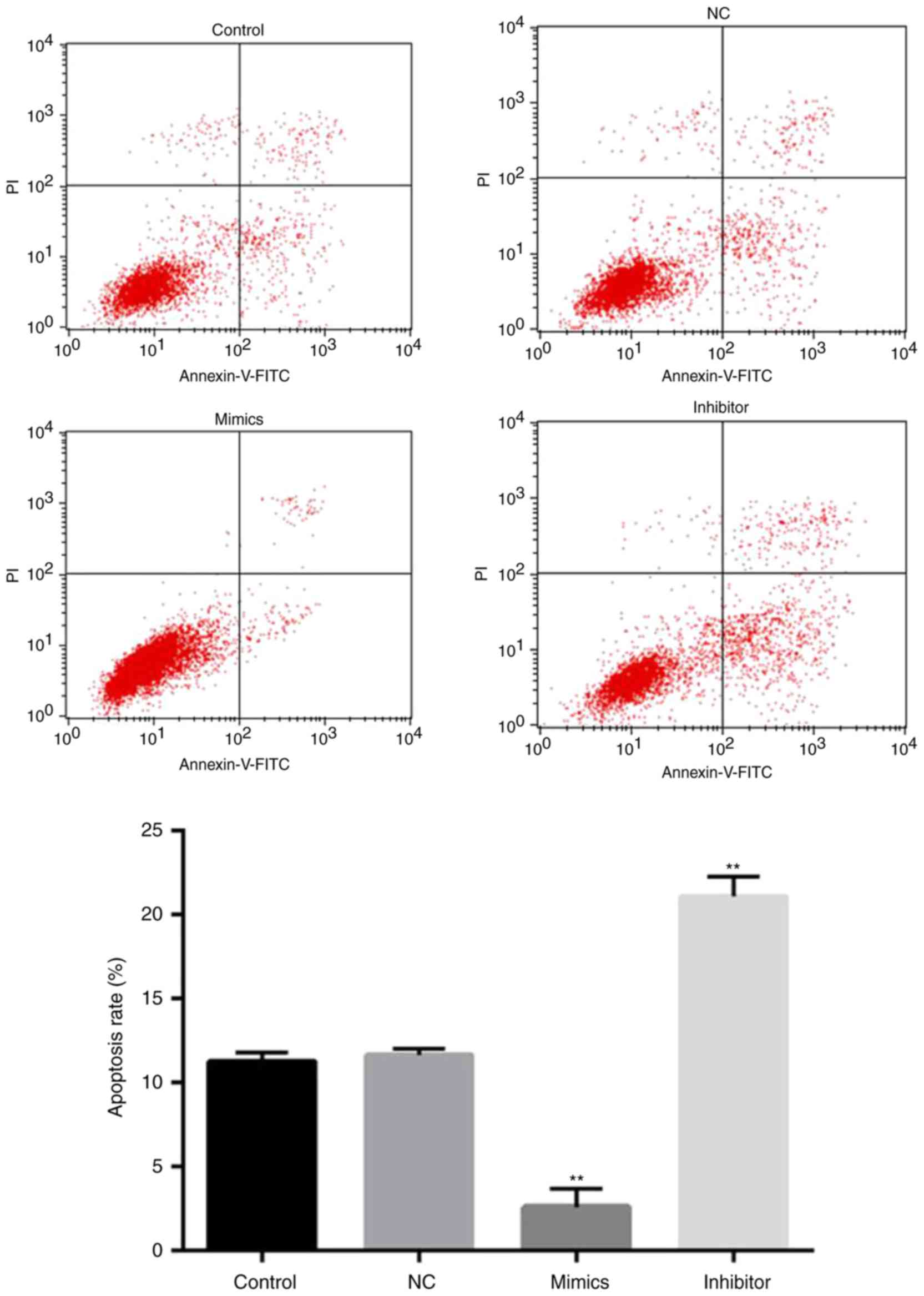
Next, whether miR-18a-5p has an effect on apoptosis
in SCC9 cells was determined by flow cytometry. The results
revealed that the apoptosis rate of SCC9 cells in the miR-18a-5p
inhibitor-transfection group was significantly upregulated compared
with the control group. However, the rate of apoptosis in the
miR-18a-5p mimic-transfection group was lower than that in the
control group. Collectively, the results indicated that miR-18a-5p
mimics inhibited the apoptosis of SCC9 cells in vitro, while
the miR-18a-5p inhibitor significantly induced SCC9 cell apoptosis
(Fig. 3).
Effect of miR-18a-5p on the invasion
and migration of SCC9 cells in vitro
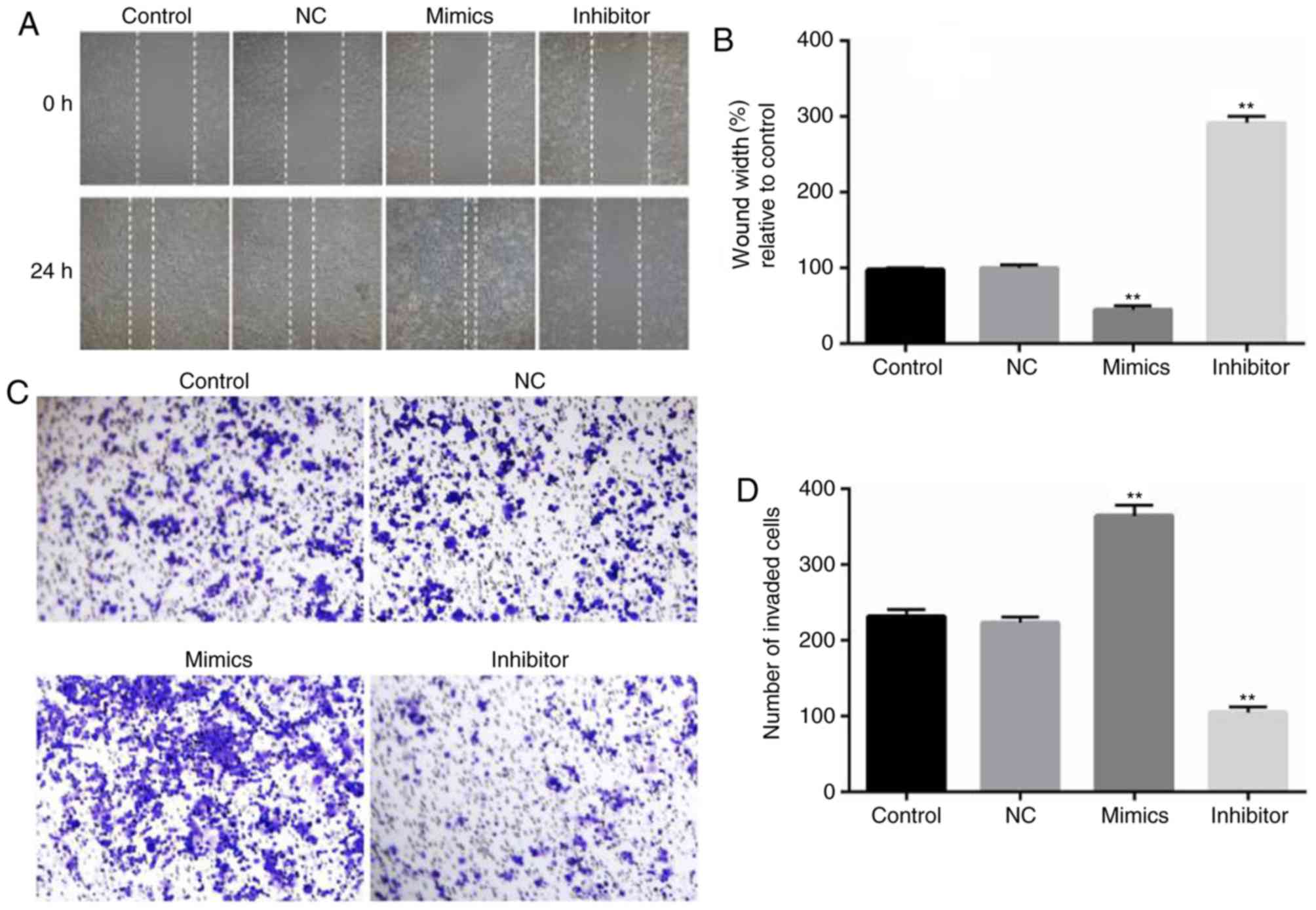
To assess the role of miR-18a-5p in the migration
and invasion of OSCC cells, wound healing and Transwell assays were
carried out. SCC9 cells were transfected with miR-18a-5p mimics or
the inhibitor for 48 h, and then wound healing and Transwell assays
were performed. The wound healing assay revealed that compared with
the control group, SCC9 cell migration ability significantly
decreased in the miR-18a-5p inhibitor-transfection group, while the
miR-18a-5p mimics significantly promoted the migration of SCC9
cells (Fig. 4A and B). As revealed
in Fig. 4C and D, the number of
invaded OSCC cells in the miR-18a-5p mimic-transfected group
significantly increased, while it was decreased in the miR-18a-5p
inhibitor-transfected group.
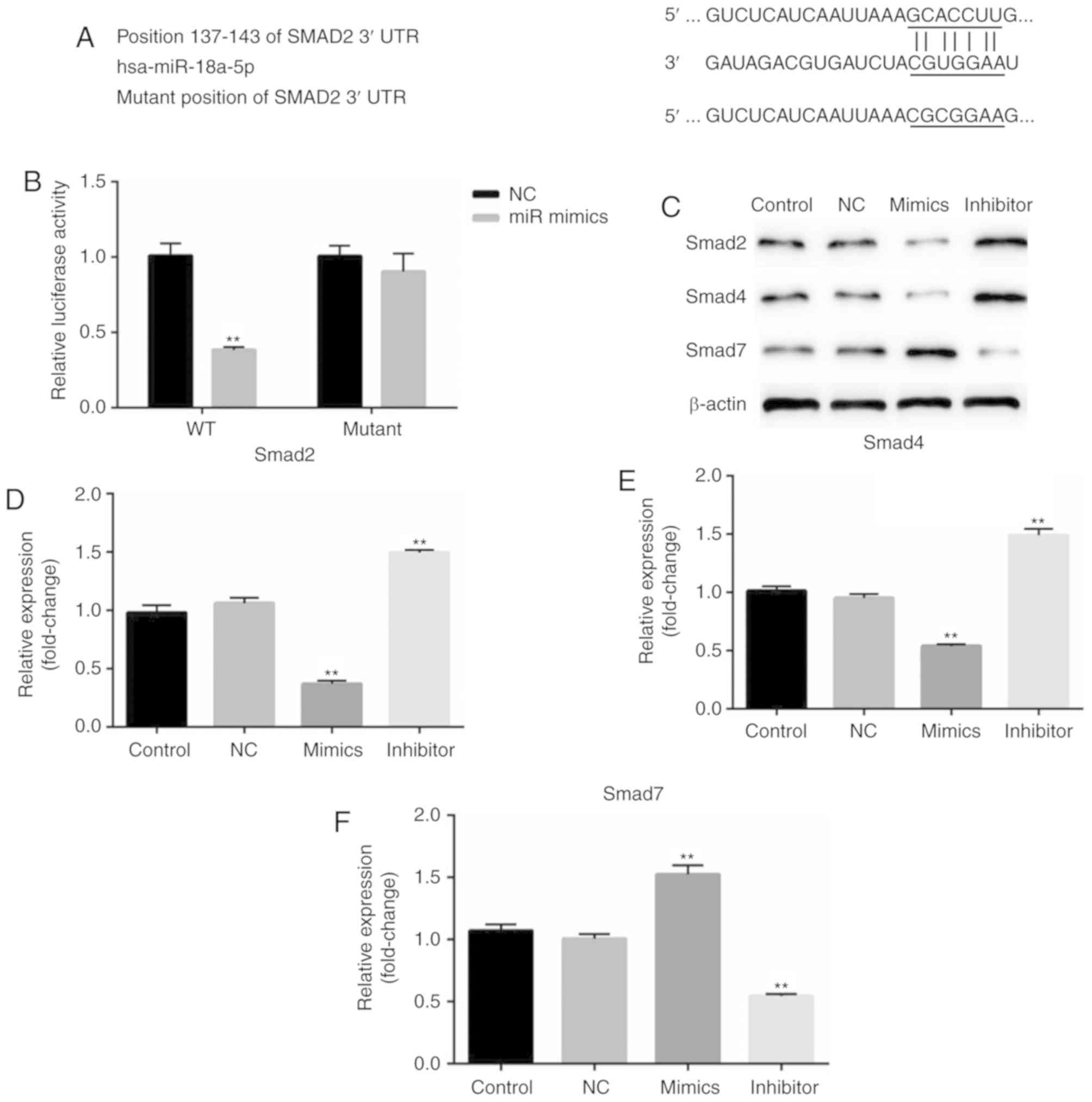
Smad2 is a target of miR-18a-5p
According to the bioinformatics software analysis,
it was observed that Smad2 was a potential target of miR-18a-5p
(Fig. 5A). Thus, it was
hypothesized that miR-18a-5p may play a role in OSCC cells by
regulating Smad2. Therefore, a luciferase reporter assay was
performed. As revealed in Fig. 5B,
it was revealed that compared with the cells co-transfected with
Smad2-WT and NC, the relative luciferase activity significantly
decreased in cells co-transfected with Smad2-WT and miR-18a-5p
mimics. While no significant differences of the relative luciferase
activity were revealed between the cells co-transfected with
Smad2-MUT and NC, and cells co-transfected with Smad2-MUT and
miR-18a-5p mimics.
In addition, the protein expression of Smad2, Smad4
and Smad7 was assessed in the present study. The results
demonstrated that Smad2 and Smad4 protein expression in the
miR-18a-5p mimic-transfected SCC9 cells was significantly
decreased, while Smad7 expression was increased. The miR-18a-5p
inhibitor had the opposite effects on the protein expression of
Smad2, Smad4 and Smad7 in SCC9 cells. These results indicated that
miR-18a-5p may play a role in the TGF-β pathway.
The role of miR-18a-5p in the TGF-β
pathway
Finally, the biological function of the involvement
of miR-18a-5p in the TGF-β pathway was observed. After 48 h of
transfection, the expression of collagen I, TGF-β, α-SMA, vimentin,
E-cadherin were assessed by western blotting method. Western blot
analysis confirmed that overexpression of miR-18a-5p resulted in
higher protein expression of collagen I, TGF-β, α-SMA, vimentin and
lower protein expression of E-cadherin. Additionally, the
expression of collagen I, TGF-β, α-SMA, and vimentin was reduced
and E-cadherin expression was enhanced in the miR-18a-5p
inhibitor-transfected SCC9 cells (Fig.
6).
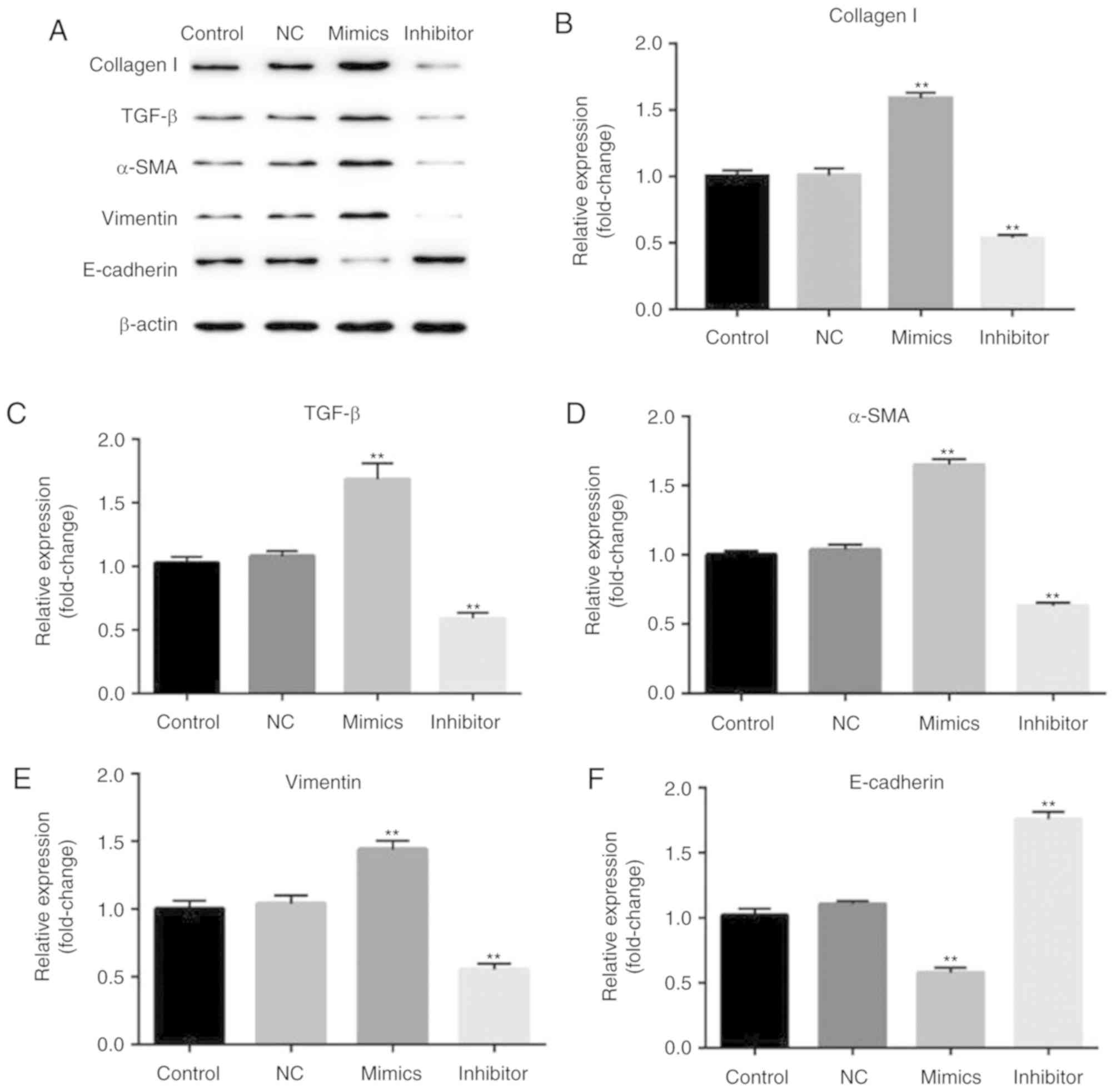 | Figure 6.Effect of miR-18a-5p on the expression
of collagen I, TGF-β, α-SMA, vimentin, and E-cadherin. A total of
48 h after transfection with miR-18a-5p mimics or the
miR-18a-5p-inhibitor group, (A) the protein level of collagen I,
TGF-β, α-SMA, vimentin, E-cadherin expression levels was assessed
using a western blot assay, and (B-F) the mRNA level of collagen I,
TGF-β, α-SMA, vimentin and E-cadherin was detected using reverse
transcription-quantitative polymerase chain reaction. **P<0.01
vs. the control. α-SMA, α-smooth muscle actin, miR-18a-5p,
microRNA-15a-5p; NC, negative control; TGF-β, transforming growth
factor-β. |
Discussion
Due to its poor prognosis and low survival rate,
OSCC has become a global health problem (13). Accumulating evidence has
demonstrated the important biological function of miRNAs in OSCC,
however the underlying mechanism of miRNAs involved in the process
of cancer is still poorly understood. The present study aimed to
identify whether miR-18a-5p has an effect on the malignant
biological behaviors of OSCC cells, and to explore the molecular
mechanism.
Previous studies have identified the higher
expression of miR-18a-5p in esophageal, colon, pancreatic and liver
cancer as well as renal cell carcinoma. In addition, miR-18a-5p has
been identified to play an important role in cancer cell
proliferation, apoptosis, migration and invasion (7). In the present study, it was revealed
that miR-18a-5p was highly expressed in SCC9 cells compared with
normal cells. Then the effect of miR-18a-5p upregulation and
miR-18a-5p downregulation was investigated on SCC9 cell viability,
migration, invasion and apoptosis. The findings demonstrated that
overexpression of miR-18a-5p could promote SCC9 cell viability,
migration, invasion and inhibit cell apoptosis. While miR-18a-5p
downregulation inhibited SCC9 cell viability, migration, invasion
and induced cell apoptosis. These results indicated that miR-18a-5p
acts as an oncogene to participate in OSCC progression and
tumorigenesis.
It was further revealed that Smad2 was the potential
target of miR-18a-5p, and in addition, it was determined that
miR-18a-5p negatively regulated the expression of Smad2 and Smad4,
and positively regulated the expression of Smad7. Studies have
confirmed that Smad2 and Smad4 are two major downstream regulators
in the TGF-β1/Smad signaling pathway and both of them act as tumor
suppressors in many human cancers, while Smad7 serves as a negative
feedback regulator of this pathway (14,15).
Numerous studies have demonstrated that the TGF-β1/Smad signaling
pathway is an important pathogenic mechanism in various cancers,
such as breast (16), prostate
(17), and gastric cancer
(18), as well as hepatocellular
carcinoma (19) and esophageal
squamous cell carcinoma (14).
Thus, we speculated that miR-18a-5p may promote OSCC cancer cell
biological progression though activation of the TGF-β1/Smad
signaling pathway and it may be an important therapeutic strategy
for OSCC. Furthermore, our results indicated that miR-18a-5p could
increase the expression of collagen I, α-SMA and vimentin, but
decrease E-cadherin expression. While miR-18a-5p inhibitor
presented the opposite effects. It is well recognized, that
collagen I as one of extracellular matrix components, plays an
important role cancer cell infiltration and metastasis (20). α-SMA has been revealed to be a
negative prognostic marker and associated with cancer metastases
(21). Vimentin is now being
perceived as a canonical prognostic marker and has been revealed to
have a high expression level in many cancers (22). Previous studies have demonstrated
that E-cadherin, a well-known tumor suppressor protein (23), is involved in the TGF-β1/Smad
signaling pathway (24). All these
findings were consistent with our present study. Thus, our findings
indicated that high expression of miR-18a-5p may indicate a poor
prognosis.
In conclusion, it was demonstrated that miR-18a-5p
upregulation promoted cell viability, migration and invasion of
OSCC cells and inhibited cell apoptosis in vitro by
activating the TGF-β1/Smad2 pathway. While miR-18a-5p
downregulation presented the opposite effects. These data indicated
that miR-18a-5p may be a promising therapeutic target for OSCC.
However, this study is only a preliminary study of the role of
miR-18a-5p in OSCC, and there are some limitations in our present
study. In order to elucidate the role of miR-18a-5p in OSCC,
numerous experiments are still required. For instance, the effect
of miR-18a-5p on apoptosis-related proteins, such as caspases,
should be determined. The level of miR-18a-5p in OSCC patients and
the relationship between the level of miR-18a-5p in OSCC patients
with the clinical characteristics of the patients should be
clarified. In addition, in vivo studies should be performed
to reveal the effect of miR-18a-5p on OSCC progression. These
issues will be addressed in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CH designed the present study, collected and
analyzed the data, performed statistical analysis, searched the
literature and prepared the manuscript. HS and LL performed
statistical analysis and interpreted the data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim JW, Park Y, Roh JL, Cho KJ, Choi SH,
Nam SY and Kim SY: Prognostic value of glucosylceramide synthase
and P-glycoprotein expression in oral cavity cancer. Int J Clin
Oncol. 21:883–889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Su CC, Lee KI, Chen MK, Kuo CY, Tang CH
and Liu SH: Cantharidin induced oral squamous cell carcinoma cell
apoptosis via the JNK-regulated mitochondria and endoplasmic
reticulum stress-related signaling pathways. PLoS One.
11:e01680952016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lai YH, Liu H, Chiang WF, Chen TW, Chu LJ,
Yu JS, Chen SJ, Chen HC and Tan BC: MiR-31-5p-ACOX1 axis enhances
tumorigenic fitness in oral squamous cell carcinoma via the
promigratory prostaglandin E2. Theranostics. 8:486–504. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alsaweed M, Hartmann PE, Geddes DT and
Kakulas F: MicroRNAs in breastmilk and the lactating breast:
Potential immunoprotectors and developmental regulators for the
infant and the mother. Int J Environ Res Public Health.
12:13981–14020. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen X, Wu L, Li D, Xu Y, Zhang L, Niu K,
Kong R, Gu J, Xu Z, Chen Z and Sun J: Radiosensitizing effects of
miR-18a-5p on lung cancer stem-like cells via downregulating both
ATM and HIF1α. Cancer Med. 7:3834–3847. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou L, Li Z, Pan X, Lai Y, Quan J, Zhao
L, Xu J, Xu W, Guan X, Li H, et al: Identification of miR-18a-5p as
an oncogene and prognostic biomarker in RCC. Am J Transl Res.
10:1874–1886. 2018.PubMed/NCBI
|
|
8
|
Li X, Luo F, Li Q, Xu M, Feng D, Zhang G
and Wu W: Identification of new aberrantly expressed miRNAs in
intestinal-type gastric cancer and its clinical significance. Oncol
Rep. 26:1431–1439. 2011.PubMed/NCBI
|
|
9
|
Hu L, Liu J, Li Z, Wang C and Nawshad A:
Transforming growth factor-β1 activates ΔNp63/c-Myc to promote oral
squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral
Radiol. 122:460–482.e4. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Heldin CH and Moustakas A: Signaling
receptors for TGF-β family members. Cold Spring Harb Perspect Biol.
8(pii): a0220532016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou XL, Chen G, Li MX, Wang HX, Hong JW,
Shen JY, Wang Q, Ge X, Ding Z and Xu LC: Targeting YOD1 by RNA
interference inhibits proliferation and migration of Human Oral
Keratinocytes through transforming growth Factor-β3 signaling
pathway. Biomed Res Int. 2018:62543082018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu L, Jiang H, Zhao J and Wen H: MiRNA-16
inhibited oral squamous carcinoma tumor growth in vitro and in vivo
via suppressing Wnt/β-catenin signaling pathway. Onco Targets Ther.
11:5111–5119. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Abudureheman A, Ainiwaer J, Hou Z, Niyaz
M, Turghun A, Hasim A, Zhang H, Lu X and Sheyhidin I: High MLL2
expression predicts poor prognosis and promotes tumor progression
by inducing EMT in esophageal squamous cell carcinoma. J Cancer Res
Clin Oncol. 144:1025–1035. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ullah I, Liao Y, Wan R, Tang L and Feng J:
Alternative splicing of SMAD4 and its function in HaCaT cells in
response to UVB irradiation. J Cancer. 9:3177–3186. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang R, Liang J, Xu GX, Ding LM, Huang HM,
Su QZ, Yan J and Li YC: Human cytomegalovirus glycoprotein B
inhibits migration of breast cancer MDA-MB-231 cells and impairs
TGF-β/Smad2/3 expression. Oncol Lett. 15:7730–7738. 2018.PubMed/NCBI
|
|
17
|
Kardooni H, Gonzalez-Gualda E, Stylianakis
E, Saffaran S, Waxman J and Kypta RM: CRISPR-mediated reactivation
of DKK3 expression attenuates TGF-β signaling in prostate cancer.
Cancers. 10(pii): E1652018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peng J, Chen W, Chen J, Yuan Y, Zhang J
and He Y: Overexpression of chloride channel-3 predicts unfavorable
prognosis and promotes cellular invasion in gastric cancer. Cancer
Manag Res. 10:1163–1175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang T, Liu W, Meng W, Zhao H, Yang Q, Gu
SJ, Xiao CC, Jia CC and Fu BS: Downregulation of miR-542-3p
promotes cancer metastasis through activating TGF-β/Smad signaling
in hepatocellular carcinoma. Onco Targets Ther. 11:1929–1939. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Su CQ, Qiu H and Zhang Y: Localization of
keratin mRNA and collagen I mRNA in gastric cancer by in situ
hybridization and hybridization electron microscopy. World J
Gastroenterol. 5:527–530. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sinn M, Denkert C, Striefler JK, Pelzer U,
Stieler JM, Bahra M, Lohneis P, Dörken B, Oettle H, Riess H and
Sinn BV: α-Smooth muscle actin expression and desmoplastic stromal
reaction in pancreatic cancer: Results from the CONKO-001 study. Br
J Cancer. 111:1917–1923. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dmello C, Sawant S, Alam H, Gangadaran P,
Mogre S, Tiwari R, D'Souza Z, Narkar M, Thorat R, Patil K, et al:
Vimentin regulates differentiation switch via modulation of keratin
14 levels and their expression together correlates with poor
prognosis in oral cancer patients. PLoS One. 12:e01725592017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Petrova YI, Schecterson L and Gumbiner BM:
Roles for E-cadherin cell surface regulation in cancer. Mol Biol
Cell. 27:3233–3244. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liang X, Zeng J, Wang L, Shen L, Ma X, Li
S, Wu Y, Ma L, Ci X, Guo Q, et al: Histone demethylase RBP2
promotes malignant progression of gastric cancer through
TGF-β1-(p-Smad3)-RBP2-E-cadherin-Smad3 feedback circuit.
Oncotarget. 6:17661–17674. 2015. View Article : Google Scholar : PubMed/NCBI
|