Introduction
Vitiligo is a common acquired disease characterized
by white spots on the skin, which affects 0.1–2% of the population
worldwide (1). Accumulating
evidence suggests that vitiligo is caused by the loss and
degradation of epidermal melanocytes (2). Several hypotheses have been proposed
for the development of this disease, including autoimmunity
(3), cytotoxic metabolites, neural
and genetic causes (4), and
induction of oxidative stress (5,6).
These factors have been suggested to explain the mechanisms
underlying the melanocyte degradation, although the exact
pathogenesis remains unknown.
Recent studies have demonstrated that vitiligo is an
autoimmune response targeting melanocytes (7,8).
Cytotoxic CD8+ T cells can specifically recognize
melanocytes, which can in turn be isolated from the lesions of
vitiligo subjects. In addition, the count of CD8+ T
cells in the peripheral blood of patients with vitiligo is
significantly increased, particularly in the advanced stages of
vitiligo (9,10). Therefore, CD8+ T cells
may serve a critical role during the processes of melanocyte loss
and degradation.
CD4+CD25+ regulatory T (Treg)
cells comprise a suppressive T cell subset that reduces the
inflammatory activity of immune cells by direct contact, enabling
the secretion of anti-inflammatory cytokines, such as
interleukin-10 (IL-10) and transforming growth factor-β (TGF-β)
(11,12). Treg cells inhibit the activity of
autoimmune T cells, namely CD4+ and CD8+ T
cells (13). Dwivedi et al
(14) have demonstrated that Treg
cells were significantly decreased in active generalized vitiligo.
In addition, Ben Ahmed et al (15) confirmed that the functional defect
of Treg cells was involved in the pathogenesis of vitiligo.
Therefore, the decrease in the number of natural Treg cells may
cause the activation of CD8+ T cells, which can in turn
damage the structure of melanocytes and lead to immune function
disorders.
MicroRNAs (miRNAs) are small conserved non-coding
RNA molecules, which have been found to serve key roles in normal
cellular processes (16). Previous
studies have proposed that miR-155 is a crucial regulator in the
process of inflammation and immunity (17,18).
In addition, miR-155 can increase the differentiation of Treg cells
by activating the transcription of forkhead box P3 (Foxp3), a
marker of Treg cells (19,20). A recent study has demonstrated that
miR-155 was dysregulated in patients with vitiligo, and that the
expression levels of the melanogenesis-associated genes in
melanocytes and keratinocytes were inhibited by this miRNA
(21). Furthermore, Yao et
al (22) demonstrated that
miR-155 regulated the differentiation of Treg cells by activating
the JAK/STAT pathway. The present study further demonstrated that
miR-155 upregulated the levels of Foxp3, a marker of Treg cell
activity. However, this result was different from the findings of
other studies. For instance, Karagiannidis et al (23) indicated that the upregulation of
Foxp3 levels by glucocorticoids increased IL-10 expression. In
addition, Ganesh et al (24) reported that IL-1β can increase the
levels of Foxp3 and TGF-β. Despite these promising studies, the
mechanisms by which miR-155 regulates the development of vitiligo
remain unclear. Thus, the present study aimed to investigate the
role of miR-155 in the development of vitiligo.
Materials and methods
Patient samples
All samples were obtained from the Wenzhou Medical
University, between April 2017 and May 2018. Peripheral blood and
skin tissues were obtained from one patient with non-segmental
vitiligo (male, 49-year-old). The disease status of the patient was
stable. In addition, the normal T cells were obtained from a
healthy donor (male, 53-years-old). The exclusion criteria were:
patients with severe liver, kidney disease, or cardiovascular
diseases; participants subjected with other associated dermatoses
during the last 6 months, such as psoriasis. The research was
approved by the Ethics Committee of Wenzhou Medical University
(Wenzhou, China; approval no. YS2019050). The patient and the
healthy donor provided informed consent for their participation in
the study.
Purification of naive T and
CD8+ T cells
Peripheral blood mononuclear cells were obtained
from the patient with vitiligo and healthy donor by Ficoll-Hypaque
density gradient centrifugation. For purification of naïve T cells
and CD8+ T cells, single cell suspensions of peripheral
blood mononuclear cells were enriched by immunomagnetic bead
selection using MACS Miltenyi system (Miltenyi Biotech, Inc.) as
previously described (25). In
addition, flow cytometry was used for sorting naïve T cells
(CD3+CD4+CD45RA+ T cells) and
CD3+CD8+ T cells. The purity of
CD3+CD4+CD45RA+ T and
CD3+CD8+ T cells was also evaluated using
flow cytometry. Naïve T cells were enriched by depletion of
magnetically labeled contaminating CD3+,
CD4+, and CD45RA+ cells. CD8+ T
cells were enriched by depletion of magnetically labeled
contaminating CD3+, CD8+ cells. The highly
enriched (90%) naïve T cells or CD8+ T cells were
subsequently stained with anti-CD3 (cat. no. 64-0037-41, 1:100
dilution), anti-CD4 (cat. no. 15-0049-42, 1:100 dilution), anti-CD8
(cat. no. MHCD0800-4, 1:100 dilution) and anti-CD45RA (cat. no.
11-0458-41, 1:100 dilution) antibodies were provided by Thermo
Fisher Scientific, Inc. Cells at a concentration of
4×107 cells/ml in staining buffer were incubated with
indicated antibodies for 30 min on ice, followed by three washes
with staining buffer.
Differentiation of naïve T cells to
Treg cells
The isolated naive T cells (4×105
cells/per well) were seeded into 6-well plates coated with anti-CD3
and anti-CD28 and cultured in RPMI 1640 medium (Thermo Fisher
Scientific) overnight. The cells were treated with all these
reagents, including IL-2 (100 U/ml, R&D Systems, Inc.), TGF-β
(10 ng/ml, R&D), and retinoic acid (10 nM, R&D Systems,
Inc.), and incubated in RPMI 1640 medium (Thermo Fisher Scientific,
Inc.) at 4°C for 30 min. After 4 days of stimulation, flow
cytometry was used to determine the purity of
CD4+CD25+FoxP3+ Treg cells
(26).
Nucleofection
A human T Cell Nucleofector® kit (Lonza
Inc.) and a nucleofector device (Lonza) were used for
nucleofection. Initially, 1×107 Treg cells were
resuspended in 100 µl Nucleofector® solution.
Subsequently, 100 pM oligonucleotides (Thermo Fisher Scientific,
Inc.) were added to the solution and mixed gently. The
oligonucleotides included an miR-155 agonist (pre-miR-155) and its
control (pre-miR-ctrl), as well as an miR-155 antagonist
(anti-miR-155) and the corresponding control (anti-miR-ctrl). The
miRNA sequences were as follows: pre-miR-155,
5′-CCCCUAUCACGAUUAGCAUUAAUU-3′; pre-miR-ctrl,
5′-AACCCCUAUCACGAUUAGCAUUAA-3′; anti-miR-155,
5′-UUAAUGCUAAUCGUGAUAGGGGUU-3′; and anti-miR-ctrl,
5′-AACCCCUAUCACGAUUAGCAUUAA-3′.
Thus, the oligonucleotides mixtures were carefully
transferred to the electroporation cuvettes and placed in the
nucleofector device, and the Treg cells were nucleofected in the
X-01 program. Finally, the cells (5×105 cells/well) were
transferred to a 12-well plate, prepared with 1.5 ml human T cell
nucleofector medium and incubated at 37°C in a 5% CO2
incubator until analysis (22).
Cell culture
Primary melanocytes were isolated from the vitiligo
patient by suction blistering and cultured in Hu 16 medium
[consisted of Ham's F12 nutrient mixture (Thermo Fisher Scientific,
Inc.) supplemented with 50 µg/ml gentamicin, 20 ng/ml fibroblast
growth factor (Sigma Aldrich; Merck KGaA), 20 µg/ml
isobutylmethylxanthine (Sigma Aldrich; Merck KGaA), 10 ng/ml
cholera toxin (Sigma Aldrich; Merck KGaA)] at 37°C in a 5%
CO2 incubator. The cell density in the culture flasks
was 5×105/ml, and the base factor (100 µg/ml of
geneticin) was added to the medium to remove keratinocytes and
fibroblasts on the third day. Subsequently, the cells were seeded
in 6-well plates at a density of 1×105 cells/ml, placed
in an incubator for 4 h and inoculated with CD8+ T
cells, Treg cells, pre-miR-155, or CD8+ T cells, Treg
cells, anti-miR-155 respectively, and incubated for 72 h at
37°C.
Flow cytometry analysis
The activation of CD8+ T cells, and the
ratio of Treg, CD4−CD8+ was evaluated with
FACSCalibur flow cytometry (BD Biosciences, Franklin Lake, NJ,
USA). The induction of cell apoptosis was detected using the
Annexin V-FITC/propidium iodide (PI) apoptosis detection kit (BD
Biosciences, Franklin Lake, NJ, USA) following the manufacturer's
protocol. Briefly, cells were harvested and washed with PBS twice.
Next, the cells were resuspended and stained with 2 µl Annexin V
and 2 µl PI for 15 min at 25°C in the dark. The number of apoptotic
cells was quantified by flow cytometry.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using the TRIzol reagent
(Thermo Fisher Scientific, Inc.) following the manufacturer's
procedure. cDNA synthesis was synthesized by using a SuperScript IV
Reverse Transcriptase kit (Thermo Fisher Scientific, Inc.). For
miR-155 analysis, cDNA was synthesized using the
PrimeScript® RT reagent kit (Takara Bio, Inc., Otsu,
Japan), miR-155 RT primers (Thermo Fisher Scientific, Inc.) and 1
µg of total RNA. qPCR was then performed using the SYBR Premix Ex
Taq II kit (Takara Bio, Inc.). The PCR conditions were as follows:
95°C for 5 min; then 45 cycles consisting of 94°C for 30 sec and 59
°C for 45 sec. The primer sequences used in qPCR are listed in
Table I. The relative levels of
the genes were normalized to those of the human β-actin gene and
evaluated by the comparative quantification cycle
(2−∆∆Cq) method (27).
 | Table I.Primers used in polymerase chain
reaction. |
Table I.
Primers used in polymerase chain
reaction.
| Gene | Forward primer | Reverse primer |
|---|
| β-actin |
5′-TGACGTGGACATCCGCAAAG-3′ |
5′-CTGGAAGGTGGACAGCGAGG-3′ |
| Foxp3 |
5′-GATCACCTCTTGGATGAGAAGG-3′ |
5′-TGTGGAAGAACTCTGGAAAGGT-3′ |
| IL-10 |
5′-GCCAGAGCCACATGCTCCTA-3′ |
5′-GATAAGGCTTGGCAACCCAAGTAA-3′ |
| TGF-β1 |
5′-GTGTGGAGCAACATGTGGAACTCTA-3′ |
5′-CGCTGAATCGAAAGCCCTGTA-3′ |
| U6 |
5′-CTCGCTTCGGCAGCACA-3′ |
|
| miR-155 |
5′-GCGCCGTTAATGCTAATCGTGAT-3′ |
|
ELISA analysis
The levels of IL-10 and TGF-β1 in the cell culture
supernatant were detected using the corresponding ELISA kits
(Neobioscience) according to the manufacturer's procedures.
Western blot analysis
Cells were lysed in RIPA buffer (Thermo Fisher
Scientific, Inc.), and then the supernatants of cell lysates were
collected. After that, BCA™ Protein Assay kit (Pierce; Thermo
Fisher Scientific, Inc.) was used to detect the concentration of
proteins in the supernatants. Total protein was separated on 12%
sodium dodecyl sulfate gels using polyacrylamide gel
electrophoresis and then transferred to polyvinylidene fluoride
membranes (EMD Millipore). The blotted membranes were blocked in 5%
BSA (Sigma Aldrich; Merck KGaA, Darmstadt, Germany) and incubated
with primary antibodies overnight at 4°C. On the following morning,
the membranes were incubated with horseradish peroxidase-conjugated
secondary antibody for 1 h at room temperature. The membranes were
washed again, and the proteins were detected using a
chemiluminescence detection kit (Thermo Fisher Scientific, Inc.),
and Image Lab™ Software (Bio-Rad Laboratories, Inc.) was used to
quantify the intensity of the bands. The primary antibodies against
Foxp3 (cat. no. ab215206, 1:1,000 dilution) and GAPDH (cat. no.
ab181602, 1:1,000 dilution), and the secondary antibody (cat. no.
ab150077, 1:5,000 dilution) used in this experiment were provided
by Abcam. GAPDH was used as a loading control.
Statistical analysis
The data are expressed as the mean ± standard
deviation. All statistical analyses were performed with GraphPad
Prism software (version 6.01; GraphPad Software, Inc.). Student's
t-test (two-sided) was applied for comparison of continuous
variables between two groups, while statistical differences among
multiple groups were analyzed by one-way analysis of variance
followed by Tukey's test. For all tests, a P<0.05 was considered
to indicate a statistically significant difference, and a P<0.01
was considered to indicate a highly significant difference.
Results
Purification of naive T and
CD8+ T cells
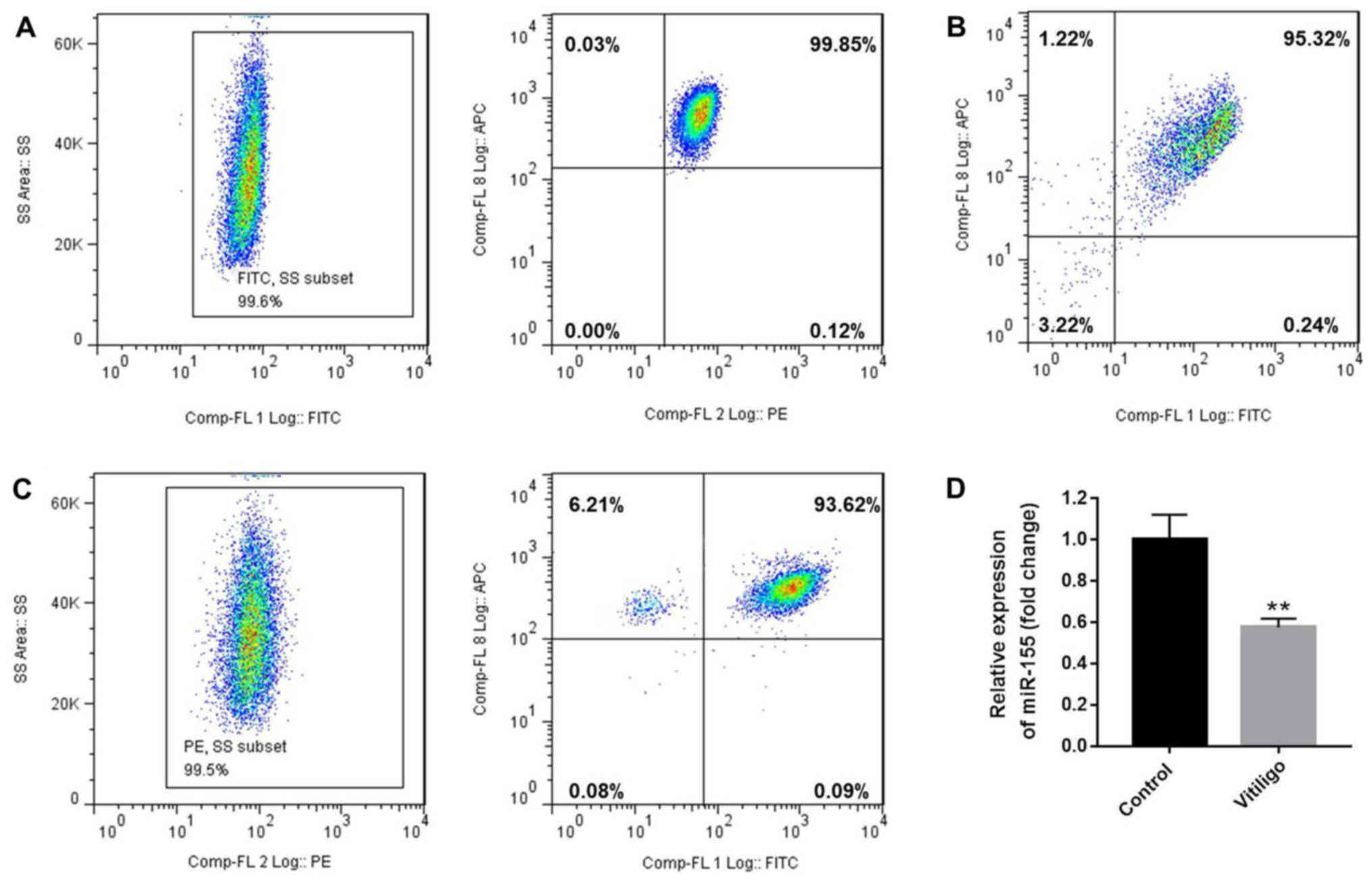
Initially, flow cytometry was used to assess the
number of naïve T cells
(CD3+CD4+CD45RA+ T cells) and
CD3+CD8+ T cells. The purity of
CD3+CD4+CD45RA+ (99.45%, Fig. 1A) and
CD3+CD8+T (Fig.
1B) cells was also evaluated by flow cytometry, and was found
to be >95%. Next, the isolated naive T cells were differentiated
to Treg cells following treatment with IL-2, TGF-β and retinoic
acid (10 nM). After 4 days of stimulation, the purity of
CD4+CD25+FoxP3+ Treg cells was
detected to be 93.15% (Fig. 1C).
In addition, it was observed that the level of miR-155 in T cells
of the patient with vitiligo was downregulated compared with that
in the healthy donor (Fig.
1D).
miR-155 increases the percentage of
Treg cells, and the secretion of IL-10 and TGF-β1 in the cell
culture medium
The study subsequently examined the effects of
miR-155 on the differentiation of Treg cells using flow cytometry.
As indicated in Fig. 2A, treatment
with anti-miR-155 significantly downregulated the level of miR-155
in Treg cells, while pre-miR-155 exhibited the opposite effect, as
compared with the corresponding control groups. In addition, the
results revealed that anti-miR-155 caused a significant decrease in
the percentage of Treg cells in the cell culture medium, while
pre-miR-155 markedly increased this percentage (Fig. 2B and C). Furthermore, it was
observed that anti-miR-155 significantly inhibited the gene and
protein levels of Foxp3, while pre-miR-155 exhibited the opposite
effects (Fig. 2D and E).
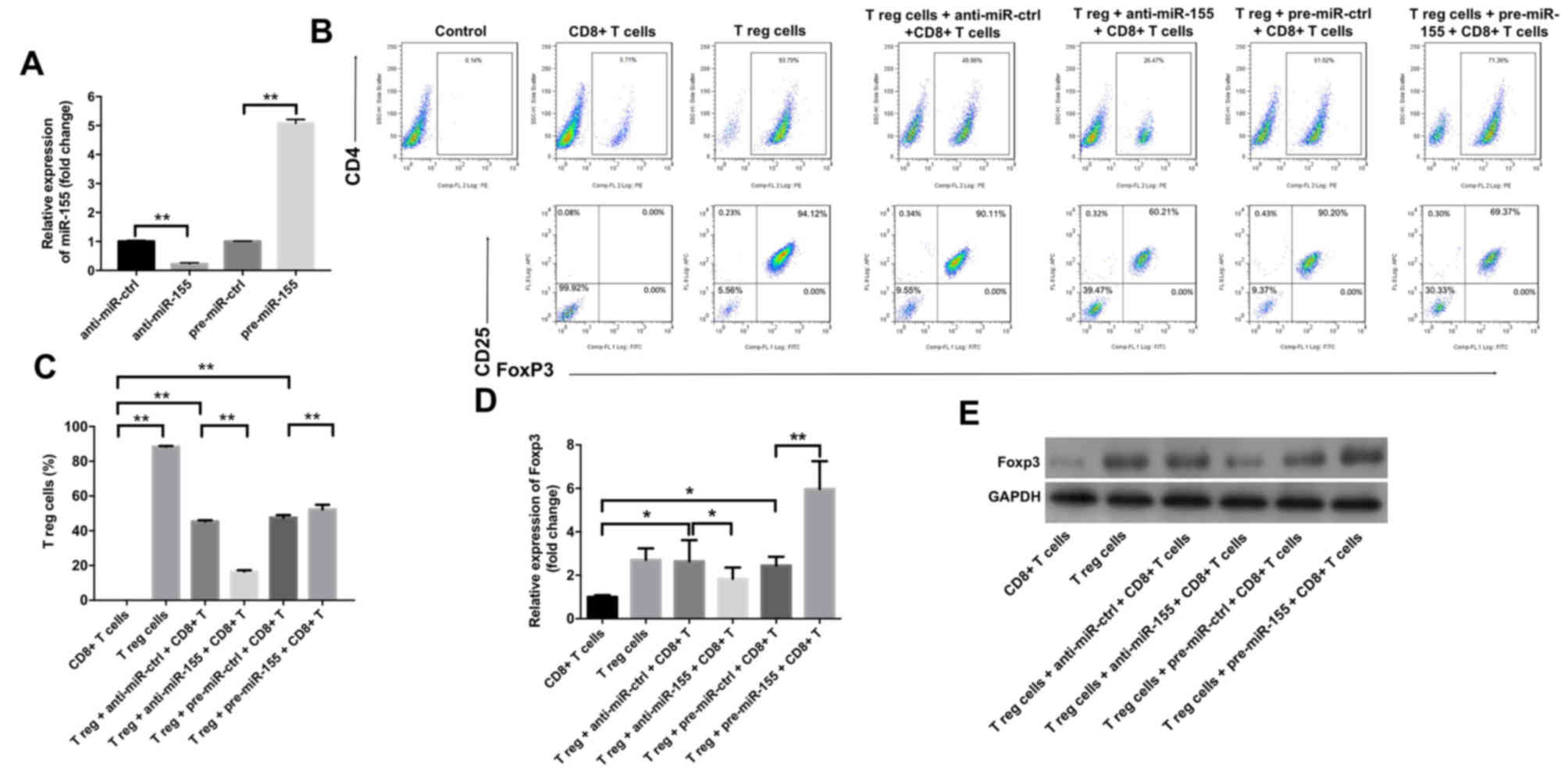 | Figure 2.miR-155 upregulated the percentage of
Treg cells in primary melanocytes. Anti-miR-ctrl, anti-miR-155,
pre-miR-ctrl and pre-miR-155 were transfected into Treg cells. (A)
The level of miR-155 in Treg cells was detected by RT-qPCR. (B)
Flow cytometry graphs and (C) percentage of Treg cells, detected
after 3 days of stimulation. Representative fluorescence-activated
cell sorting images from a single case are shown. (D) mRNA and (E)
protein expression levels of Foxp3 in T cells, detected by RT-qPCR
and western blot analysis, respectively, at 3 days after
transfection. Collective results from three independent experiments
are shown. *P<0.05 and **P<0.01. miR-155, microRNA-155; Treg,
regulatory T; FoxP3, forkhead box P3; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; ctrl,
control. |
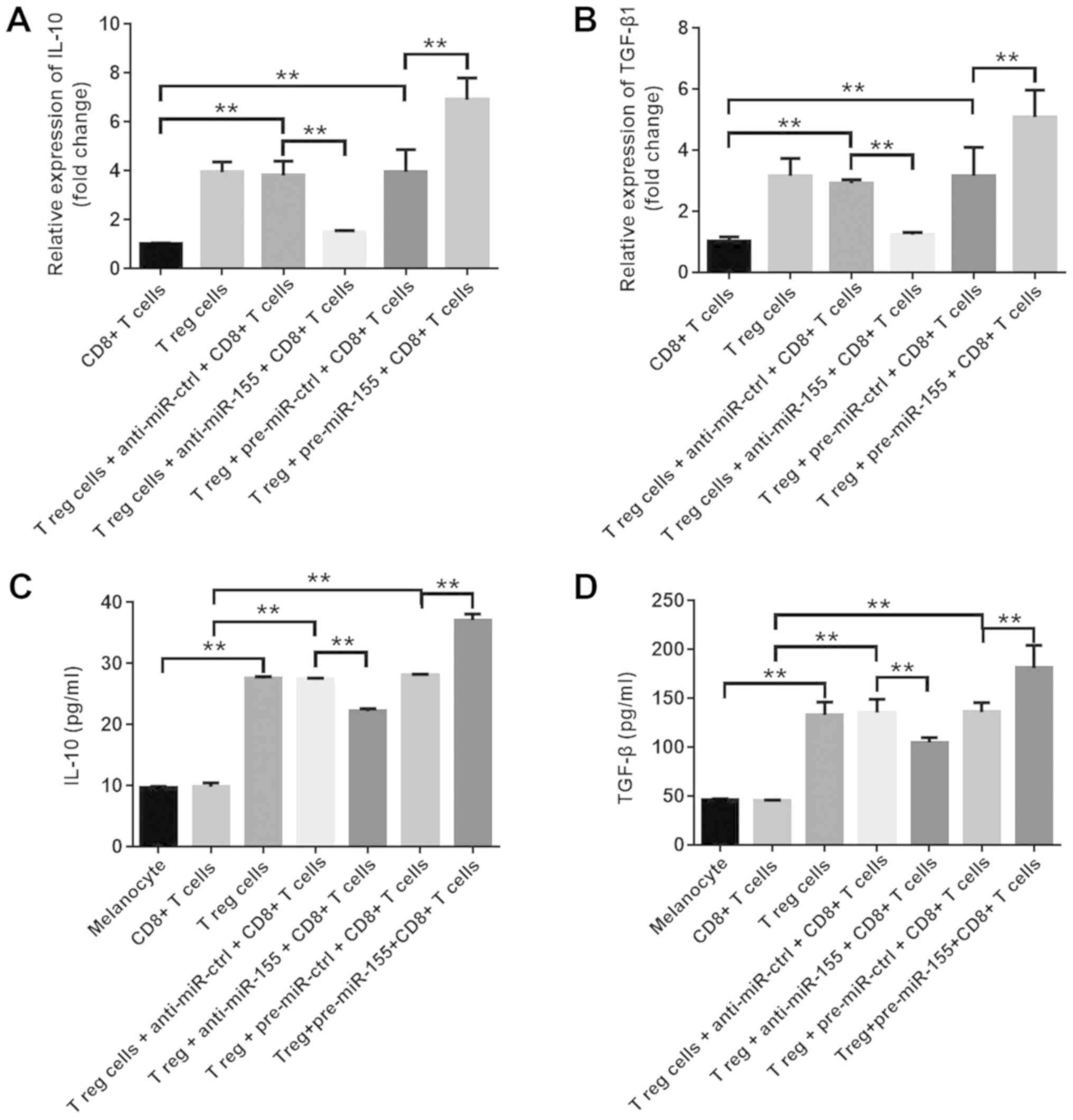
In order to investigate the effects of miR-155 on
the function of Treg cells, the mRNA levels of IL-10 and TGF-β1
were assessed in T cells, and the extracellular secretions of these
cytokines in the culture medium were also examined. The results
indicated that pre-miR-155 significantly increased IL-10 and TGF-β1
mRNA expression levels, while anti-miR-155 markedly downregulated
the levels of these cytokines (Fig. 3A
and B). Furthermore, the extracellular secretions of IL-10 and
TGF-β1 were significantly increased in the pre-miR-155 group, which
was consistent with the previous findings, while they were
downregulated in the anti-miR-155 group (Fig. 3C and D). These data suggested that
miR-155 increased the percentage of Treg cells, and promoted the
secretion of IL-10 and TGF-β1 in the cell culture medium.
miR-155 decreases the percentage of
CD8+ T cells by inducing apoptosis in the cell culture
medium
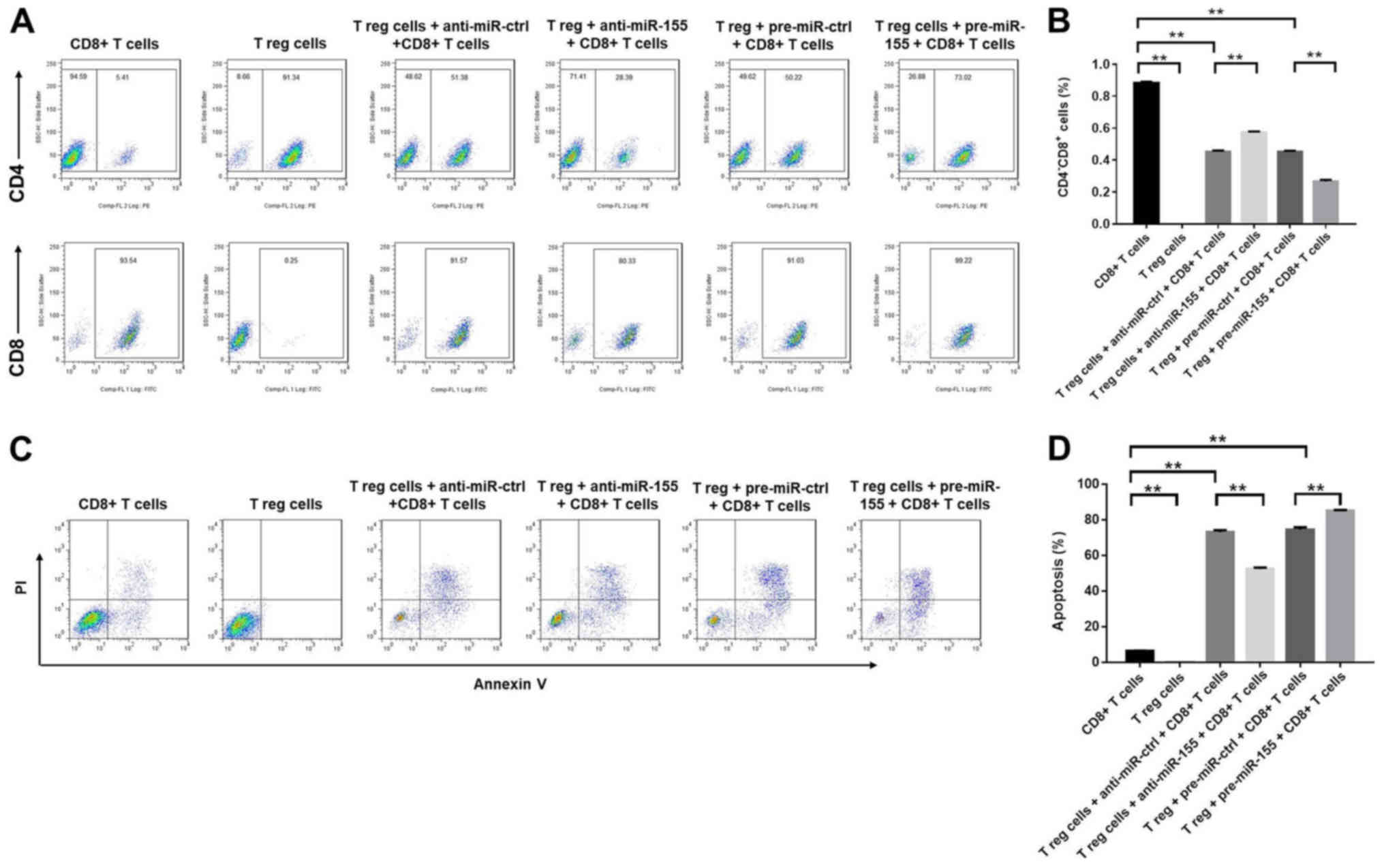
The effects of miR-155 on CD8+ T cells
were subsequently evaluated using flow cytometry. The application
of anti-miR-155 significantly increased the percentage of
CD8+ cells, while pre-miR-155 exhibited the opposite
effect (Fig. 4A and B). In
addition, Treg cells induced the apoptosis of CD8+ T
cells, which was further enhanced by treatment with pre-miR-155,
and reduced by treatment with anti-miR-155 (Fig. 4C and D). In conclusion, the results
demonstrated that miR-155 decreased the percentage of
CD8+ T cells by promoting the induction of
apoptosis.
Treg cells and/or miR-155 inhibit the
induction of melanocyte apoptosis by CD8+ T cells
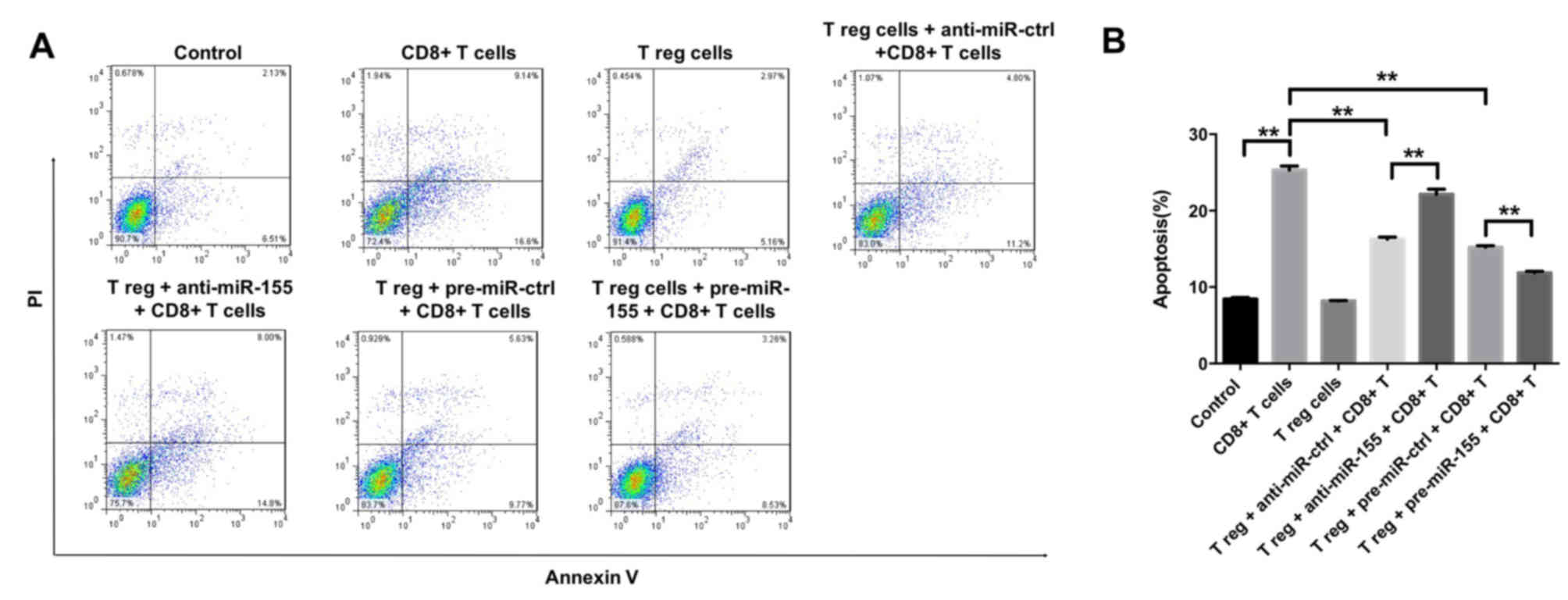
To further investigate the effects of miR-155 on
melanocytes, the induction of melanocyte apoptosis was detected by
flow cytometry, following successful transfection with miR-155
mimics and subsequent 3 days of cell incubation. It was observed
that CD8+ T cells were able to induce apoptosis in
melanocytes, which was partly reversed by the function of Treg
cells (Fig. 5A and B). In
addition, the CD8+ T cell-induced apoptosis was markedly
inhibited by the application of pre-miR-155 and significantly
enhanced by anti-miR-155 (Fig. 5A and
B). Taken together, the data suggested that Treg cells and/or
miR-155 were able to inhibit the induction of melanocyte apoptosis
by CD8+ T cells.
Discussion
It has recently been reported that miR-155 can
modulate the expression of melanogenesis-associated genes both in
keratinocytes and melanocytes, suggesting its important role during
the pathogenesis of vitiligo (21). The present study revealed that
miR-155 was able to protect melanocytes from CD8+ T
lymphocytes by regulating the activity of Treg cells. The
overexpression of miR-155 promoted the differentiation and function
of Treg cells. It was further demonstrated that miR-155 was able to
inhibit the differentiation of CD8+ T cells and decrease
the apoptotic rate of the melanocytes.
Cell-mediated autoimmunity is associated with the
degradation of melanocytes in vitiligo (28). Previous studies have reported an
apparent increase in the number of CD8+ T cells and a
significant reduction in the number of Treg cells in patients with
generalized vitiligo, which indicates that the infiltration of
CD8+ T cells and the deregulation of natural Treg cells
may be closely associated with the pathogenesis of this disease
(29,30). In the present study, it was
demonstrated that the anti-miR-155 group exhibited a decrease in
the percentage of Treg cells and an increase in the percentage of
CD8+ T cells. In addition, transfection of the cells
with anti-miR-155 significantly increased the apoptotic rate of the
melanocytes. A study by Le Poole and Mehrotra (31) indicated that a considerably low
number of Treg cells was able to effectively interfere with
depigmentation when transferred into depigmenting mice. The present
study is in accordance with these previous findings, demonstrating
that the pre-miR-155 group can decrease the apoptotic rate of
melanocytes by increasing the number of Treg cells. The current
study further revealed that miR-155 exerted a positive regulation
on the differentiation of Treg cells.
Tregs can mediate their suppressive activity by a
cellular contact dependent mechanism or by suppressor cytokines,
including TGF-β1 and IL-10 (23,24).
In the current study, it was observed that miR-155 increased the
percentage of Treg cells, and promoted the secretion of IL-10 and
TGF-β1 in the cell culture medium. Nevertheless, a previous study
by Gracias et al (32)
reported that miR-155 overexpression augmented anti-viral
CD8+ T cell responses in C57Bl/6 mice. The contrary
results were observed in the present study, which may be due to the
different species investigated. In addition, TGF-β1 and IL-10 have
been reported to exhibit an inhibitory effect on autoimmune
responses, and a decrease in TGF-β1 and IL-10 levels in the skin
compromised the local immune suppressing function, leading to an
autoimmune reaction against the melanocytes in the patients with
vitiligo (23,24).
A previous study has demonstrated that
CD8+ T cells induce the apoptosis of autologous
melanocytes at the perilesional margins of vitiligo patients
(33). Several previous studies
have also reported that cytotoxic CD8+ T cells can
specifically recognize melanocytes in patients with vitiligo
(10,11). Therefore, the cytotoxic effect of
CD8+ T cells on melanocytes has been suggested as a key
factor during the pathogenesis of vitiligo (34,35).
In addition, CD69 and CD137 serve an important role in
CD8+ T cell activation (36). CD69 was reported to be an early
surface marker of activated T cells, while CD137 was only expressed
on the surface of activated T cells (37). These data suggested that miR-155
was able to inhibit the activation of CD8+ T cells by
regulating the differentiation of Treg cells.
However, there are certain limitations in the
present study. The study included solely cellular assays, and the
experiments and conclusions were based on samples obtained from
only one patient. Therefore, further experiments are required to
clarify the function of miR-155 during the pathogenesis of
vitiligo.
In conclusion, the present study indicated that
miR-155 positively regulated the number of Treg cells, which
prevented the degradation of melanocytes from CD8+ T
cells. Therefore, it is proposed that miR-155 may serve as a
potential therapeutic target for the treatment of patients with
vitiligo.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from The National
Natural Science Foundation of China (grant nos. 81703105, 81571395
and 81771531) and Wenzhou Science & Technology Bureau of China
(grant no. Y20190576).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ML, ZJL, JL and FL analyzed and interpreted the
patient data, and were major contributors in the development of the
first draft of the present manuscript. QZ, ZML and YW participated
in experiment design, tissue collection and experiment execution.
KW and YX participated in experiment design, tissue collection, and
reviewed and approved the final draft of the manuscript prior to
submission.
Ethics approval and consent to
participate
Ethics approval for the present study was provided
by the Wenzhou Medical University Ethics Committee. Informed
consent was obtained from the patient and healthy donor.
Patient consent for publication
Informed consent was obtained from the patient and
healthy donor.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Krüger C and Schallreuter KU: A review of
the worldwide prevalence of vitiligo in children/adolescents and
adults. Int J Dermatol. 51:1206–1212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben
S, Riccardi SL, Cole JB, Gowan K, Holland PJ, Bennett DC, et al:
Genome-wide association analyses identify 13 new susceptibility
loci for generalized vitiligo. Nat Genet. 44:676–680. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ezzedine K, Eleftheriadou V, Whitton M and
van Geel N: Vitiligo. Lancet. 386:74–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gey A, Diallo A, Seneschal J,
Leaute-Labreze C, Boralevi F, Jouary T, Taieb A and Ezzedine K:
Autoimmune thyroid disease in vitiligo: multivariate analysis
indicates intricate pathomechanisms. Br J Dermatol. 168:756–761.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi Q, Zhang W, Guo S, Jian Z, Li S, Li K,
Ge R, Dai W, Wang G, Gao T and Li C: Oxidative stress-induced
overexpression of miR-25: The mechanism underlying the degeneration
of melanocytes in vitiligo. Cell Death Differ. 23:496–508. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu W, Zhao Y, Kong Y, Zhang W, Ma W, Li W
and Wang K: Geniposide prevents H2 O2 -induced oxidative damage in
melanocytes by activating the PI3K-Akt signalling pathway. Clin Exp
Dermatol. 43:667–674. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Patel S, Rauf A, Khan H, Meher BR and
Hassan SSU: A holistic review on the autoimmune disease vitiligo
with emphasis on the causal factors. Biomed Pharmacother.
92:501–508. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Strassner JP and Harris JE: Understanding
mechanisms of autoimmunity through translational research in
vitiligo. Curr Opin Immunol. 43:81–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van den Boorn JG, Konijnenberg D,
Dellemijn TA, van der Veen JP, Bos JD, Melief CJ, Vyth-Dreese FA
and Luiten RM: Autoimmune destruction of skin melanocytes by
perilesional T cells from vitiligo patients. J Invest Dermatol.
129:2220–2232. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wankowicz-Kalinska A, van den Wijngaard
RM, Tigges BJ, Westerhof W, Ogg GS, Cerundolo V, Storkus WJ and Das
PK: Immunopolarization of CD4+ and CD8+ T cells to Type-1-like is
associated with melanocyte loss in human vitiligo. Lab Invest.
83:683–695. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kleinewietfeld M and Hafler DA: Regulatory
T cells in autoimmune neuroinflammation. Immunol Rev. 259:231–244.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Askenasy N, Kaminitz A and Yarkoni S:
Mechanisms of tregulatory cell function. Autoimmun Rev. 7:370–375.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lan Q, Zhou X, Fan H, Chen M, Wang J,
Ryffel B, Brand D, Ramalingam R, Kiela PR, Horwitz DA, et al:
Polyclonal CD4+Foxp3+ Treg cells induce TGFβ-dependent tolerogenic
dendritic cells that suppress the murine lupus-like syndrome. J Mol
Cell Biol. 4:409–419. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dwivedi M, Laddha NC, Arora P, Marfatia YS
and Begum R: Decreased regulatory T-cells and CD4(+) /CD8(+) ratio
correlate with disease onset and progression in patients with
generalized vitiligo. Pigment Cell Melanoma Res. 26:586–591. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ben Ahmed M, Zaraa I, Rekik R,
Elbeldi-Ferchiou A, Kourda N, Belhadj Hmida N, Abdeladhim M, Karoui
O, Ben Osman A, Mokni M and Louzir H: Functional defects of
peripheral regulatory T lymphocytes in patients with progressive
vitiligo. Pigment Cell Melanoma Res. 25:99–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pauley KM, Cha S and Chan EK: MicroRNA in
autoimmunity and autoimmune diseases. J Autoimmun. 32:189–194.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Malmhäll C, Alawieh S, Lu Y, Sjöstrand M,
Bossios A, Eldh M and Rådinger M: MicroRNA-155 is essential for
T(H)2-mediated allergen-induced eosinophilic inflammation in the
lung. J Allergy Clin Immunol. 133:1429–1438, 1438.e1421-1427. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dudda JC, Salaun B, Ji Y, Palmer DC,
Monnot GC, Merck E, Boudousquie C, Utzschneider DT, Escobar TM,
Perret R, et al: MicroRNA-155 is required for effector CD8+ T cell
responses to virus infection and cancer. Immunity. 38:742–753.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liston A, Lu LF, O'Carroll D, Tarakhovsky
A and Rudensky AY: Dicer-dependent microRNA pathway safeguards
regulatory T cell function. J Exp Med. 205:1993–2004. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kempinska-Podhorodecka A, Milkiewicz M,
Wasik U, Ligocka J, Zawadzki M, Krawczyk M and Milkiewicz P:
Decreased expression of vitamin D receptor affects an immune
response in primary biliary cholangitis via the VDR-miRNA155-SOCS1
pathway. Int J Mol Sci. 18:E2892017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Šahmatova L, Tankov S, Prans E, Aab A,
Hermann H, Reemann P, Pihlap M, Karelson M, Abram K, Kisand K, et
al: MicroRNA-155 is dysregulated in the skin of patients with
vitiligo and inhibits melanogenesis-associated genes in melanocytes
and keratinocytes. Acta Derm Venereol. 96:742–747. 2016.PubMed/NCBI
|
|
22
|
Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X
and Liao YH: MicroRNA-155 modulates treg and Th17 cells
differentiation and Th17 cell function by targeting SOCS1. PLoS
One. 7:e460822012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Karagiannidis C, Akdis M, Holopainen P,
Woolley NJ, Hense G, Ruckert B, Mantel PY, Menz G, Akdis CA, Blaser
K and Schmidt-Weber CB: Glucocorticoids upregulate FOXP3 expression
and regulatory T cells in asthma. J Allergy Clin Immunol.
114:1425–1433. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ganesh BB, Bhattacharya P, Gopisetty A,
Sheng J, Vasu C and Prabhakar BS: IL-1β promotes TGF-β1 and IL-2
dependent Foxp3 expression in regulatory T cells. PLoS One.
6:e219492011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Polanczyk MJ, Walker E, Haley D,
Guerrouahen BS and Akporiaye ET: Blockade of TGF-β signaling to
enhance the antitumor response is accompanied by dysregulation of
the functional activity of CD4 + CD25 + Foxp3 + and CD4 + CD25 -
Foxp3 + T cells. J Transl Med. 17:2192019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mucida D, Pino-Lagos K, Kim G, Nowak E,
Benson MJ, Kronenberg M, Noelle RJ and Cheroutre H: Retinoic acid
can directly promote TGF-beta-mediated Foxp3(+) Treg cell
conversion of naive T cells. Immunity. 30:471–472. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Klarquist J, Eby JM, Henning SW, Li M,
Wainwright DA, Westerhof W, Luiten RM, Nishimura MI and Le Poole
IC: Functional cloning of a gp100-reactive T-cell receptor from
vitiligo patient skin. Pigment Cell Melanoma Res. 29:379–384. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nigam PK, Patra PK, Khodiar PK and Gual J:
A study of blood CD3+, CD4+, and CD8+ T cell levels and CD4+:CD8+
ratio in vitiligo patients. Indian J Dermatol Venereol Leprol.
77:1112011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lili Y, Yi W, Ji Y, Yue S, Weimin S and
Ming L: Global activation of CD8+ cytotoxic T lymphocytes
correlates with an impairment in regulatory T cells in patients
with generalized vitiligo. PLoS One. 7:e375132012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Le Poole IC and Mehrotra S: Replenishing
regulatory T cells to halt depigmentation in vitiligo. J Investig
Dermatol Symp Proc. 18:S38–S45. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gracias DT, Stelekati E, Hope JL,
Boesteanu AC, Doering TA, Norton J, Mueller YM, Fraietta JA, Wherry
EJ, Turner M and Katsikis PD: The microRNA miR-155 controls CD8(+)
T cell responses by regulating interferon signaling. Nat Immunol.
14:593–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu J, Zhou M, Wan Y and Xu A: CD8+ T cells
from vitiligo perilesional margins induce autologous melanocyte
apoptosis. Mol Med Rep. 7:237–241. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Glassman SJ: Vitiligo, reactive oxygen
species and T-cells. Clin Sci (Lond). 120:99–120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang L, Yang S, Lei J, Hu W, Chen R, Lin F
and Xu AE: Role of chemokines and the corresponding receptors in
vitiligo: A pilot study. J Dermatol. 45:31–38. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guan C, Li Q, Song X, Xu W, Li L and Xu A:
Antroquinonol exerts immunosuppressive effect on CD8(+) T Cell
proliferation and activation to resist depigmentation induced by
H2O2. Oxid Med Cell Longev. 2017:93030542017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Arneth BM: Activation of CD4 and CD8 T
cell receptors and regulatory T cells in response to human
proteins. PeerJ. 6:e44622018. View Article : Google Scholar : PubMed/NCBI
|