Introduction
Acute pancreatitis (AP) is a common acute abdominal
disease with high mortality and mortality rates. In the US, 220,000
patients with AP were hospitalized in 2013; of these cases, 1,661
were fatal (1). Additionally,
there were also reports that the overall mortality rate in patients
with acute pancreatitis was ~5%, with the mortality rate in
patients with necrotizing pancreatitis ~17% (2). At present, the pathogenesis of AP
remains unclear; however, trypsinogen activation is considered as
the first step in the development of AP (3). Dawra et al (4) hypothesized that activation of
trypsinogen in acinar cells results in the death of acinar cells,
with this process ultimately leading to pancreatic damage. The rat
pancreatic acinar cell line AR42J secretes digestive enzymes and
has therefore been widely used as an in vitro model of AP
(5). Ma et al (6) used taurolithocholic acid 3-sulfate
(TLC-S) to significantly induce trypsinogen activation in AR42J
cells, and identified differentially expressed protein kinases by
microarray analysis. Furthermore, Yang et al (7) reported that the JNK signaling pathway
can promote trypsinogen activation. Although the development of AP
is associated with gene regulation, it has been reported that
changes in certain genes (TMEM173, XIAP, BHLHA15, CTSB)
affect the severity of AP (8–12).
Early growth factor 1 (Egr1) is an immediate early gene that
contains three zinc finger domains (13). The expression level of Egr1 mRNA
increased after 30 min of AP model establishment (14), whereas early pancreatitis may be
caused by trypsinogen activation (15). In addition, reduced level of
inflammation was measured in an Egr1-knockout mouse model of AP
(16). These data confirmed the
role of trypsinogen activation and Egr1 in AP and suggested novel
targets that may also influence the treatment of the disease.
MicroRNAs (miRNAs) are small single-stranded RNAs
(21–25 nucleotides in length) that are relatively conserved in
biological evolution. Although these molecules do not encode
proteins, they mediate numerous cellular processes (17). Previous studies reported that
certain miRNAs are associated with AP. For example, miR-216a
upregulation promotes the development of AP via the Akt and TGF-β
pathway in mice (18), and miR-155
upregulation can inhibit zonula occludens-1 expression and
aggravate AP (19). miRNAs can
downregulate gene expression by binding to complementary sites of
target mRNAs, and subsequently degrade or inhibit these mRNAs
(20). Based on the negative
regulation of mRNAs by miRNAs, the present study aimed to identify
the mRNAs regulated by miRNAs that could affect trypsinogen
activation. According to miRNA and mRNA microarrays, the results
from the present study suggested that the expression of miR-92a-3p
and Erg1 was decreased and increased, respectively, in AR42J cells
treated with TLC-S. In addition, the TargetScan tool, which could
predict the binding of miRNA seed regions to mRNAs, identified that
miR-92a-3p may bind to the 3′untranslated region (UTR) sequence of
Egr1 mRNA. Since our previous research (unpublished data) reported
that small interfering (si)-Egr1 can reduce trypsinogen activation,
the present study investigated whether miR-92a-3p could mediate
trypsinogen activation in AR42J cells by modulating Egr1.
Materials and methods
Cell culture and mRNA and miRNA
microarrays
The AR42J cell line was obtained from The Cell Bank
of Type Culture Collection of the Chinese Academy of Sciences.
Cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 µg/ml streptomycin and 100 U/ml penicillin (Beyotime Institute
of Biotechnology), and placed at 37°C in a humidified incubator
containing 5% CO2. The cells in the TLC-S group were
treated with 200 µM TLC-S (Sigma-Aldrich; Merck KGaA) for 40 min at
37°C (21,22), whereas cells in the control group
were left untreated. Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Gene expression analyses (rat microarray v2.0; cat. no.
Agilent-062716; Agilent Technologies, Inc.) and miRNA expression
analyses (Affymetrix® GeneChip® miRNA 4.0
Array; cat. no. 902446, Affymetrix, Inc.; Thermo Fisher Scientific,
Inc.) were performed by BioCloud, Inc. (Iyunbio).
Integrated analysis of miRNA-mRNA data
and literature review
Gene names were integrated by referring to the
abbreviations in the Rat Genome Database (https://rgd.mcw.edu/). Differences in mRNA/miRNA
expression profiles between RNA samples from TLC-S-treated cells
and RNA samples from control cells were analyzed using Student's
t-tests and fold change (FC) assessment. Differentially expressed
mRNAs were defined as those with a t-test false discovery rate
<0.05 and |log(FC)|>1. Differentially expressed miRNAs had a
FC>1.5 or FC<2/3. TargetScan version 7.2 (http://www.targetscan.org) was used to predict the
differentially expressed miRNA target genes. Furthermore, to
establish a miRNA-mRNA regulatory network, Cytoscape software
(version 3.6.1; http://cytoscape.org/) was used to identify the
downregulated miRNAs and the upregulated target gene mRNAs. To
obtain potentially valuable mRNAs, a literature review was
performed in order to determine the association between mRNAs from
the regulatory network and AP. First, a literature review algorithm
was designed using Perl 4 software (http://www.perl.org/). Then, data from the literature
were retrieved from the PubMed database (https://www.ncbi.nlm.nih.gov/pubmed/) using ActivePerl
5.16.2 software (ActiveState Software, Inc.), and titles and
abstracts from the literature were searched using the keywords
‘acute pancreatitis’ and the mRNAs involved in AP were identified.
Subsequently, the number of statistical documents was determined.
Eventually, the target literature was manually screened to ensure
the accuracy of the research content.
Cell transfection experiments
AR42J cells (1×105 cells) were seeded
into six-well plates in complete DMEM and cultured overnight at
37°C. Medium was removed and Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.) was used for cell transfection according to
manufacturer's instructions. Cells were transfected with miR-92a-3p
mimic, miR-92a-3p mimic-NC, miR-92a-3p inhibitor, miR-92a-3p
inhibitor-NC, si-Egr1 and si-Egr1-NC (Shanghai GenePharma Co.,
Ltd.) at final concentrations of 50 nM using 0.25% Lipofectamine
2000. After 4–6 h of transfection, the culture medium was replaced
with fresh medium, and then the cells were collected by continuous
culture for 48 h. Each transfected group was then treated with 200
µM TLC-S at 37°C for 40 min before collecting the cells. The
sequence of the oligonucleotides used were as follows: miR-92a-3p
mimic, 5′-UAUUGCACUUGUCCCGGCCUGGGCCGGGACAAGUGCAAUAUU-3′; miR-92a-3p
mimic-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′; miR-92a-3p inhibitor,
5′-CAGGCCGGGACAAGUGCAAUA-3′; miR-92a-3p inhibitor-NC,
5′-CAGUACUUUUGUGUAGUACAA-3′; si-Egr1,
5′-CCAGGACUUAAAGGCUCUUTTAAGAGCCUUUAAGUCCUGGTT-3′; si-Egr1-NC,
5′-UUCUCCGAACGUGUCACGUTT-3′.
3′UTR sequence of Egr1 mRNA
According to the rat EGR1 gene (gene ID:
24330), TargetScan was used to predict the miRNAs that may bind to
Egr1 mRNA, and analyze their potential binding sites and the
context score percentiles.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from AR42J cell lines (1×106
cells) was isolated using Axygen reagent (Axygen; Corning Inc.),
and first strand cDNA was synthesized using ReverTraAce qPCR RT kit
(cat. no. FSQ-101; Toyobo Life Science) according to the
manufacturer's protocol. qPCR was performed using SYBR®
Green Real-time PCR Master Mix (Toyobo Life Science) under the
following conditions: 95°C for 10 min, followed by 40 cycles of
95°C for 2 sec and 60°C for 30 sec, and a final extension at 72°C
for 10 min. The primers for Egr1, GAPDH, miR-92a-3p and U6 were
synthesized by Guangzhou RiboBio Co., Ltd. The relative expression
levels of Egr1 and miR-92a-3p were normalized to endogenous control
and were expressed as 2−∆∆Cq (23). The RT-qPCR primers were as follows:
miR-92a-3p, forward 5′-CGCGTATTGCACTTGTCCC-3′, reverse
5′-AGTGCAGGGTCCGAGGTATT-3′; U6, forward
5′-AGAGAAGATTAGCATGGCCCCTG-3′, reverse 5′-AGTGCAGGGTCCGAGGTATT-3′;
Egr1, forward 5′-ACTGGAGGAGATGATGCTGCTGAG-3′, reverse
5′-CCGCTGCTGCTGCTGCTG-3′; and GAPDH, forward
5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse
5′-GCCATCAGCCACAGTTTC-3′.
Western blotting
Cells were lysed using RIPA buffer (cat. no. P0013K;
Beyotime Institute of Biotechnology), and the protein concentration
was measured using a Bicinchoninic Acid Protein Assay kit (cat. no.
P0011; Beyotime Institute of Biotechnology). Proteins (40 µg) were
separated via 8% SDS-PAGE and transferred onto 0.45-µm PVDF
membranes. Membranes were blocked with in 5% skimmed milk dissolved
in PBST for 1 h at room temperature. Membranes were incubated with
primary antibodies against Egr1 (1:500; cat. no. 4153; Cell
Signaling Technology, Inc.) and β-actin (1:2,000; cat. no. PR-0255;
ZSGB-BIO; OriGene Technologies, Inc.) at 4°C overnight. Membranes
were then incubated with horseradish peroxidase secondary antibody
(1:2,000; cat. no. ZB-2301; ZSGB-BIO; OriGene Technologies, Inc.)
for 1 h at room temperature. An ECL kit (cat. no. P0018; Beyotime
Biotech, Inc.) was used to detect the signal on the membrane. Data
were analyzed by densitometry using Image Lab software (version
2.0.1; Bio-Rad Laboratories, Inc.) and normalized to expression of
the internal control β-actin.
Detection of trypsinogen activation by
flow cytometry
Cells (1×106) were harvested and
centrifuged at 500 × g for 5 min at room temperature and the
supernatant was discarded. Cell pellet was washed twice with 1X
HEPES and centrifuged at 500 × g for 5 min at room temperature.
Cells were then resuspended in BziPAR-Rhodamine 110 (Invitrogen;
Thermo Fisher Scientific, Inc.) working solution and allowed to
react in the dark at 37°C for 20 min. BziPAR-Rhodamine 110 is a
trypsin substrate; after enzymatic cleavage, fluorescence is
increased (24). After
centrifugation at 500 × g for 5 min at room temperature,
supernatant was discarded and cells were gently resuspended with 1X
HEPES and analyzed by flow cytometry (Calibur II, BD Biosciences).
The cells were collected using CellQuest software (version 6.0; BD
Biosciences), and the experimental data were analyzed using the
Kaluza Analysis software program (version 2.1; Beckman Coulter,
Inc.).
Statistical analysis
Experiments were repeated at least three times. All
data are presented as the mean ± SD. Comparisons between groups
were made using Student's t-test or one-way ANOVA followed by
Bonferroni test. Statistical analysis was performed using SPSS 17.0
software (SPSS, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Screening of differentially expressed
mRNAs and miRNAs
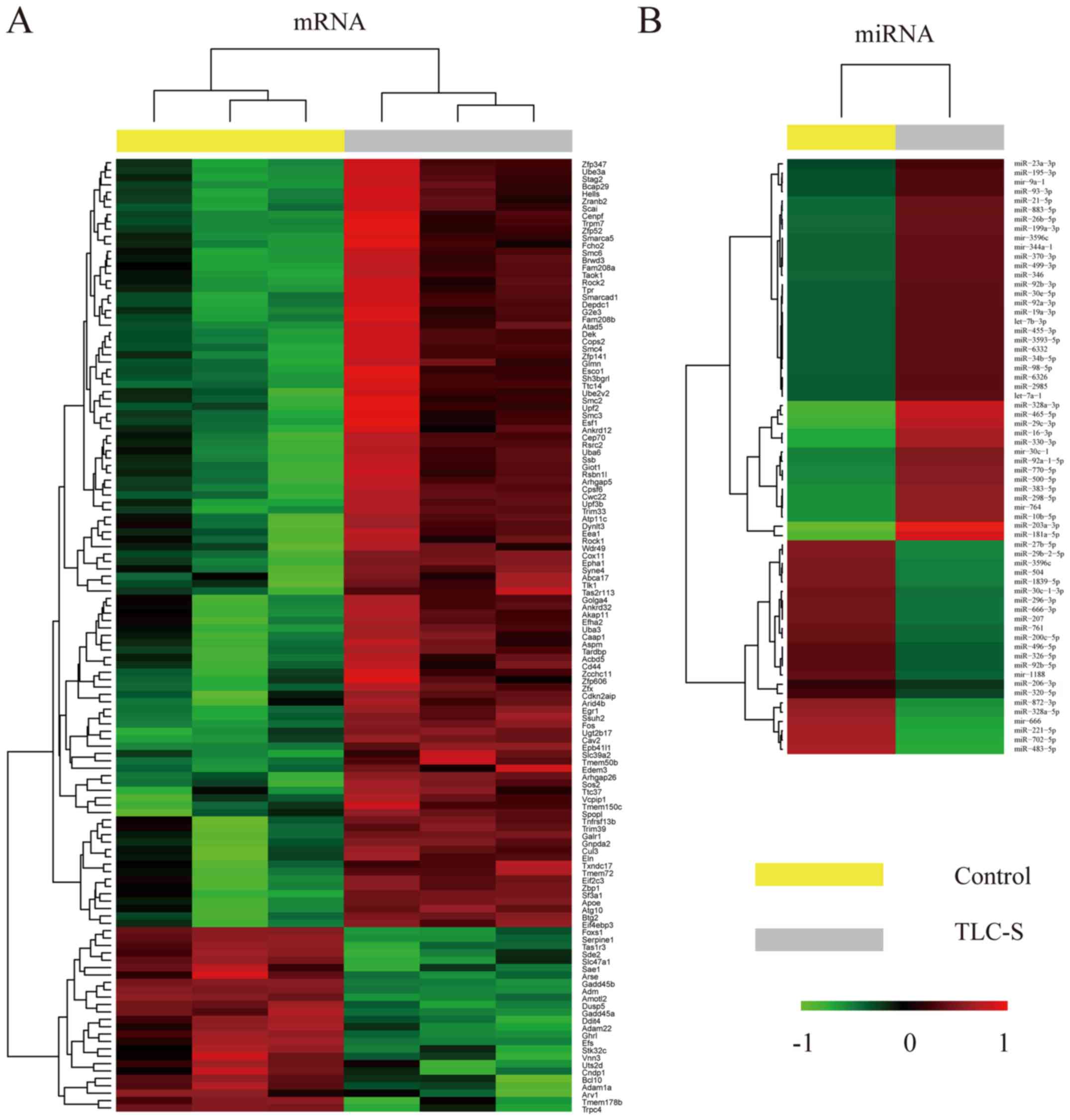
Analysis of mRNA and miRNA microarrays revealed
significant differences in mRNA and miRNA expression levels between
TLC-S-treated AR42J cells and control group. In total, 129 mRNAs
and 64 miRNAs were found to be differentially expressed (Fig. 1).
Construction of the miRNA-mRNA
regulatory network
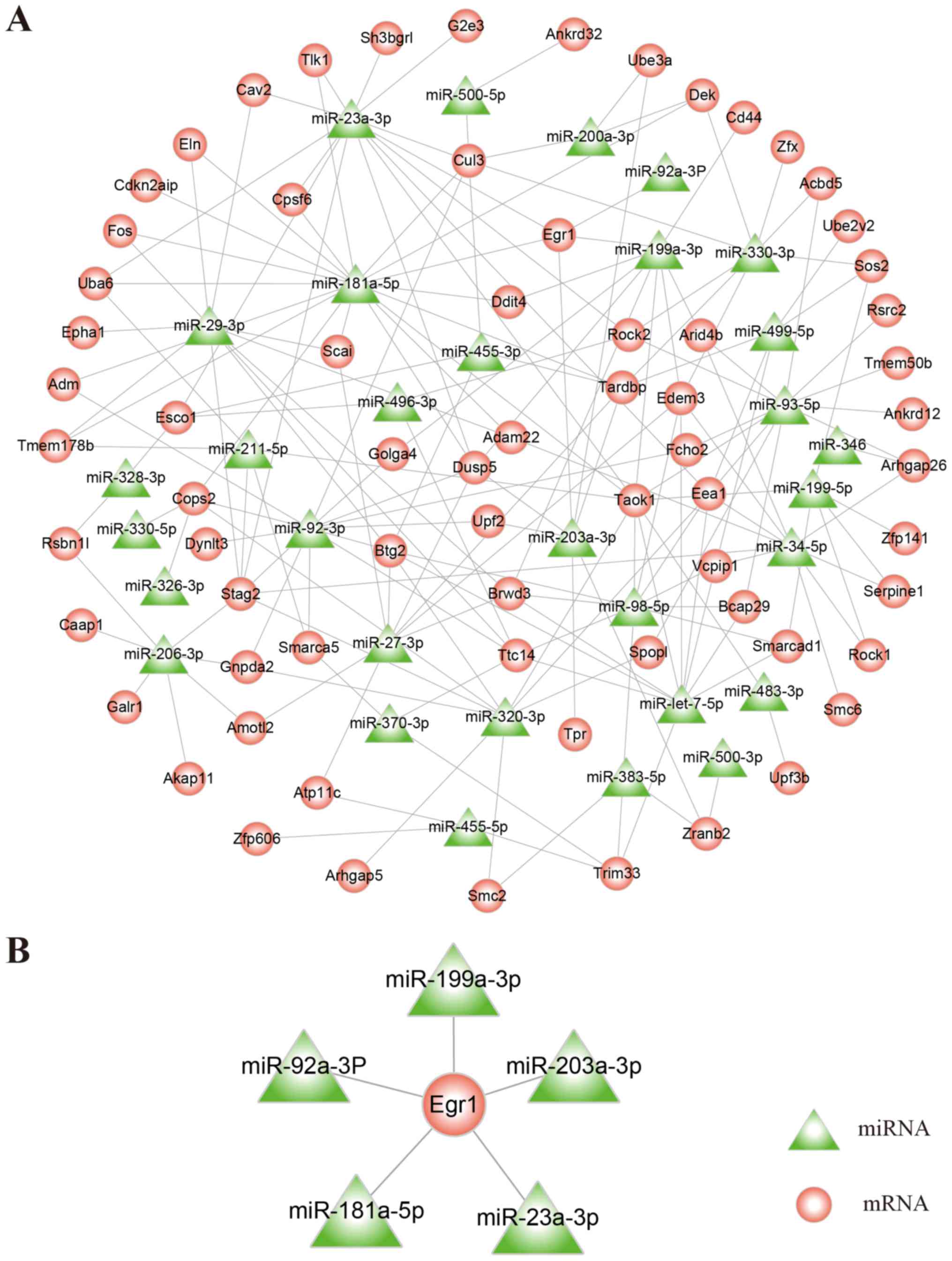
Based on TargetScan bioinformatics predictions and
differentially expressed mRNAs and miRNAs, Cytoscape software was
used to construct a regulatory network containing upregulated mRNAs
and downregulated miRNAs (Fig.
2A). Through a literature review, the results demonstrated that
Egr1 was associated with AP. Subsequently, five miRNAs
(miR-199a-3p, miR-203a-3p, miR-23a-3p, miR-181a-5p and miR-92a-3p)
that exhibited regulatory association with Egr1 were identified
(Fig. 2B).
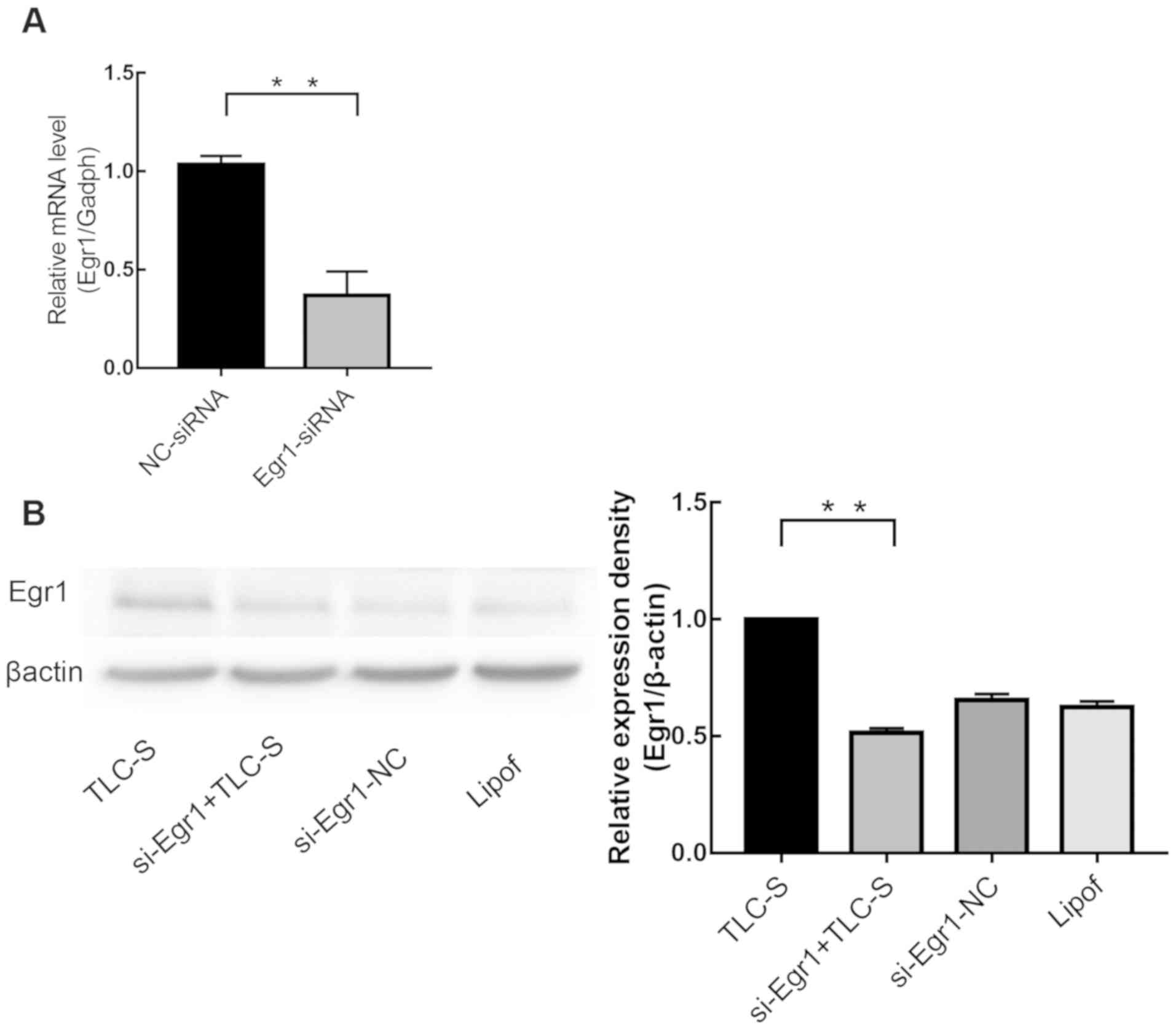
si-Egr1 transfection efficiency
Following 4–6 h transfection of AR42J cells with
si-Egr1, Egr1 expression was analyzed by RT-qPCR (Fig. 3A) and western blotting (Fig. 3B). The results demonstrated that
si-Egr1-transfected cells presented significantly reduced Egr1
expression level, whereas there was no significant difference
between the siRNA-NC group and the lipofectamine-treated group
(P>0.05).
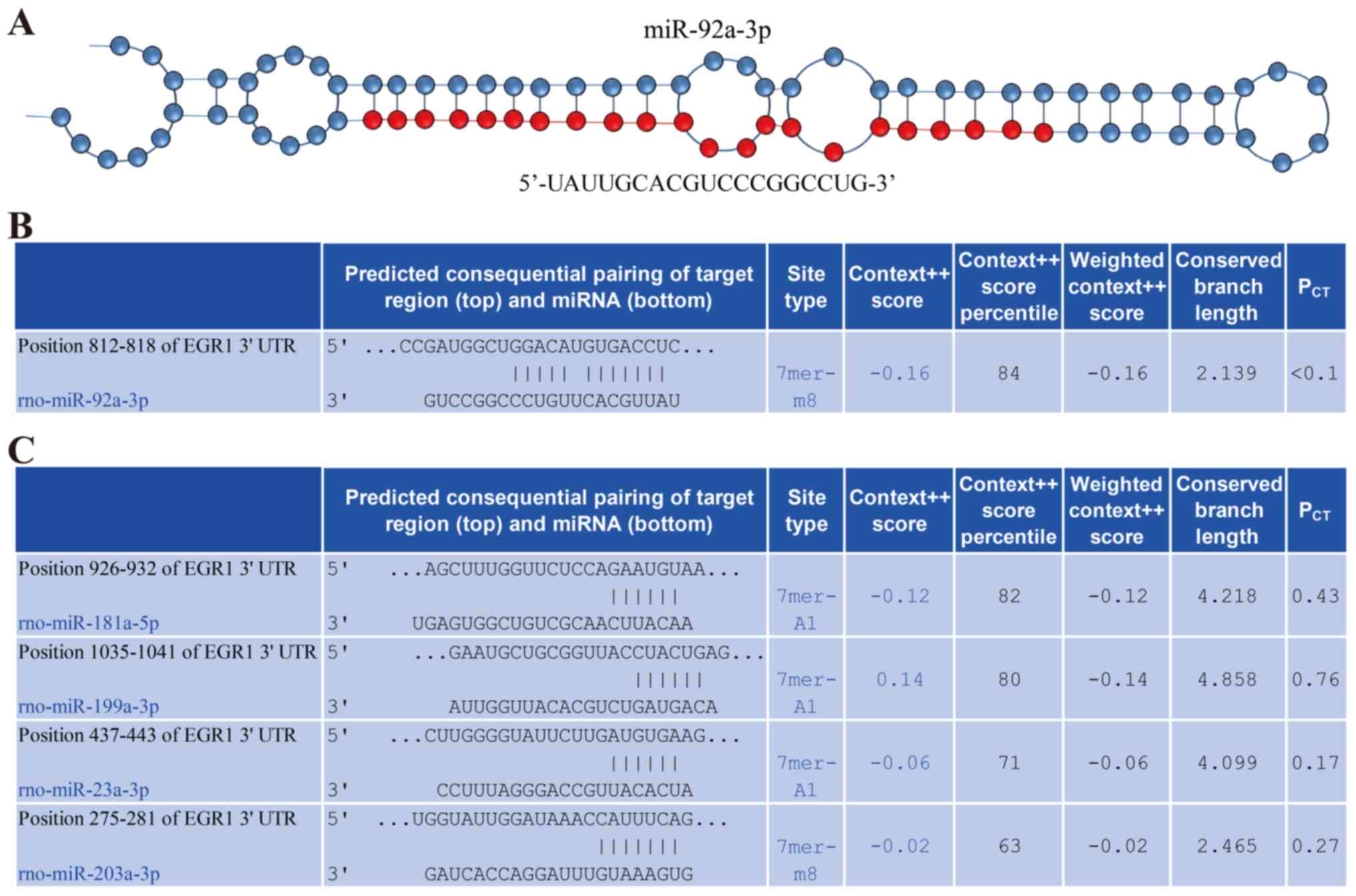
Potential miR-92a-3p binding site was
identified in the Egr1 3′UTR sequence
The structure of miR-92a-3p is presented in Fig. 4A. Bioinformatics analysis using
TargetScan revealed that the 3′UTR sequence of Egr1 contained a
potential binding site for miR-92a-3p (Fig. 4B). Furthermore, potential binding
sites for miR-181a-5p, miR-203a-3p, miR-199a-3p and miR-23a-3p were
also found in the Egr1 3′UTR sequence (Fig. 4C). In addition, the context score
percentiles of miR-181a-5p, miR-203a-3p, miR-199a-3p and miR-23a-3p
were reduced compared with that of miR-92a-3p, which indicated that
miR-92a-3p was more likely to bind Egr1 compared with the other
miRNAs. The present bioinformatics analysis suggested that
miR-92a-3p may regulate Egr1 expression.
miR-92a-3p downregulates Egr1
expression
In order to experimentally confirm the
bioinformatics analyses, the present study investigated whether
miR-92a-3p could affect Egr1 expression in AR42J cells. miR-92a-3p
mimic or inhibitor was transfected into AR42J cells to enhance and
decrease the effect of miR-92a-3p, respectively. Subsequently,
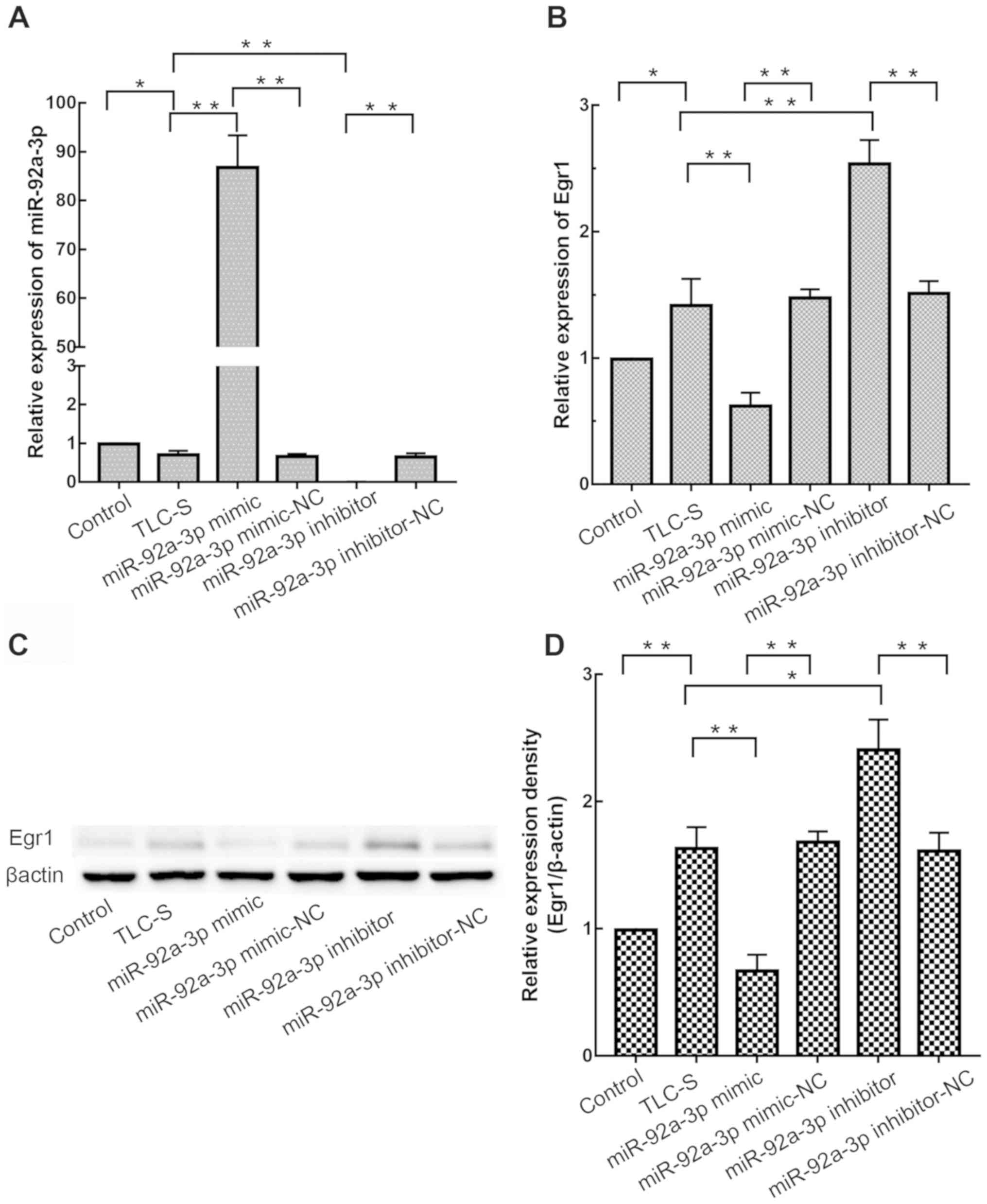
cells were treated with TLC-S, and miR-92a-3p (Fig. 5A) and Egr1 (Fig. 5B) expression were measured by
RT-qPCR. As presented in Fig. 5B,
compared with the TLC-S group, Egr1 expression in the miR-92a-3p
mimic group was significantly decreased (P<0.01). Furthermore,
compared with the TLC-S group, Egr1 expression was significantly
increased in miR-92a-3p inhibitor group (P<0.01). There was no
significant difference in Egr1 expression between miR-92a-3p
mimic-NC group and miR-92a-3p inhibitor-NC group (P>0.05). Egr1
protein level was consistent with RT-qPCR results (Fig. 5C and D). These results indicated
that miR-92a-3p downregulated Egr1 expression in AR42J cells.
miR-92a-3p regulates trypsinogen
activation via Egr1
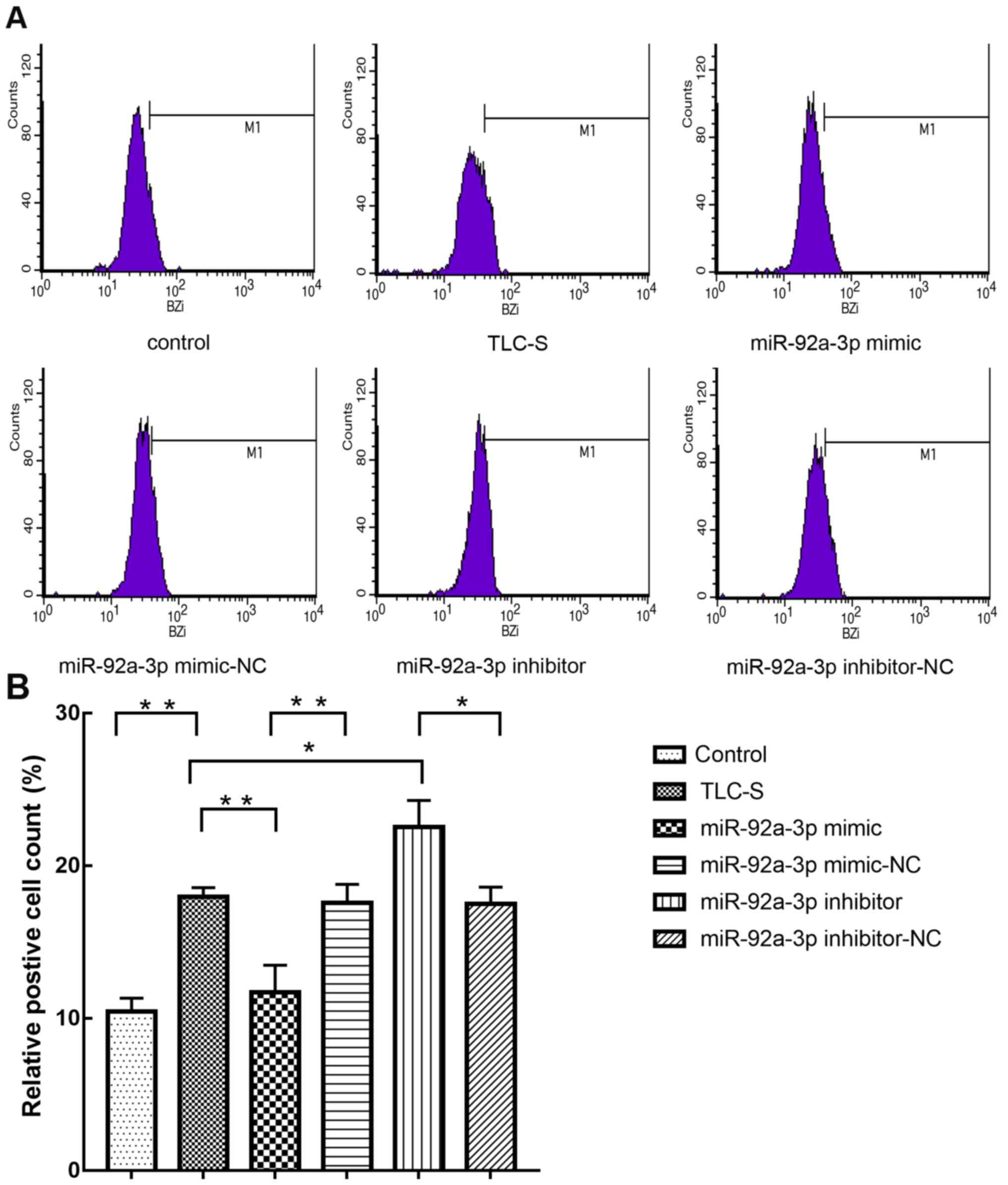
BziPAR-Rhodamine 110 has been reported to be an
excellent substrate for serine proteinases, and results from
fluorescence analysis demonstrated that trypsin fluorescence is
further increased following enzymatic cleavage of the fluorescent
substrates (25). In the present
experiment, flow cytometry was used to detect the percentage of
green fluorescent cells in order to assess the level of trypsinogen
activation. The present results suggested that trypsinogen
activation in the miR-92a-3p mimic group was significantly
decreased compared with the TLC-S group (Fig. 6). Furthermore, compared with the
TLC-S group, the miR-92a-3p inhibitor group exhibited significantly
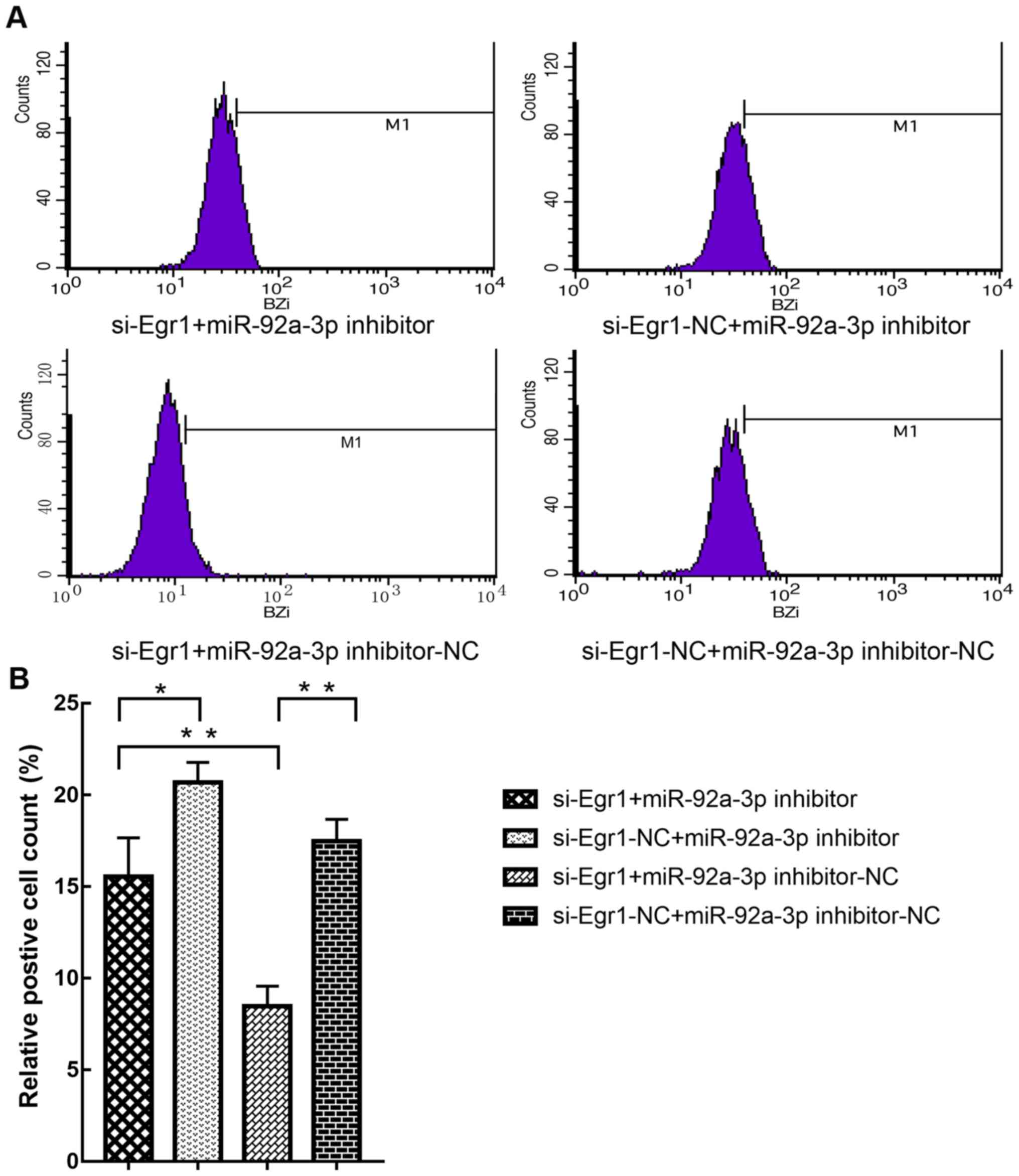
increased trypsinogen activation. To further determine whether
miR-92a-3p regulation of trypsinogen activation was mediated by
Egr1, AR42J cells were transfected with si-Egr1 and miR-92a-3p
inhibitor, and the results demonstrated that the decrease in
trypsinogen activation induced by si-Egr1 was reversed (Fig. 7). These results suggested that
miR-92a-3p may regulate trypsinogen activation via Egr1.
Discussion
AP is a common acute abdominal disease, the
incidence of which has recently increased (26,27).
Premature activation of digestive proteases in the pancreas is the
first step that leads to autodigestion of pancreatic tissue and
therefore to the development of AP. Ohmuraya and Yamamura (28) demonstrated that autophagy
accelerates lysosomal hydrolase activation of trypsinogen, which
subsequently causes AP, in an autophagy-related protein 5 knockout
mice model. Halangk et al (29) used a CTSB-deficient mice model to
induce pancreatitis and demonstrated that trypsinogen activity in
the pancreas of CTSB knockout mice was 80% lower than that of wild
animals. In addition, the degree of acinar tissue necrosis was 50%
compared with control animals. These previous studies provided the
first conclusive evidence indicating that inhibition of trypsinogen
activation can reduce the extent of AP. Previous in vitro
cell experiments used different treatments (TLC-S, cerulein and
TNF-α) to induce trypsinogen activation or AR42J cell apoptosis in
order to elucidate the underlying mechanisms of AP development and
to determine the genes or pathways of interest (14,16,30,31).
Studies using AP animal models will be performed.
In the present study, AR42J cells treated with TLC-S
were used to establish an in vitro trypsinogen activation
model of AP. Microarray and bioinformatics assays were also used to
construct mRNA and miRNA interaction networks based on Egr1 as an
entry point, and to study the role of miR-92a-3p binding with Egr1
in AP.
Egr1 is located on the q23-31 ‘cytokine aggregation’
region of human chromosome 5 and is an early gene that serves
crucial roles in regulating cell cycle, proliferation and
differentiation, and damage repair (32). Abnormal expression of Egr1 was
reported to be associated with various types of disease, including
ischaemic injury, atherosclerosis, inflammation, cardiovascular
pathogenesis and cancer (33).
Egr1 serves a key role in cell damage (34). In unstimulated cells, Egr1 is
rarely expressed; however, when cells are stimulated by
extracellular signaling molecules (growth factors, hormones and
neurotransmitters), Egr1 is immediately synthesized (35,36).
It has been demonstrated that Egr1 is early
synthesized in pancreatic acinar cells following AP induction by
cerulein injection (14). Sandoval
et al (16) used TLC-S to
induce AP in rats and AR42J cells and reported that Egr1 is an
early proinflammatory gene. Furthermore, the expression of
inflammation-associated genes, including monocyte chemoattractant
protein 1, macrophage inflammatory protein 1 and intercellular
adhesion molecule 1, and lung inflammation were reduced in
Egr1-deficient mice, and it was demonstrated that Egr1 activity can
affect the severity of AP (37–39).
Stimulation of Egr-1 synthesis is transient, which indicates that
cells are adapting to prevent constitutive synthesis of Egr-1. The
inhibitory domains NGFI-A binding protein 1 and 2 (NAB1 and NAB2)
are comprised between the Egr1 activation domain and the DNA
binding domain. Both NAB1 and NAB2 block the biological activity of
Egr-1 (40,41). Achieving Egr-1 overexpression has
therefore been a challenge, and experimental methods attempting to
do so were not always successful (42). In addition, the results of the
present study indicated that miR-92a-3p may regulate trypsinogen
activation via Egr1, suggesting the importance of Egr1 in AP.
Previous work from our laboratory suggested that
high Egr1 expression is associated with the development of AP and
that AR42J cell transfection with si-Egr1 reduces trypsinogen
activation (unpublished data). By using microarray analysis and
literature review, the present study demonstrated that Egr1 was
associated with AP. Furthermore, microarrays and bioinformatics
analyses demonstrated that miR-92a-3p regulated Egr1 expression. It
has been demonstrated that miR-92a-3p is associated with cancer
invasion (43) and insulin
secretion (44); however, its role
in pancreatitis has not yet been reported. In the present study,
the results from miRNA microarray and RT-qPCR demonstrated that
miR-92a-3p levels were decreased in the trypsinogen activation
in vitro model of AP. In addition, miR-92a-3p mimic and
inhibitor were used to study the function of miR-92a-3p. The
present results demonstrated that trypsinogen activation was
significantly increased in the miR-92a-3p mimic group but was
significantly decreased in the miR-92a-3p inhibitor group.
Present bioinformatics analyses suggested that Egr1
may be a target gene of miR-92a-3p. The results of the present
study suggested that AR42J cells transfected with miR-92a-3p mimics
expressed low Egr1 mRNA and protein levels, whereas miR-92a-3p
inhibitors induced overexpression of Egr1 mRNA and protein levels.
These data suggested that miR-92a-3p overexpression may
downregulate Egr1 expression. Furthermore, the effect of AR42J cell
transfection with si-Egr1 and miR-92a-3p inhibitor on trypsinogen
activation were investigated. The present results suggested that
transfection with miR-92a-3p inhibitor could reverse the
si-Egr1-induced decrease in trypsinogen activation. The present
findings suggested that miR-92a-3p may regulate Egr1 expression,
reducing the transcription and translation levels of
trypsinogen-associated genes in AR42J cells. Further studies are
required to investigate the role and underlying mechanisms of genes
that could potentially interact with Egr1. In conclusion, the
results of the present study suggested that miR-92a-3p was
significantly reduced in AR42J cells treated with TLC-S compared
with control group. The present data indicated that a decrease in
miR-92a-3p may not completely inhibit Egr1 gene and may lead to
trypsinogen activation.
Acknowledgements
The authors would like to thank Ms Xinyi Liu (Harbin
Medical University) and Ms Xuanxuan Zou (University of Chinese
Academy of Sciences) for their help with bioinformatics
analysis.
Funding
The present work was supported by the National
Natural Science Foundation of China (grant no. 81570581).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DX and WZ designed the study. BG and YH provided
study material. YZ, ZL, DZ and BM performed the experiments and
assembled the data. XZ, BG, YH and YZ analyzed and interpreted the
data. XZ performed experiments and wrote the manuscript. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Krishna SG, Kruger AJ, Patel N, Hinton A,
Yadav D and Conwell DL: Cholecystectomy during index admission for
acute biliary pancreatitis lowers 30-day readmission rates.
Pancreas. 47:996–1002. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pandol SJ, Saluja AK, Imrie CW and Banks
PA: Acute pancreatitis: Bench to the bedside. Gastroenterology.
132:1127–1151. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gao B, Wang D, Sun W, Meng X, Zhang W and
Xue D: Differentially expressed microRNA identification and target
gene function analysis in starvation-induced autophagy of AR42J
pancreatic acinar cells. Mol Med Rep. 14:590–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dawra R, Sah RP, Dudeja V, Rishi L,
Talukdar R, Garg P and Saluja AK: Intra-acinar trypsinogen
activation mediates early stages of pancreatic injury but not
inflammation in mice with acute pancreatitis. Gastroenterology.
141:2210–2217.e2. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sato H, Siow RC, Bartlett S, Taketani S,
Ishii T, Bannai S and Mann GE: Expression of stress proteins heme
oxygenase-1 and −2 in acute pancreatitis and pancreatic islet
betaTC3 and acinar AR42J cells. FEBS Lett. 405:219–223. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma B, Wu L, Lu M, Gao B, Qiao X, Sun B,
Xue D and Zhang W: Differentially expressed kinase genes associated
with trypsinogen activation in rat pancreatic acinar cells treated
with taurolithocholic acid 3-sulfate. Mol Med Rep. 7:1591–1596.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang Z, Yang W, Lu M, Li Z, Qiao X, Sun B,
Zhang W and Xue D: Role of the c-Jun N-terminal kinase signaling
pathway in the activation of trypsinogen in rat pancreatic acinar
cells. Int J Mol Med. 41:1119–1126. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao Q, Wei Y, Pandol SJ, Li L and
Habtezion A: STING signaling promotes inflammation in experimental
acute pancreatitis. Gastroenterology. 154:1822–1835.e2. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Y, Chen XD, Yu J, Chi JL, Long FW,
Yang HW, Chen KL, Lv ZY, Zhou B, Peng ZH, et al: Deletion Of XIAP
reduces the severity of acute pancreatitis via regulation of cell
death and nuclear factor-κB activity. Cell Death Dis. 8:e26852017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kowalik AS, Johnson CL, Chadi SA, Weston
JY, Fazio EN and Pin CL: Mice lacking the transcription factor
Mist1 exhibit an altered stress response and increased sensitivity
to caerulein-induced pancreatitis. Am J Physiol Gastrointest Liver
Physiol. 292:G1123–G1132. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sendler M, Weiss FU, Golchert J, Homuth G,
van den Brandt C, Mahajan UM, Partecke LI, Döring P, Gukovsky I,
Gukovskaya AS, et al: Cathepsin B-mediated activation of
trypsinogen in endocytosing macrophages increases severity of
pancreatitis in mice. Gastroenterology. 154:704–718.e10. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
An F, Zhan Q, Xia M, Jiang L, Lu G, Huang
M, Guo J and Liu S: From moderately severe to severe
hypertriglyceridemia induced acute pancreatitis: Circulating miRNAs
play role as potential biomarkers. PLoS One. 9:e1110582014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gashler A and Sukhatme VP: Early growth
response protein 1 (Egr-1): prototype of a zinc-finger family of
transcription factors. Prog Nucleic Acid Res Mol Biol. 50:191–224.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaufmann A, Rössler OG and Thiel G:
Expression of the transcription factor Egr-1 in pancreatic acinar
cells following stimulation of cholecystokinin or Gαq-coupled
designer receptors. Cell Physiol Biochem. 33:1411–1425. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhan X, Wan J, Zhang G, Song L, Gui F,
Zhang Y, Li Y, Guo J, Dawra RK, Saluja AK, et al: Elevated
intracellular trypsin exacerbates acute pancreatitis and chronic
pancreatitis in mice. Am J Physiol Gastrointest Liver Physiol.
316:G816–G825. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sandoval J, Pereda J, Pérez S, Finamor I,
Vallet-Sánchez A, Rodríguez JL, Franco L, Sastre J and López-Rodas
G: Epigenetic regulation of early- and late-response genes in acute
pancreatitis. J Immunol. 197:4137–4150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Ning X, Cui W, Bi M, Zhang D and
Zhang J: Transforming growth factor (TGF)-β-induced microRNA-216a
promotes acute pancreatitis via Akt and TGF-β pathway in mice. Dig
Dis Sci. 60:127–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tian R, Wang RL, Xie H, Jin W and Yu KL:
Overexpressed miRNA-155 dysregulates intestinal epithelial apical
junctional complex in severe acute pancreatitis. World J
Gastroenterol. 19:8282–8291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ambros V: MicroRNAs and developmental
timing. Curr Opin Genet Dev. 21:511–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gerasimenko JV, Flowerdew SE, Voronina SG,
Sukhomlin TK, Tepikin AV, Petersen OH and Gerasimenko OV: Bile
acids induce Ca2+ release from both the endoplasmic
reticulum and acidic intracellular calcium stores through
activation of inositol trisphosphate receptors and ryanodine
receptors. J Biol Chem. 281:40154–40163. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Voronina SG, Barrow SL, Gerasimenko OV,
Petersen OH and Tepikin AV: Effects of secretagogues and bile acids
on mitochondrial membrane potential of pancreatic acinar cells:
Comparison of different modes of evaluating DeltaPsim. J Biol Chem.
279:27327–27338. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song Z, Huang Y, Liu C, Lu M, Li Z, Sun B,
Zhang W and Xue D: miR-352 participates in the regulation of
trypsinogen activation in pancreatic acinar cells by influencing
the function of autophagic lysosomes. Oncotarget. 9:10868–10879.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sherwood MW, Prior IA, Voronina SG, Barrow
SL, Woodsmith JD, Gerasimenko OV, Petersen OH and Tepikin AV:
Activation of trypsinogen in large endocytic vacuoles of pancreatic
acinar cells. Proc Natl Acad Sci USA. 104:5674–5679. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vaz J, Akbarshahi H and Andersson R:
Controversial role of toll-like receptors in acute pancreatitis.
World J Gastroenterol. 19:616–630. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kylänpää L, Rakonczay Z Jr and O'Reilly
DA: The clinical course of acute pancreatitis and the inflammatory
mediators that drive it. Int J Inflam. 2012:3606852012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ohmuraya M and Yamamura K: Autophagy and
acute pancreatitis: A novel autophagy theory for trypsinogen
activation. Autophagy. 4:1060–1062. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Halangk W, Lerch MM, Brandt-Nedelev B,
Roth W, Ruthenbuerger M, Reinheckel T, Domschke W, Lippert H,
Peters C and Deussing J: Role of cathepsin B in intracellular
trypsinogen activation and the onset of acute pancreatitis. J Clin
Invest. 106:773–781. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen WD, Zhang JL, Wang XY, Hu ZW and Qian
YB: The JAK2/STAT3 signaling pathway is required for inflammation
and cell death induced by cerulein in AR42J cells. Eur Rev Med
Pharmacol Sci. 23:1770–1777. 2019.PubMed/NCBI
|
|
31
|
Meng S, Wang H, Xue D and Zhang W:
Screening and validation of differentially expressed extracellular
miRNAs in acute pancreatitis. Mol Med Rep. 16:6412–6418. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sukhatme VP, Cao XM, Chang LC, Tsai-Morris
CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB,
Curran T, et al: A zinc finger-encoding gene coregulated with c-fos
during growth and differentiation, and after cellular
depolarization. Cell. 53:37–43. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan L, Wang Y, Liang J, Liu Z, Sun X and
Cai K: MiR-301b promotes the proliferation, mobility, and
epithelial-to-mesenchymal transition of bladder cancer cells by
targeting EGR1. Biochem Cell Biol. 95:571–577. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan SF, Fujita T, Lu J, Okada K, Shan Zou
Y, Mackman N, Pinsky DJ and Stern DM: Egr-1, a master switch
coordinating upregulation of divergent gene families underlying
ischemic stress. Nat Med. 6:1355–1361. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thiel G and Cibelli G: Regulation of life
and death by the zinc finger transcription factor Egr-1. J Cell
Physiol. 193:287–292. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alagappan D, Balan M, Jiang Y, Cohen RB,
Kotenko SV and Levison SW: Egr-1 is a critical regulator of
EGF-receptor-mediated expansion of subventricular zone neural stem
cells and progenitors during recovery from hypoxia-hypoglycemia.
ASN Neuro. 5:183–193. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ji B, Chen XQ, Misek DE, Kuick R, Hanash
S, Ernst S, Najarian R and Logsdon CD: Pancreatic gene expression
during the initiation of acute pancreatitis: identification of
EGR-1 as a key regulator. Physiol Genomics. 14:59–72. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wan H, Yuan Y, Liu J and Chen G:
Pioglitazone, a PPAR-γ activator, attenuates the severity of
cerulein-induced acute pancreatitis by modulating early growth
response-1 transcription factor. Transl Res. 160:153–161. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gong LB, He L, Liu Y, Chen XQ and Jiang B:
Expression of early growth response factor-1 in rats with
cerulein-induced acute pancreatitis and its significance. World J
Gastroenterol. 11:5022–5024. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Russo MW, Sevetson BR and Milbrandt J:
Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated
transcription. Proc Natl Acad Sci USA. 92:6873–6877. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Svaren J, Sevetson BR, Apel ED, Zimonjic
DB, Popescu NC and Milbrandt J: NAB2, a corepressor of NGFI-A
(Egr-1) and Krox20, is induced by proliferative and differentiative
stimuli. Mol Cell Biol. 16:3545–3553. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qu Z, Wolfraim LA, Svaren J, Ehrengruber
MU, Davidson N and Milbrandt J: The transcriptional corepressor
NAB2 inhibits NGF-induced differentiation of PC12 cells. J Cell
Biol. 142:1075–1082. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G,
Zhao Q, Wu D, Gong W, Du M, et al: LncRNA MT1JP functions as a
ceRNA in regulating FBXW7 through competitively binding to
miR-92a-3p in gastric cancer. Mol Cancer. 17:872018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Setyowati Karolina D, Sepramaniam S, Tan
HZ, Armugam A and Jeyaseelan K: miR-25 and miR-92a regulate insulin
I biosynthesis in rats. RNA Biol. 10:1365–1378. 2013. View Article : Google Scholar : PubMed/NCBI
|